Genetic Heterogeneity Correlated with Phenotypic Variability in 48 Patients with Cystic Fibrosis
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussions
Genotype–Phenotype Correlations in Patients with Cystic Fibrosis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Online Inherithance of Man (OMIM). Available online: https://omim.org/entry/602421?search=CFTR&highlight=cftr (accessed on 6 March 2025).
- Diab Cáceres, L.; Zamarrón de Lucas, E. Cystic fibrosis: Epidemiology, clinical manifestations, diagnosis and treatment. Med. Clin. 2023, 161, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Savant, A.; Lyman, B.; Bojanowsk, C.; Upadia, J. Cystic Fibrosis. In GeneReviews® [Internet]; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1250/ (accessed on 7 March 2025).
- Crespo-Lessmann, A.; Bernal, S.; Del Río, E.; Rojas, E.; Martínez-Rivera, C.; Marina, N.; Pallarés-Sanmartín, A.; Pascual, S.; García-Rivero, J.L.; Padilla-Galo, A. Association of the CFTR gene with asthma and airway mucus hypersecretion. PLoS ONE 2021, 16, e0251881. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Kotsimbos, T. Respiratory Infection and Inflammation in Cystic Fibrosis: A Dynamic Interplay among the Host, Microbes, and Environment for the Ages. Int. J. Mol. Sci. 2023, 24, 4052. [Google Scholar] [CrossRef] [PubMed]
- Cystic Fibrosis Mutations Database. Available online: http://www.genet.sickkids.on.ca/ (accessed on 6 March 2025).
- Rueda-Nieto, S.; Mondejar-Lopez, P.; Mira-Escolano, M.P.; Cutillas-Tolín, A.; Maceda-Roldán, L.A.; Arense-Gonzalo, J.J.; Palomar-Rodríguez, J.A. Analysis of the genotypic profile and its relationship with the clinical manifestations in people with cystic fibrosis: Study from a rare disease registry. Orphanet. J. Rare Dis. 2022, 17, 222. [Google Scholar] [CrossRef]
- Van Rens, J.; Fox, A.; Krasnyk, M.; Orenti, A.; Zolin, A.; Jung, A.; Naehrlich, L. The European Cystic Fibrosis Society Patient Registry’s Data Quality programme. In 10th European Conference on Rare Diseases & Orphan Products (ECRD 2020). Orphanet. J. Rare Dis. 2020, 15 (Suppl. S1). [Google Scholar]
- Mei-Zahav, M.; Orenti, A.; Jung, A.; Kerem, E. Variability in disease severity among cystic fibrosis patients carrying residual-function variants: Data from the European Cystic Fibrosis Society Patient Registry. ERJ Open Res. 2025, 11, 00587-2024. [Google Scholar] [CrossRef]
- Petrova, N.; Balinova, N.; Marakhonov, A.; Vasilyeva, T.; Kashirskaya, N.; Galkina, V.; Ginter, E.; Kutsev, S.; Zinchenko, R. Ethnic Differences in the Frequency of CFTR Gene Mutations in Populations of the European and North Caucasian Part of the Russian Federation. Front. Genet. 2021, 12, 678374. [Google Scholar] [CrossRef]
- Estivill, X.; Bancells, C.; Ramos, C. Geographic distribution and regional origin of 272 cystic fibrosis mutations in European populations. The Biomed CF Mutation Analysis Consortium. Hum. Mutat. 1997, 10, 135–154. [Google Scholar] [CrossRef]
- WHO Human Genetics Programme. The Molecular Genetic Epidemiology of Cystic Fibrosis: Report of a Joint Meeting of WHO/IECFTN/ICF(M)A/ECFS, Genoa, Italy, 19 June 2002. World Health Organization. 2004. Available online: https://iris.who.int/handle/10665/68702 (accessed on 19 March 2025).
