Expression of miRNAs in Pre-Schoolers with Autism Spectrum Disorders Compared with Typically Developing Peers and Its Effects after Probiotic Supplementation
Abstract
:1. Introduction
2. Methods
2.1. Subjects and Plasma Collection
2.2. Zonulin and Lactoferrin Assays
2.3. miRNA Extraction, Reverse Transcription, and Real-Time PCR
3. Statistics
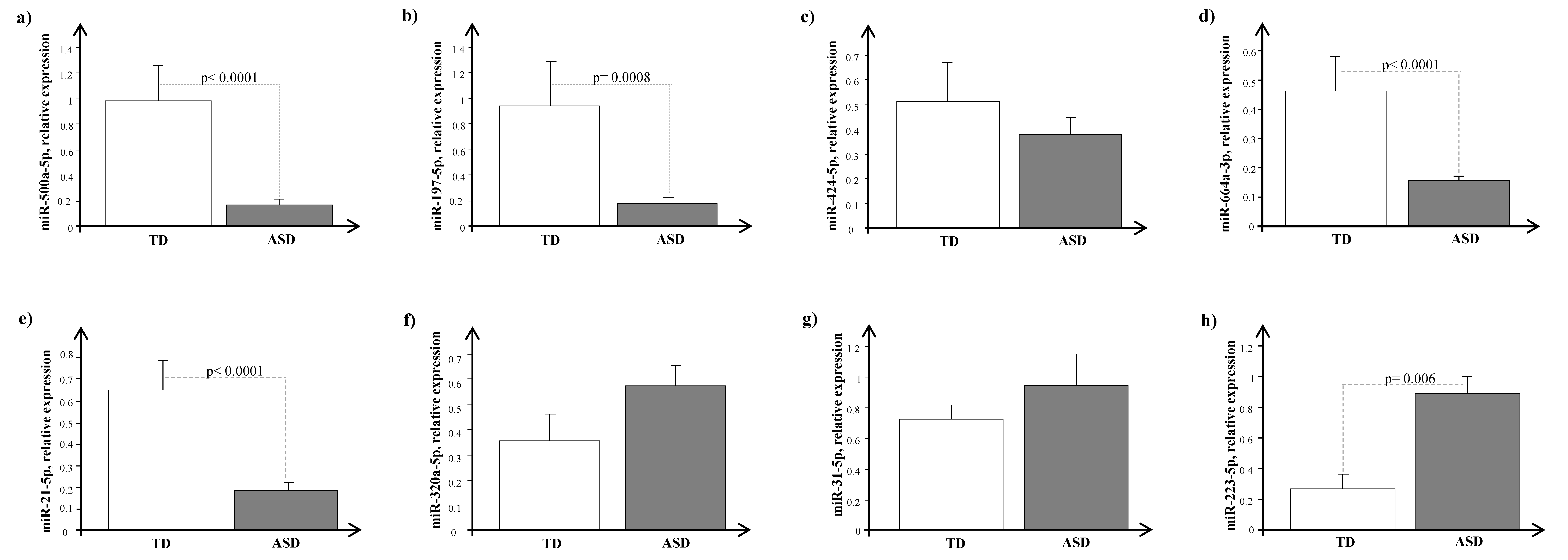
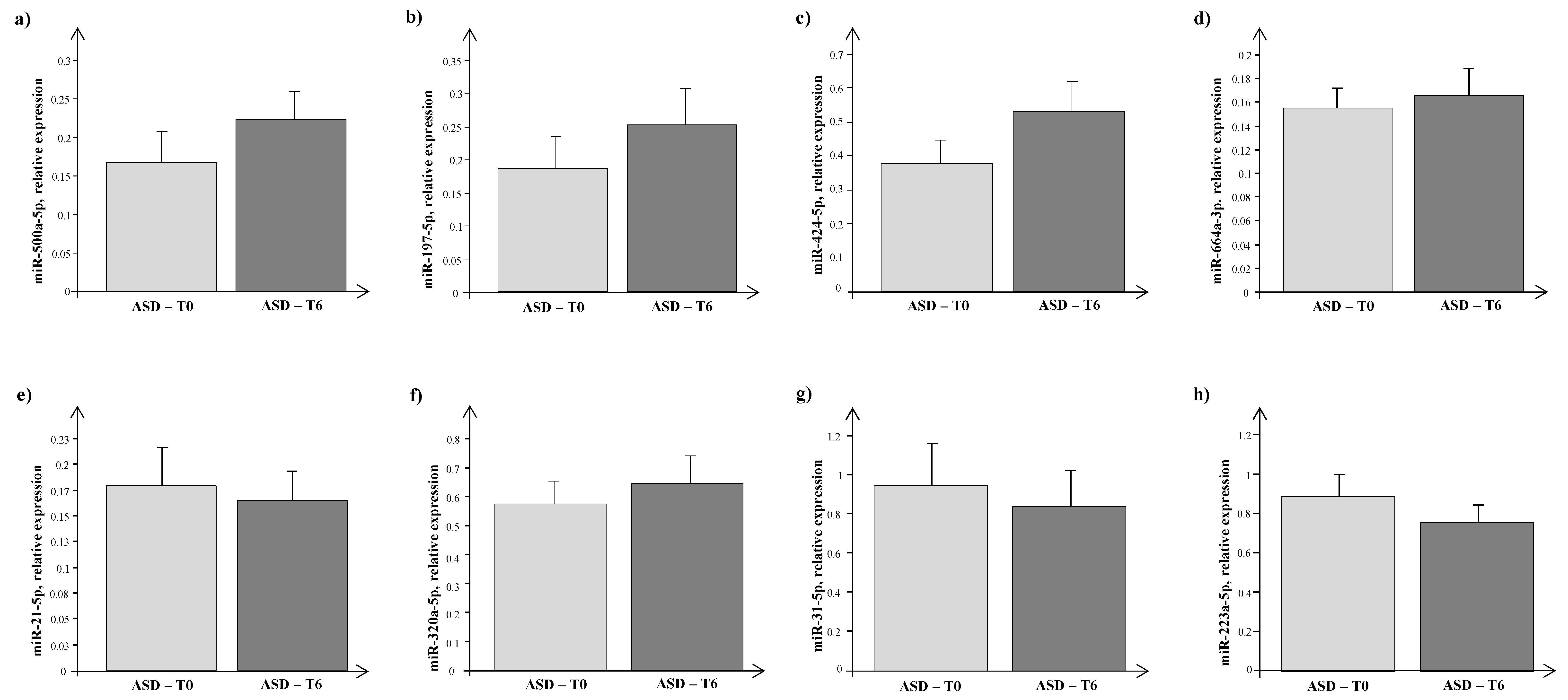
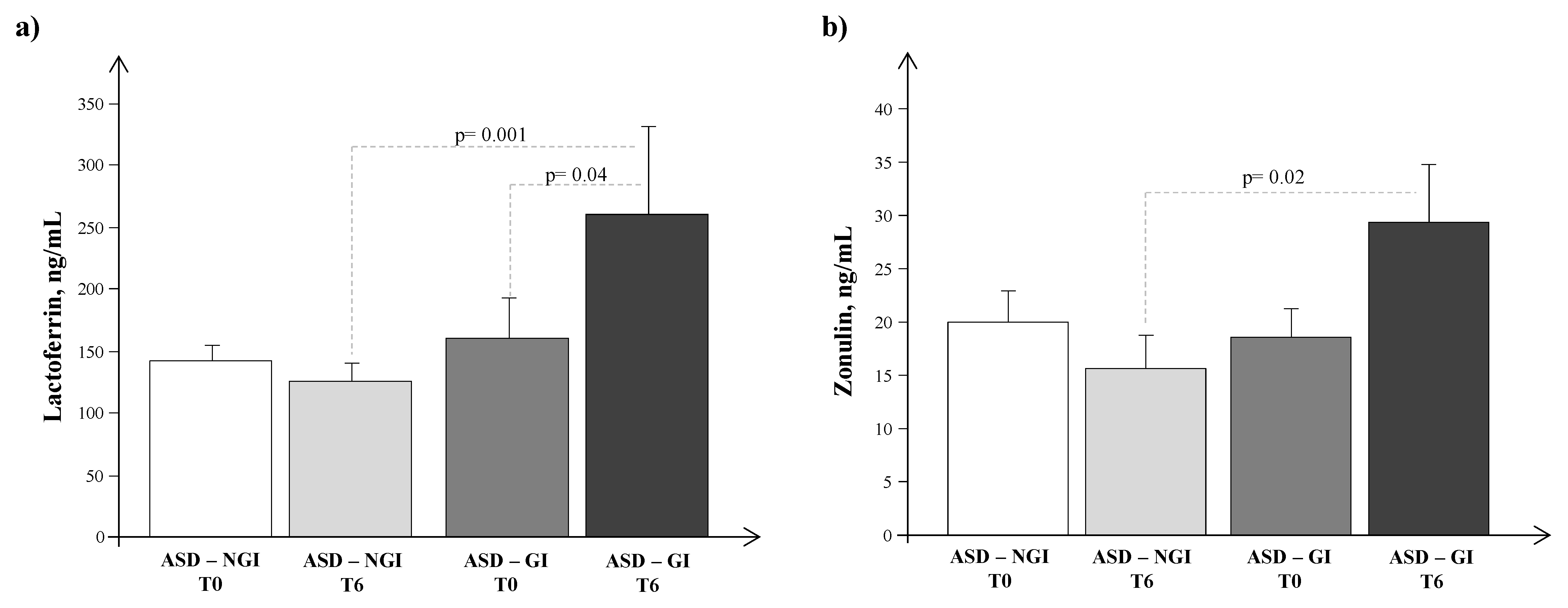
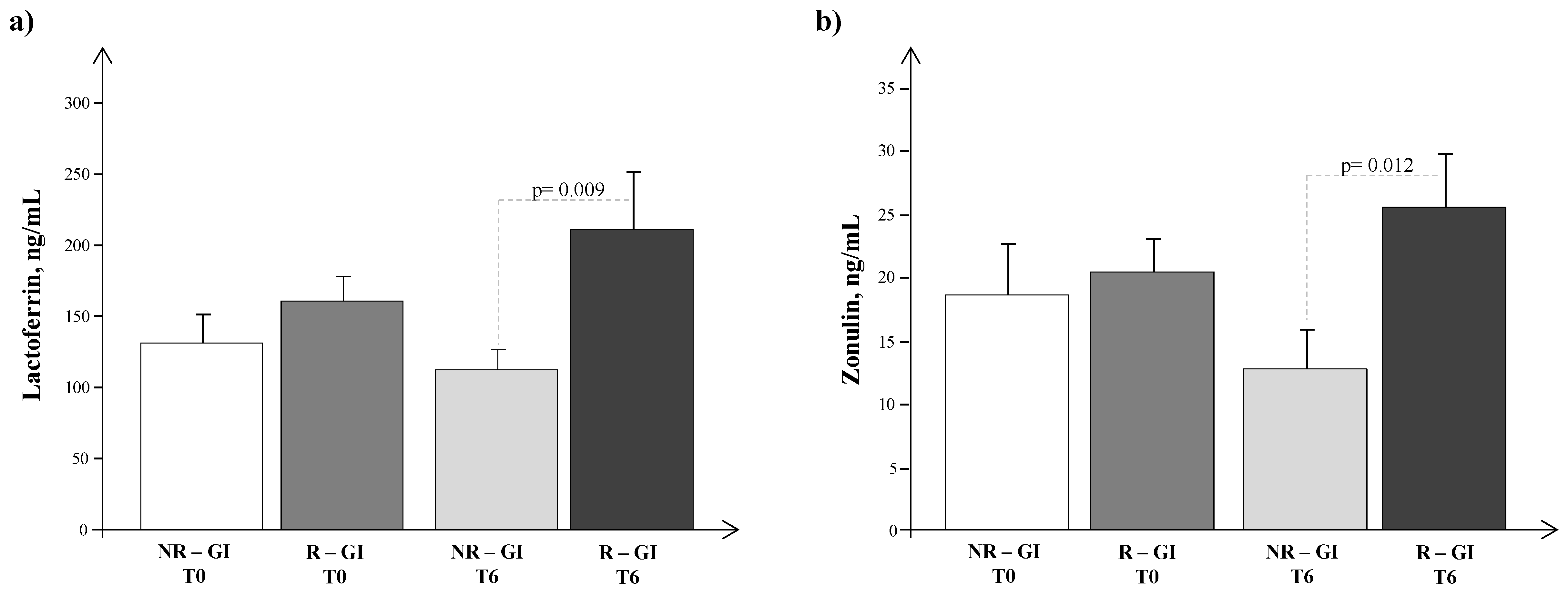
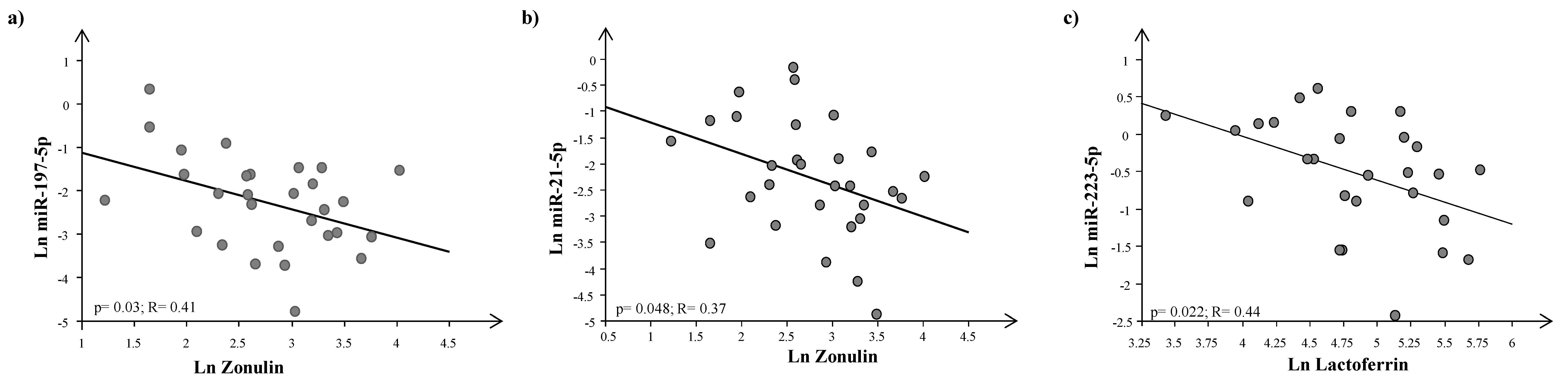
4. Results
5. Discussion
6. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Publishing: Arlignton, VA, USA, 2013. [Google Scholar]
- Zeidan, J.; Fombonne, E.; Scorah, J.; Ibrahim, A.; Durkin, M.S.; Saxena, S.; Yusuf, A.; Shih, A.; Elsabbagh, M. Global prevalence of autism: A systematic review update. Autism Res. 2022, 15, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Calderoni, S. Sex/gender differences in children with autism spectrum disorder: A brief overview on epidemiology, symptom profile, and neuroanatomy. J. Neurosci. Res. 2023, 101, 739–750. [Google Scholar] [CrossRef]
- Bougeard, C.; Picarel-Blanchot, F.; Schmid, R.; Campbell, R.; Buitelaar, J. Prevalence of Autism Spectrum Disorder and Co-morbidities in Children and Adolescents: A Systematic Literature Review. Front. Psychiatry 2021, 12, 744709. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.F.; Tillmann, J.; Ahmad, J.; Crawley, D.; San Jose Caceres, A.; Holt, R.; Charman, T.; Banaschewski, T.; Buitelaar, J.; Simonoff, E.; et al. How do core autism traits and associated symptoms relate to quality of life? Findings from the Longitudinal European Autism Project. Autism 2021, 25, 389–404. [Google Scholar] [CrossRef]
- Cortese, S.; Solmi, M.; Michelini, G.; Bellato, A.; Blanner, C.; Canozzi, A.; Eudave, L.; Farhat, L.C.; Højlund, M.; Köhler-Forsberg, O.; et al. Candidate diagnostic biomarkers for neurodevelopmental disorders in children and adolescents: A systematic review. World Psychiatry 2023, 22, 129–149. [Google Scholar] [CrossRef] [PubMed]