- Abeliovich, D.; Lavon, I.P.; Lerer, I.; Cohen, T.; Springer, C.; Avital, A.; Cutting, G.R. Screening for five mutations detects 97% of cystic fibrosis (CF) chromosomes and predicts a carrier frequency of 1:29 in the Jewish Ashkenazi population. Am. J. Hum. Genet. 1992, 51, 951–956. [Google Scholar]
- Messaoud, T.; Bel Haj Fredj, S.; Bibi, A.; Elion, J.; Férec, C.; Fattoum, S. Epidémiologie moléculaire de la mucoviscidose en Tunisie [Molecular epidemiology of cystic fibrosis in Tunisia]. Ann. Biol. Clin. 2005, 63, 627–630. [Google Scholar]
- Marson, F.A.L.; Bertuzzo, C.S.; Ribeiro, J.D. Classification of CFTR mutation classes. Lancet Respir. Med. 2016, 4, e37–e38. [Google Scholar] [CrossRef]
- Stanke, F.; Tümmler, B. Classification of CFTR mutation classes. Lancet Respir. Med. 2016, 4, e36. [Google Scholar] [CrossRef]
- De Boeck, K.; Amaral, M.D. Progress in therapies for cystic fibrosis. Lancet Respir. Med. 2016, 4, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Zemanick, E.T.; Emerman, I.; McCreary, M.; Mayer-Hamblett, N.; Warden, M.N.; Odem-Davis, K.; VanDevanter, D.R.; Ren, C.L.; Young, J.; Konstan, M.W.; et al. Heterogeneity of CFTR modulator-induced sweat chloride concentrations in people with cystic fibrosis. J. Cyst. Fibros. 2024, 23, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Pacheco, M. CFTR modulators: The changing face of cystic fibrosis in the era of precision medicine. Front. Pharmacol. 2019, 10, 1662. [Google Scholar] [CrossRef] [PubMed]
- Butnariu, L.I.; Țarcă, E.; Cojocaru, E.; Rusu, C.; Moisă, Ș.M.; Leon Constantin, M.M.; Gorduza, E.V.; Trandafir, L.M. Genetic Modifying Factors of Cystic Fibrosis Phenotype: A Challenge for Modern Medicine. J. Clin. Med. 2021, 10, 5821. [Google Scholar] [CrossRef]
- Mésinèle, J.; Ruffin, M.; Guillot, L.; Corvol, H. Modifier Factors of Cystic Fibrosis Phenotypes: A Focus on Modifier Genes. Int. J. Mol. Sci. 2022, 23, 14205. [Google Scholar] [CrossRef]
- Varkki, S.D.; Aaron, R.; Chapla, A.; Danda, S.; Medhi, P.; Jansi Rani, N.; Paul, G.R. CFTR mutations and phenotypic correlations in people with cystic fibrosis: A retrospective study from a single centre in south India. Lancet Reg. Health Southeast Asia 2024, 27, 100434. [Google Scholar] [CrossRef]
- Dupuis, A.; Keenan, K.; Ooi, C.Y.; Dorfman, R.; Sontag, M.K.; Naehrlich, L.; Castellani, C.; Strug, L.J.; Rommens, J.M.; Gonska, T. Prevalence of meconium ileus marks the severity of mutations of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene. Genet. Med. 2016, 18, 333–340. [Google Scholar] [CrossRef]
- Frenţescu, L.; Brownsell, E.; Hinks, J.; Malone, G.; Shaw, H.; Budişan, L.; Bulman, M.; Schwarz, M.; Pop, L.; Filip, M.; et al. The study of cystic fibrosis transmembrane conductance regulator gene mutations in a group of patients from Romania. J. Cyst. Fibros. 2008, 7, 423–428. [Google Scholar] [CrossRef]
- Sciuca, S.; Turcu, O.; Posselt, H.G.; Fergelot, P.; Hedtfeld, S.; Sylvie Labatut, S.; Oberkanins, C.; Pühringer, H.; Reboul, M.P.; Tümmler, B. CFTR mutations of patients with cystic fibrosis from Republic of Moldova. MJHS 2015, 3, 27–30. Available online: https://repository.usmf.md/bitstream/20.500.12710/2445/1/Mutatiile_CFTR_la_pacientii_cu_fibroza_chistica_din_RM.pdf (accessed on 13 March 2025).[Green Version]
- Kasmi, I.; Kasmi, G.; Basholli, B.; Sefa, H.S.; Vevecka, E. The Spectrum and Frequency of Cystic Fibrosis Mutations in Albanian Patients. Balk. J. Med. Genet. 2024, 27, 31–36. [Google Scholar] [CrossRef]
- Petrova, G.; Yaneva, N.; Hrbková, J.; Libik, M.; Savov, A.; Macek, M., Jr. Identification of 99% of CFTR gene mutations in Bulgarian, Bulgarian Turk, and Roma cystic fibrosis patients. Mol. Genet. Genom. Med. 2019, 7, e696. [Google Scholar] [CrossRef]
- Křenková, P.; Piskáčková, T.; Holubová, A.; Balaščaková, M.; Krulišová, V.; Čamajová, J.; Turnovec, M.; Libik, M.; Norambuena, P.; Štambergová, A.; et al. Distribution of CFTR mutations in the Czech population: Positive impact of integrated clinical and laboratory expertise, detection of novel/de novo alleles and relevance for related/derived populations. J. Cyst. Fibros. 2013, 12, 532–537. [Google Scholar] [CrossRef]
- Apostol, P.; Cimponeriu, D.; Radu, I.; Gavrila, L. The analysis of some CFTR gene mutations in a small group of cf patients from southern part of Romania. Anal. Univ. Oradea Fac. Biol. 2009, XVI, 8–11. [Google Scholar]
- Dobre, M.; Chesaru, B.; Romila, A.; Tutunaru, D.; Gurău, G. Cystic Fibrosis in Romanian Children [Internet]. 2015. Available online: https://www.researchgate.net/publication/279202066_Cystic_fibrosis_in_Romanian_children (accessed on 19 March 2025).
- Popa, I.; Pop, L.; Popa, Z.; Schwarz, M.J.; Hambleton, G.; Malone, G.M.; Haworth, A.; Super, M. Cystic fibrosis mutations in Romania. Eur. J. Pediatr. 1997, 156, 212–213. [Google Scholar] [CrossRef]
- Zolin, A.; Orenti, A.; Jung, A.; van Rens, J.; Adamoli, A.; Fox, A.; Krasnyk, M.; Lorca Mayor, S.; Naehrlich, L.; Gkolia, P.; et al. European Cystic Fibrosis Society Pacient Registry. ECFSPR Annual Report 2021. 2023. Available online: https://www.ecfs.eu/sites/default/files/Annual%20Report_2021_09Jun2023.pdf (accessed on 12 March 2025).
- Ideozu, J.E.; Liu, M.; Riley-Gillis, B.M.; Paladugu, S.R.; Rahimov, F.; Krishnan, P.; Tripathi, R.; Dorr, P.; Levy, H.; Singh, A.; et al. Diversity of CFTR variants across ancestries characterized using 454,727 UK biobank whole exome sequences. Genome Med. 2024, 16, 43. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.L.; Schrijver, I. Cystic Fibrosis: A Review of Associated Phenotypes, Use of Molecular Diagnostic Approaches, Genetic Characteristics, Progress, and Dilemmas. J. Mol. Diagn. 2016, 18, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Casals, T.; Nunes, V.; Palacio, A.; Giménez, J.; Gaona, A.; Ibáñez, N.; Morral, N.; Estivill, X. Cystic fibrosis in Spain: High frequency of mutation G542X in the Mediterranean coastal area. Hum. Genet. 1993, 91, 66–70. [Google Scholar] [CrossRef]
- Rosa, K.M.D.; Lima, E.D.S.; Machado, C.C.; Rispoli, T.; Silveira, V.D.; Ongaratto, R.; Comaru, T.; Pinto, L.A. Genetic and phenotypic traits of children and adolescents with cystic fibrosis in Southern Brazil. J. Bras. Pneumol. 2018, 44, 498–504. [Google Scholar] [CrossRef]
- McHugh, D.R.; Steele, M.S.; Valerio, D.M.; Miron, A.; Mann, R.J.; LePage, D.F.; Conlon, R.A.; Cotton, C.U.; Drumm, M.L.; Hodges, C.A. A G542X cystic fibrosis mouse model for examining nonsense mutation directed therapies. PLoS ONE 2018, 13, e0199573. [Google Scholar] [CrossRef]
- Viotti Perisse, I.; Fan, Z.; Van Wettere, A.; Liu, Y.; Leir, S.H.; Keim, J.; Regouski, M.; Wilson, M.D.; Cholewa, K.M.; Mansbach, S.N.; et al. Sheep models of F508del and G542X cystic fibrosis mutations show cellular responses to human therapeutics. FASEB Bioadv. 2021, 3, 841–854. [Google Scholar] [CrossRef] [PubMed]
- ClinVar Database. Available online: https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 29 March 2025).
- Ooi, C.Y.; Durie, P.R. Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in pancreatitis. J. Cyst. Fibros. 2012, 11, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Bienvenu, T.; Viel, M.; Leroy, C.; Cartault, F.; Lesure, J.F.; Renouil, M. Spectrum of CFTR mutations on Réunion Island: Impact on neonatal screening. Hum. Biol. 2005, 77, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Indika, N.L.R.; Vidanapathirana, D.M.; Dilanthi, H.W.; Kularatnam, G.A.M.; Chandrasiri, N.D.P.D.; Jasinge, E. Phenotypic spectrum and genetic heterogeneity of cystic fibrosis in Sri Lanka. BMC Med. Genet. 2019, 20, 89. [Google Scholar] [CrossRef]
- Available online: https://cftr2.org/mutations_history (accessed on 29 March 2025).
- Zielenski, J.; Bozon, D.; Markiewicz, D.; Aubin, G.; Simard, F.; Rommens, J.M.; Tsui, L.C. Analysis of CFTR transcripts in nasal epithelial cells and lymphoblasts of a cystic fibrosis patient with 621 + 1G-->T and 711 + 1G-->T mutations. Hum. Mol. Genet. 1993, 2, 683–687. [Google Scholar] [CrossRef]
- De Braekeleer, M.; Allard, C.; Leblanc, J.P.; Simard, F.; Aubin, G. Genotype-phenotype correlation in five cystic fibrosis patients homozygous for the 621 + 1G-->T mutation. J. Med. Genet. 1997, 34, 788–789. [Google Scholar] [CrossRef]
- Petrova, N.V.; Kashirskaya, N.Y.; Vasilyeva, T.A.; Kondratyeva, E.I.; Zhekaite, E.K.; Voronkova, A.Y.; Sherman, V.D.; Galkina, V.A.; Ginter, E.K.; Kutsev, S.I.; et al. Analysis of CFTR Mutation Spectrum in Ethnic Russian Cystic Fibrosis Patients. Genes 2020, 11, 554. [Google Scholar] [CrossRef]
- Sosnay, P.R.; Raraigh, K.S.; Gibson, R.L. Molecular Genetics of Cystic Fibrosis Transmembrane Conductance Regulator: Genotype and Phenotype. Pediatr. Clin. North Am. 2016, 63, 585–598. [Google Scholar] [CrossRef]
- Angelicheva, D.; Boteva, K.; Jordanova, A.; Savov, A.; Kufardjieva, A.; Tolun, A.; Telatar, M.; Akarsubaşi, A.; Köprübaşi, F.; Aydoğdu, S.; et al. Cystic fibrosis patients from the Black Sea region: The 1677delTA mutation. Hum. Mutat. 1994, 3, 353–357. [Google Scholar] [CrossRef]
- Tkemaladze, T.; Kvaratskhelia, E.; Ghughunishvili, M.; Rtskhiladze, I.; Zaalishvili, Z.; Nakaidze, N.; Lentze, M.J.; Abzianidze, E.; Skrahina, V.; Rolfs, A. Additional evidence on the phenotype produced by combination of CFTR 1677delTA alleles and their relevance in causing CFTR-related disease. SAGE Open Med. Case Rep. 2023, 11, 2050313X231177163. [Google Scholar] [CrossRef]
- Amato, F.; Bellia, C.; Cardillo, G.; Castaldo, G.; Ciaccio, M.; Elce, A.; Lembo, F.; Tomaiuolo, R. Extensive molecular analysis of patients bearing CFTR-related disorders. J. Mol. Diagn. 2012, 14, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Cohn, J.A.; Friedman, K.J.; Noone, P.G.; Knowles, M.R.; Silverman, L.M.; Jowell, P.S. Relation between mutations of the cystic fibrosis gene and idiopathic pancreatitis. N. Engl. J. Med. 1998, 339, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Genome Aggregation Database. Available online: https://gnomad.broadinstitute.org/ (accessed on 25 March 2025).