- Chaste, P.; Leboyer, M. Autism risk factors: Genes, environment, and gene-environment interactions. Dialogues Clin. Neurosci. 2012, 14, 281–292. [Google Scholar] [CrossRef]
- Pugsley, K.; Scherer, S.W.; Bellgrove, M.A.; Hawi, Z. Environmental exposures associated with elevated risk for autism spectrum disorder may augment the burden of deleterious de novo mutations among probands. Mol. Psychiatry 2022, 27, 710–730. [Google Scholar] [CrossRef]
- Wiśniowiecka-Kowalnik, B.; Nowakowska, B.A. Genetics and epigenetics of autism spectrum disorder-current evidence in the field. J. Appl. Genet. 2019, 60, 37–47. [Google Scholar] [CrossRef]
- Stoccoro, A.; Conti, E.; Scaffei, E.; Calderoni, S.; Coppedè, F.; Migliore, L.; Battini, R. DNA Methylation Biomarkers for Young Children with Idiopathic Autism Spectrum Disorder: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 9138. [Google Scholar] [CrossRef]
- Loke, Y.J.; Hannan, A.J.; Craig, J.M. The role of epigenetic change in autism spectrum disorders. Front. Neurol. 2015, 6, 107. [Google Scholar] [CrossRef]
- Mazzio, E.A.; Soliman, K.F.A. Basic concepts of epigenetics impact of environmental signals on gene expression. Epigenetics 2012, 7, 119–130. [Google Scholar] [CrossRef]
- Im, H.-I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef]
- Rajgor, D.; Hanley, J. The ins and outs of miRNA-mediated gene silencing during neuronal synaptic plasticity. Non-Coding RNA 2016, 2, 1. [Google Scholar] [CrossRef]
- Anitha, A.; Thanseem, I. microRNA and Autism. In microRNA: Medical Evidence. Advances in Experimental Medicine and Biology; Santulli, G., Ed.; Springer: Cham, Switzerland, 2015; Volume 888. [Google Scholar] [CrossRef]
- Vasu, M.M.; Sumitha, P.S.; Rahna, P.; Thanseem, I.; Anitha, A. microRNAs in Autism Spectrum Disorders. Curr. Pharm. Des. 2019, 25, 4368–4378. [Google Scholar] [CrossRef]