- Makukh, H.; Krenková, P.; Tyrkus, M.; Bober, L.; Hancárová, M.; Hnateyko, O.; Macek, M., Jr. A high frequency of the Cystic Fibrosis 2184insA mutation in Western Ukraine: Genotype-phenotype correlations, relevance for newborn screening and genetic testing. J. Cyst. Fibros. 2010, 9, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Dörk, T.; Mekus, F.; Schmidt, K.; Bosshammer, J.; Fislage, R.; Heuer, T.; Dziadek, V.; Neumann, T.; Kälin, N.; Wulbrand, U.; et al. Detection of more than 50 different CFTR mutations in a large group of German cystic fibrosis patients. Hum. Genet. 1994, 94, 533–542. [Google Scholar] [CrossRef]
- Stuhrmann, M.; Dörk, T.; Frühwirth, M.; Golla, A.; Skawran, B.; Antonin, W.; Ebhardt, M.; Loos, A.; Ellemunter, H.; Schmidtke, J. Detection of 100% of the CFTR mutations in 63 CF families from Tyrol. Clin. Genet. 1997, 52, 240–246. [Google Scholar] [CrossRef]
- Ivady, G.; Madar, L.; Nagy, B.; Gonczi, F.; Ajzner, E.; Dzsudzsak, E.; Dvořáková, L.; Gombos, E.; Kappelmayer, J.; Macek, M.; et al. Distribution of CFTR mutations in Eastern Hungarians: Relevance to genetic testing and to the introduction of newborn screening for cystic fibrosis. J. Cyst. Fibros. 2011, 10, 217–220. [Google Scholar] [CrossRef]
- Kolesar, P.; Minarik, G.; Baldovic, M.; Ficek, A.; Kovacs, L.; Kadasi, L. Mutation analysis of the CFTR gene in Slovak cystic fibrosis patients by DHPLC and subsequent sequencing: Identification of four novel mutations. Gen. Physiol. Biophys. 2008, 27, 299–305. [Google Scholar][Green Version]
- Buratti, E.; Chivers, M.; Královicová, J.; Romano, M.; Baralle, M.; Krainer, A.R.; Vorechovsky, I. Aberrant 5′ splice sites in human disease genes: Mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007, 35, 4250–4263. [Google Scholar] [CrossRef]
- Zhang, M.Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998, 7, 919–932. [Google Scholar] [CrossRef]
- Terzic, M.; Jakimovska, M.; Fustik, S.; Jakovska, T.; Sukarova-Stefanovska, E.; Plaseska-Karanfilska, D. Cystic Fibrosis Mutation Spectrum in North Macedonia: A Step Toward Personalized Therapy. Balk. J. Med. Genet. 2019, 22, 35–40. [Google Scholar] [CrossRef]
- Yang, B.; Wang, X.; Zhang, W.; Li, H.; Wang, B. Compound heterozygous mutations in CFTR causing CBAVD in Chinese pedigrees. Mol. Genet. Genom. Med. 2018, 6, 1097–1103. [Google Scholar] [CrossRef]
- Kilinç, M.O.; Ninis, V.N.; Dağli, E.; Demirkol, M.; Ozkinay, F.; Arikan, Z.; Coğulu, O.; Hüner, G.; Karakoç, F.; Tolun, A. Highest heterogeneity for cystic fibrosis: 36 mutations account for 75% of all CF chromosomes in Turkish patients. Am. J. Med. Genet. 2002, 113, 250–257. [Google Scholar] [CrossRef]
- Akinsal, E.C.; Baydilli, N.; Dogan, M.E.; Ekmekcioglu, O. Comorbidity of the congenital absence of the vas deferens. Andrologia 2018, 50, e12994. [Google Scholar] [CrossRef] [PubMed]
- Adler, A.I.; Shine, B.S.; Chamnan, P.; Haworth, C.S.; Bilton, D. Genetic determinants and epidemiology of cystic fibrosis-related diabetes: Results from a British cohort of children and adults. Diabetes Care 2008, 31, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Nowak, J.K.; Szczepanik, M.; Wojsyk-Banaszak, I.; Mądry, E.; Wykrętowicz, A.; Krzyżanowska-Jankowska, P.; Drzymała-Czyż, S.; Nowicka, A.; Pogorzelski, A.; Sapiejka, E.; et al. Cystic fibrosis dyslipidaemia: A cross-sectional study. J. Cyst. Fibros. 2019, 18, 566–571. [Google Scholar] [CrossRef] [PubMed]
- McCague, A.F.; Raraigh, K.S.; Pellicore, M.J.; Davis-Marcisak, E.F.; Evans, T.A.; Han, S.T.; Lu, Z.; Joynt, A.T.; Sharma, N.; Castellani, C.; et al. Correlating Cystic Fibrosis Transmembrane Conductance Regulator Function with Clinical Features to Inform Precision Treatment of Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1116–1126. [Google Scholar] [CrossRef]
- Petrova, N.V.; Marakhonov, A.V.; Vasilyeva, T.A.; Kashirskaya, N.Y.; Ginter, E.K.; Kutsev, S.I.; Zinchenko, R.A. Comprehensive genotyping reveals novel CFTR variants in cystic fibrosis patients from the Russian Federation. Clin. Genet. 2019, 95, 444–447. [Google Scholar] [CrossRef]
- Claustres, M.; Thèze, C.; des Georges, M.; Baux, D.; Girodon, E.; Bienvenu, T.; Audrezet, M.P.; Dugueperoux, I.; Férec, C.; Lalau, G.; et al. CFTR-France, a national relational patient database for sharing genetic and phenotypic data associated with rare CFTR variants. Hum. Mutat. 2017, 38, 1297–1315. [Google Scholar] [CrossRef]
- Gaitch, N.; Hubert, D.; Gameiro, C.; Burgel, P.R.; Houriez, F.; Martinez, B.; Honoré, I.; Chapron, J.; Kanaan, R.; Dusser, D.; et al. CFTR and/or pancreatitis susceptibility genes mutations as risk factors of pancreatitis in cystic fibrosis patients? Pancreatology 2016, 16, 515–522. [Google Scholar] [CrossRef]
- Smits, R.M.; Oud, M.S.; Vissers, L.E.L.M.; Lugtenberg, D.; Braat, D.D.M.; Fleischer, K.; Ramos, L.; D’Hauwers, K.W.M. Improved detection of CFTR variants by targeted next-generation sequencing in male infertility: A case series. Reprod. Biomed. Online 2019, 39, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Kerem, B.S.; Zielenski, J.; Markiewicz, D.; Bozon, D.; Gazit, E.; Yahav, J.; Kennedy, D.; Riordan, J.R.; Collins, F.S.; Rommens, J.M.; et al. Identification of mutations in regions corresponding to the two putative nucleotide (ATP)-binding folds of the cystic fibrosis gene. Proc. Natl. Acad. Sci. USA 1990, 7, 8447–8451. [Google Scholar] [CrossRef] [PubMed]
- Hull, J.; Shackleton, S.; Harris, A. Abnormal mRNA splicing resulting from three different mutations in the CFTR gene. Hum. Mol. Genet. 1993, 2, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Sosnay, P.R.; Ramalho, A.S.; Douville, C.; Franca, A.; Gottschalk, L.B.; Park, J.; Lee, M.; Vecchio-Pagan, B.; Raraigh, K.S.; et al. Experimental assessment of splicing variants using expression minigenes and comparison with in silico predictions. Hum. Mutat. 2014, 35, 1249–1259. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Shelton, C.; LaRusch, J.; Whitcomb, D.C. Pancreatitis Overview. In GeneReviews® [Internet]; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK190101/ (accessed on 25 March 2025).