- Kichukova, T.; Petrov, V.; Popov, N.; Minchev, D.; Naimov, S.; Minkov, I.; Vachev, T. Identification of serum microRNA signatures associated with autism spectrum disorder as promising candidate biomarkers. Heliyon 2021, 7, e07462. [Google Scholar] [CrossRef] [PubMed]
- Le Couteur, A.; Lord, C.; Rutter, M. Autism Diagnostic Interview—Revised; Western Psychological Services: Los Angeles, CA, USA, 2003. [Google Scholar]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.M.; Bercik, P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology 2009, 136, 2003–2014. [Google Scholar] [CrossRef]
- Rhee, S.H.; Pothoulakis, C.; Mayer, E.A. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Bezawada, N.; Phang, T.H.; Hold, G.L.; Hansen, R. Autism Spectrum Disorder and the Gut Microbiota in Children: A Systematic Review. Ann. Nutr. Metab. 2020, 24, 1–14. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Gut microbiota: A missing link in psychiatry. World Psychiatry 2020, 19, 111–112. [Google Scholar] [CrossRef] [PubMed]
- Iannone, L.F.; Preda, A.; Blottiere, H.M.; Clarke, G.; Albani, D.; Belcastro, V.; Carotenuto, M.; Cattaneo, A.; Citraro, R.; Ferraris, C.; et al. Microbiota-gut brain axis involvement in neuropsychiatric disorders. Expert. Rev. Neurother. 2019, 19, 1037–1050. [Google Scholar] [CrossRef]
- Santocchi, E.; Guiducci, L.; Fulceri, F.; Billeci, L.; Buzzigoli, E.; Apicella, F.; Calderoni, S.; Grossi, E.; Morales, M.A.; Muratori, F. Gut to brain interaction in Autism Spectrum Disorders: A randomized controlled trial on the role of probiotics on clinical, biochemical and neurophysiological parameters. BMC Psychiatry 2016, 16, 183. [Google Scholar] [CrossRef] [PubMed]
- Santocchi, E.; Guiducci, L.; Prosperi, M.; Calderoni, S.; Gaggini, M.; Apicella, F.; Tancredi, R.; Billeci, L.; Mastromarino, P.; Grossi, E.; et al. Effects of Probiotic Supplementation on Gastrointestinal, Sensory and Core Symptoms in Autism Spectrum Disorders: A Randomized Controlled Trial. Front. Psychiatry 2020, 11, 550593. [Google Scholar] [CrossRef] [PubMed]
- Holingue, C.; Newill, C.; Lee, L.C.; Pasricha, P.J.; Daniele Fallin, M. Gastrointestinal symptoms in autism spectrum disorder: A review of the literature on ascertainment and prevalence. Autism Res. 2018, 11, 24–36. [Google Scholar] [CrossRef]
- Deng, W.; Wang, S.; Li, F.; Wang, F.; Xing, Y.P.; Li, Y.; Lv, Y.; Ke, H.; Li, Z.; Lv, P.J.; et al. Gastrointestinal symptoms have a minor impact on autism spectrum disorder and associations with gut microbiota and short-chain fatty acids. Front. Microbiol. 2022, 13, 1000419. [Google Scholar] [CrossRef]
- Maenner, M.J.; Arneson, C.L.; Levy, S.E.; Kirby, R.S.; Nicholas, J.S.; Durkin, M.S. Brief report: Association between behavioral features and gastrointestinal problems among children with autism spectrum disorder. J. Autism Dev. Disord. 2012, 42, 1520–1525. [Google Scholar] [CrossRef] [PubMed]
- Mazefsky, C.A.; Schreiber, D.R.; Olino, T.M.; Minshew, N.J. The association between emotional and behavioral problems and gastrointestinal symptoms among children with high-functioning autism. Autism 2013, 18, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Fulceri, F.; Morelli, M.; Santocchi, E.; Cena, H.; Del Bianco, T.; Narzisi, A.; Calderoni, S.; Muratori, F. Gastrointestinal symptoms and behavioral problems in preschoolers with Autism Spectrum Disorder. Dig. Liver Dis. 2016, 48, 248–254. [Google Scholar] [CrossRef]
- Prosperi, M.; Santocchi, E.; Balboni, G.; Narzisi, A.; Bozza, M.; Fulceri, F.; Apicella, F.; Igliozzi, R.; Cosenza, A.; Tancredi, R.; et al. Behavioral Phenotype of ASD Preschoolers with Gastrointestinal Symptoms or Food Selectivity. J. Autism Dev. Disord. 2017, 47, 3574–3588. [Google Scholar] [CrossRef]