- Sosnay, P.R.; Siklosi, K.R.; Van Goor, F.; Kaniecki, K.; Yu, H.; Sharma, N.; Ramalho, A.S.; Amaral, M.D.; Dorfman, R.; Zielenski, J.; et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013, 45, 1160–1167. [Google Scholar] [CrossRef]
- Kanavakis, E.; Efthymiadou, A.; Strofalis, S.; Doudounakis, S.; Traeger-Synodinos, J.; Tzetis, M. Cystic fibrosis in Greece: Molecular diagnosis, haplotypes, prenatal diagnosis and carrier identification amongst high-risk individuals. Clin. Genet. 2003, 63, 400–409. [Google Scholar] [CrossRef]
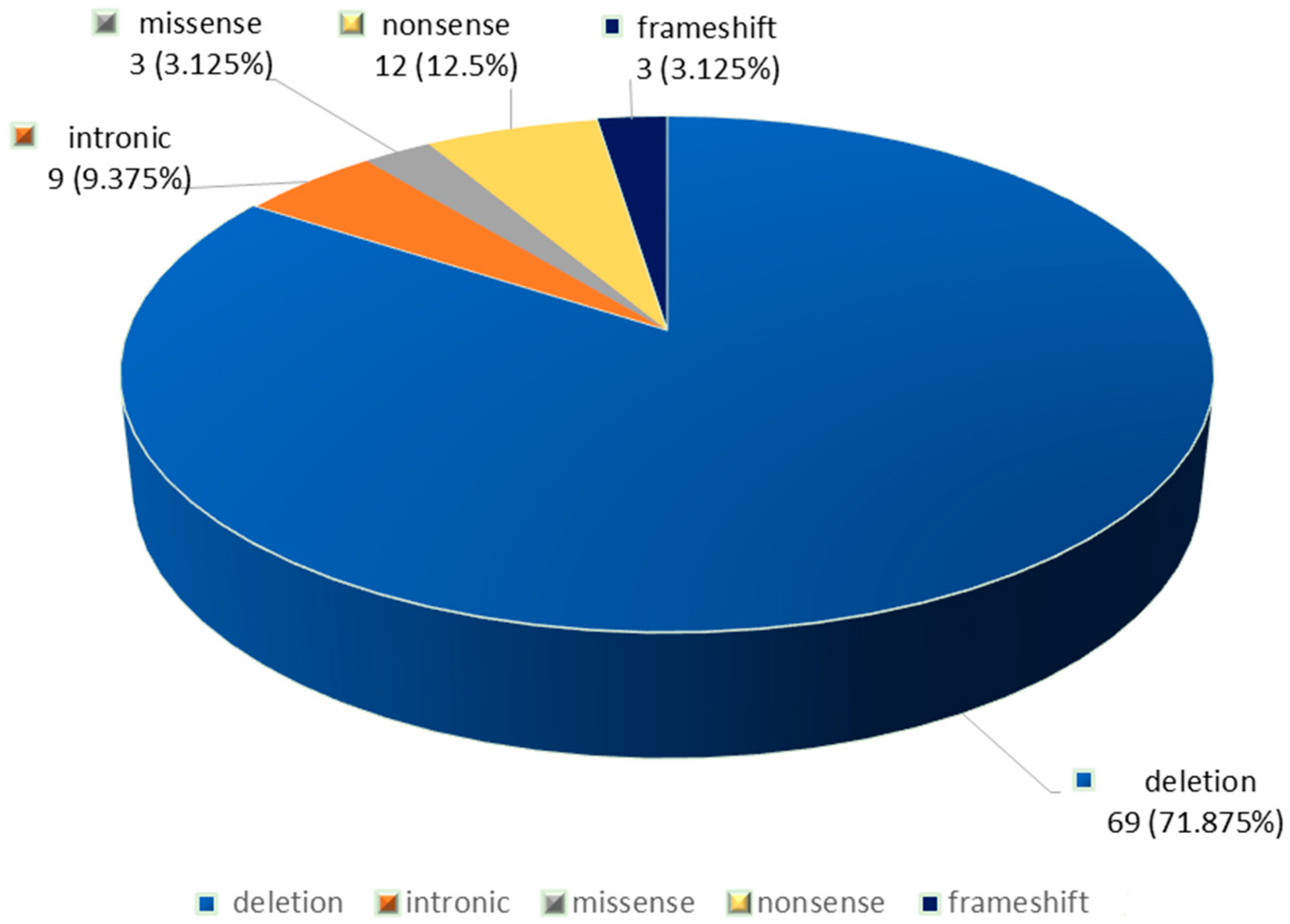
| Allele1 | Allele 2 | No of Cases |
|---|---|---|
| F508del | F508del | 22 |
| F508del | 621+1G>T (c.489+1 G>T) | 3 |
| F508del | G542X | 3 |
| F508del | 1677delTA | 3 |
| G542X | 185+1G->T (c.53+1G>T) | 3 |
| F508del | 2184insA (c.2052dup) (p.Gln685fs) (Q685fs) | 2 |
| F508del | c.917dupA p.(Asn306Lysfs*2) | 2 |
| F508del | c.3849G->A (c.3717G>A) (p.Arg1239=) | 2 |
| F508del | R1158X | 1 |
| F508del | K598*(c.1792A>T) (p.Lys598Ter) | 1 |
| F508del | c.1040G>A (p.Arg347His) (R347H) | 1 |
| F508del | c.54-5940_273+10250del (CFTRdele2,3 (21kb)) | 1 |
| F508del | 1717-1G>A | 1 |
| F508del | c2589_2599delAATTTGGTGCT c.2589_2599del (p.Ile864fs)(I864fs) | 1 |
| F508del | c.1487G>A, p.(Trp496Ter) | 1 |
| F508del | c.(53+1_54-1)_(164+1_165-1)del (CFTRdele2) | 1 |
| CFTR Variant | No. Allele (%) (Our Study) | Frentescu et al. [24] (Romania) | Sciuca et al. [25] (Rep. Moldova) | Kasmi et al. [26] (Albania) | Petrova et al. [27] (Bulgaria | Křenková et al. [28] Czech Rep.) | Ruenda -Nieto et al. [7] (Spain) |
|---|---|---|---|---|---|---|---|
| F508del | 67 (69.79%) | 144 (56.3%) | 79 (57.4%) | 203 (83.19%) | 154 (55%) | 809 (67.42%) | 142 (37.0%) |
| G542X | 6 (6.25%) | 10 (3.9%) | 4 (3.3%) | 4 (1.63%) | 11 (3.93%) | 24 (2%) | 31 (8.1%) |
| 621+1G>T (c.489+1G>T) | 3 (3.12%) | 2 (0.8%) | 1 (0.8%) | 6 (2.45%) | 4 (1.43%) | 5 (0.43%) | 1 (0.3%) |