- Restrepo, B.; Angkustsiri, K.; Taylor, S.L.; Rogers, S.J.; Cabral, J.; Heath, B.; Hechtman, A.; Solomon, M.; Ashwood, P.; Amaral, D.G.; et al. Developmental-behavioral profiles in children with autism spectrum disorder and co-occurring gastrointestinal symptoms. Autism Res. 2020, 13, 1778–1789. [Google Scholar] [CrossRef]
- Horvath, K.; Perman, J.A. Autistic disorder and gastrointestinal disease. Curr. Opin. Pediatr. 2002, 14, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.B.; Audhya, T.; McDonough-Means, S.; Rubin, R.A.; Quig, D.; Geis, E.; Gehn, E.; Loresto, M.; Mitchell, J.; Atwood, S.; et al. Nutritional and metabolic status of children with autism vs. neurotypical children, and the association with autism severity. Nutr. Metab. 2011, 8, 34. [Google Scholar] [CrossRef]
- Fowlie, G.; Cohen, N.; Ming, X. The Perturbance of Microbiome and Gut-Brain Axis in Autism Spectrum Disorders. Int. J. Mol. Sci. 2018, 19, 2251. [Google Scholar] [CrossRef] [PubMed]
- Cekici, H.; Sanlier, N. Current nutritional approaches in managing autism spectrum disorder: A review. Nutr. Neurosci. 2019, 22, 145–155. [Google Scholar] [CrossRef]
- Peretti, S.; Mariano, M.; Mazzocchetti, C.; Mazza, M.; Pino, M.C.; Verrotti Di Pianella, A.; Valenti, M. Diet: The keystone of autism spectrum disorder? Nutr. Neurosci. 2018, 22, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Tas, A.A. Dietary strategies in Autism Spectrum Disorder (ASD). Prog. Nutr. 2018, 20, 554–562. [Google Scholar]
- Prosperi, M.; Santocchi, E.; Guiducci, L.; Frinzi, J.; Morales, M.A.; Tancredi, R.; Muratori, F.; Calderoni, S. Interventions on Microbiota: Where Do We Stand on a Gut–Brain Link in Autism? A Systematic Review. Nutrients 2022, 14, 462. [Google Scholar] [CrossRef]
- Vasiliu, O. The current state of research for psychobiotics use in the management of psychiatric disorders–A systematic literature review. Front. Psychiatry 2023, 14, 1074736. [Google Scholar] [CrossRef]
- Lord, C.; Charman, T.; Havdahl, A.; Carbone, P.; Anagnostou, E.; Boyd, B.; Carr, T.; de Vries, P.J.; Dissanayake, C.; Divan, G.; et al. The Lancet Commission on the future of care and clinical research in autism. Lancet 2021, 399, 271–334. [Google Scholar] [CrossRef]
- Fasano, A.; Not, T.; Wang, W.; Uzzau, S.; Berti, I.; Tommasini, A.; Goldblum, S.E. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet 2000, 355, 1518–1519. [Google Scholar] [CrossRef]
- Schneider, C.K.; Melmed, R.D.; Barstow, L.E.; Enriquez, F.J.; Ranger-Moore, J.; Ostrem, J.A. Oral human immunoglobulin for children with autism and gastrointestinal dysfunction: A prospective, open label study. J. Autism Dev. Disord. 2006, 36, 1053–1064. [Google Scholar] [CrossRef]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism-comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef]
- Wiggins, L.D.; Barger, B.; Moody, E.; Soke, G.; Pandey, J.; Levy, S. Brief Report: The ADOS Calibrated Severity Score Best Measures Autism Diagnostic Symptom Severity in Pre-School Children. J. Autism Dev. Disord. 2019, 49, 2999–3006. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Rutter, M.; DiLavore, P.; Risi, S.; Gotham, K.; Bishop, S. Autism Diagnostic Observation Schedule (ADOS-2), 2nd ed.; Manual (Part I), Modules 1–4; Western Psychological Services: Torrance, CA, USA, 2012. [Google Scholar]
- Cabiati, M.; Randazzo, E.; Salvadori, C.; Peroni, D.; Federico, G.; Del Ry, S. Circulating microRNAs associated with C-type natriuretic peptide in childhood obesity. Peptides 2020, 133, 170387. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Jiaoming, L.; Xiang, W.; Yanhui, L.; Shu, J.; Maling, G.; Qing, M. Analyzing the interactions of mRNAs, miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA networksglioblastoma. J. Neurooncol. 2018, 137, 493–502. [Google Scholar] [CrossRef]
- Yoshino, Y.; Roy, B.; Dwivedi, Y. Differential and unique patterns of synaptic miRNA expression in dorsolateral prefrontal cortex of depressed subjects. Neuropsychopharmacology 2020, 46, 900–910. [Google Scholar] [CrossRef]