| 1677delTA | 3 (3.12%) | 1 (0.4%) | 1 (0.8%) | - | 3 (1.07%) | - | - |
| 185+1G ->T (c.53+1G>T) | 3 (3.12%) | - | 1 (0.8%) | - | - | 2 (0.17%) | - |
| 2184insA | 2 (2.08%) | - | 4 (3.3%) | - | 8 (2.89%) | 5 (0.42%) | - |
| c.917dupA p.(Asn306Lysfs*2) | 2 (2.08%) | - | - | - | - | - | |
| 3849G>A (c.3717G>A) (p.Arg1239=) | 2 (2.08%) | - | 2 (1.6%) | - | - | - | 1 (0.3%) |
| R1158X (c.3472C>T) | 1 (1.04%) | - | - | 1 (0.40%) | 1 (0.36%) | 1 (0.08) | 1 (0.3%) |
| K598* (c.1792A>T) (p.Lys598Ter) | 1 (1.04%) | - | - | - | - | - | - |
| R347H (c.1040G>A) | 1 (1.04%) | - | - | - | - | 1 (0.08%) | - |
| c.54-5940_273+10250del (CFTRdele2,3 (21kb) | 1 (1.04%) | 4 (1.6%) | 2 (1.6%) | 1 (0.40%) | 2 (0.71) | 69 (5.75%) | - |
| 1717-1G>A (c.1585-2A>T) | 1 (1.04%) | 1 (0.4%) | - | - | - | 4 (0.33) | 1 (0.3%) |
| c2589_2599delAATTTGGTGCT c.2589_2599del (p.Ile864fs) | 1 (1.04%) | - | - | - | - | 1 (0.08) | - |
| R496H (c.1487G>A) p.(Trp496Ter) | 1 (1.04%) | - | - | - | - | - | |
| c.(53+1_54-1)_(164+1_165-1)del (CFTRdele2) | 1 (1.04%) | - | - | - | - | - | - |
| No total allele | 96 | 256 | 122 | 244 | 277 | 1200 | 384 |
| Criteria | Homozygous No. (%) | Compound Heterozygous No. (%) |
|---|---|---|
| Female | 12/22 (54.5%) | 12/26 (46.15%) |
| Male | 10/22 (45.5%) | 14/26 (53.84%) |
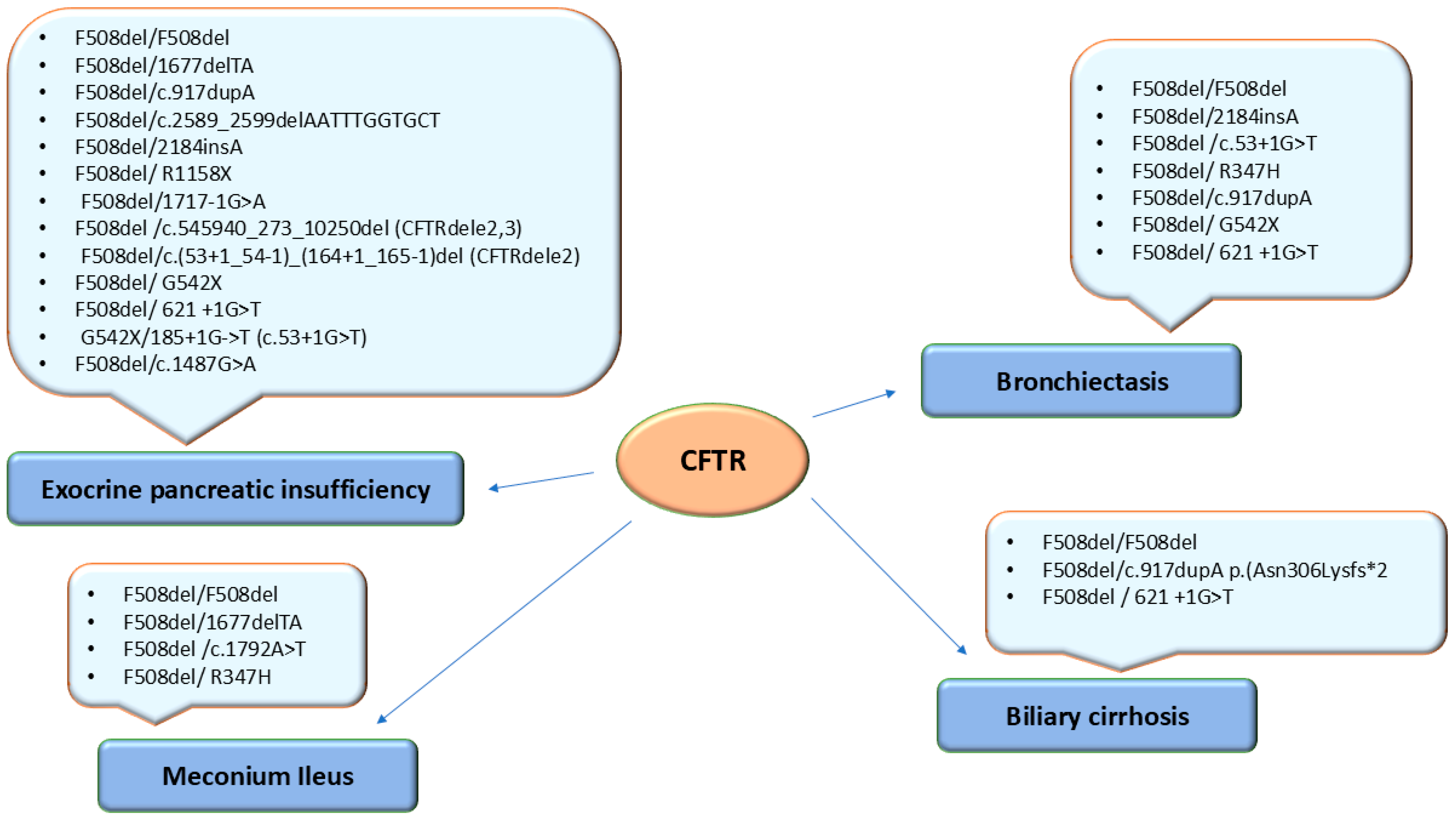
| Respiratory manifestations | ||
| Bronchiectasis | 5/22 (22.72%) | 9/26 (34.61%) |
| R-URTIs a | 10/22 (45.45%) | 13/26 (50%) |
| Gastrointestinal and nutritional manifestations | ||
| Hepatocytolysis | 14/22 (63.63%) | 18/26 (69.23%) |
| Biliary cirrhosis | 3/22 (13.63%) | 2/26 (7.69%) |
| Liver fibrosis | 1/22 (4.54%) | - |
| Gallbladder stones | 2/22 (9.09%) | 1/26 (3.84%) |
| EPI b | 22/22 (100%) | 19/26 (73.07%) |
| CF-related pancreatitis | 1/22 (4.54%) | - |
| CF-related GI manifestations | - | 1/26 (3.84%) |
| Growth failure | 19/22 (86.36%) | 26/26 (100%) |
| Metabolic manifestations | ||
| Dyslipidemia | 2/22 (9.09%) | 3/26 (11.53%) |
| Hepatic steatosis | 5/22 (22.72%) | 6/26 (23.07%) |
| CFRD | 3/22 (13.63%) | 3/26 (11.53%) |
| 25-OH vitamin D deficiency | 13/22 (59.09%) | 14/26 (53.84%) |
| Surgical manifestations | ||
| Meconium ileus | 6/22 (27.27%) | 3/26 (11.53%) |
| Subocclusive syndrome | 2/22 (9.09%) | 1/26 (3.84%) |
| Rectal prolapse | 2/22 (9.09%) | 4/26 (15.38%) |
| Renal and urological manifestations | ||
| Urolithiasis | - | 1/26(3.84%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donos, M.A.; Butnariu, L.I.; Anton Păduraru, D.T.; Murgu, A.M.; Rusu, C.; Pânzaru, M.C.; Popescu, R.; Țarcă, E.; Cojocaru, E.; Ghiga, G.; et al. Genetic Heterogeneity Correlated with Phenotypic Variability in 48 Patients with Cystic Fibrosis. J. Clin. Med. 2025, 14, 5362. https://doi.org/10.3390/jcm14155362
Donos MA, Butnariu LI, Anton Păduraru DT, Murgu AM, Rusu C, Pânzaru MC, Popescu R, Țarcă E, Cojocaru E, Ghiga G, et al. Genetic Heterogeneity Correlated with Phenotypic Variability in 48 Patients with Cystic Fibrosis. Journal of Clinical Medicine. 2025; 14(15):5362. https://doi.org/10.3390/jcm14155362
Chicago/Turabian StyleDonos, Mădălina Andreea, Lăcrămioara Ionela Butnariu, Dana Teodora Anton Păduraru, Alina Mariela Murgu, Cristina Rusu, Monica Cristina Pânzaru, Roxana Popescu, Elena Țarcă, Elena Cojocaru, Gabriela Ghiga, and et al. 2025. "Genetic Heterogeneity Correlated with Phenotypic Variability in 48 Patients with Cystic Fibrosis" Journal of Clinical Medicine 14, no. 15: 5362. https://doi.org/10.3390/jcm14155362
APA StyleDonos, M. A., Butnariu, L. I., Anton Păduraru, D. T., Murgu, A. M., Rusu, C., Pânzaru, M. C., Popescu, R., Țarcă, E., Cojocaru, E., Ghiga, G., & Trandafir, L. M. (2025). Genetic Heterogeneity Correlated with Phenotypic Variability in 48 Patients with Cystic Fibrosis. Journal of Clinical Medicine, 14(15), 5362. https://doi.org/10.3390/jcm14155362