- Brennan, G.P.; Henshall, D.C. MicroRNAs as regulators of brain function and targets for treatment of epilepsy. Nat. Rev. Neurol. 2020, 16, 506–519. [Google Scholar] [CrossRef]
- Hu, Z.; Gao, S.; Lindberg, D.; Panja, D.; Wakabayashi, Y.; Li, K.; Kleinman, J.E.; Zhu, J.; Li, Z. Temporal dynamics of miRNAs in human DLPFC and its association with miRNA dysregulation in schizophrenia. Transl. Psychiatry 2019, 9, 196. [Google Scholar] [CrossRef]
- Siedlecki-Wullich, D.; Català-Solsona, J.; Fábregas, C.; Hernández, I.; Clarimon, J.; Lleó, A.; Boada, M.; Saura, C.A.; Rodríguez-Álvarez, J.; Miñano-Molina, A.J. Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer’s disease. Alzheimer’s Res. Ther. 2019, 11, 46. [Google Scholar] [CrossRef]
- Kosik, K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006, 7, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Torres, N.; Guzmán-Torres, K.; García-Cerro, S.; Pinilla Bermúdez, G.; Cruz-Baquero, C.; Ochoa, H.; García-González, D.; Canal-Rivero, M.; Crespo-Facorro, B.; Ruiz-Veguilla, M. miRNAs as biomarkers of autism spectrum disorder: A systematic review and meta-analysis. Eur. Child. Adolesc. Psychiatry 2023. Epub ahead of print. [Google Scholar] [CrossRef]
- Simon, D.J.; Madison, J.M.; Conery, A.L.; Thompson-Peer, K.L.; Soskis, M.; Ruvkun, G.B.; Kaplan, J.M.; Kim, J.K. The microRNA miR-1regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 2008, 133, 903–915. [Google Scholar] [CrossRef]
- Südhof, T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef]
- Hu, Z.; Hom, S.; Kudze, T.; Tong, X.-J.; Choi, S.; Aramuni, G.; Zhang, W.; Kaplan, J.M. Neurexin and neuroligin mediate retrograde synaptic inhibition in C. elegans. Science 2012, 337, 980–984. [Google Scholar] [CrossRef]
- Sarachana, T.; Zhou, R.; Chen, G.; Manji, H.K.; Hu, V.W. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2010, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.S.; Fregeac, J.; Bole-Feysot, C.; Cagnard, N.; Iyer, A.; Anink, J.; Aronica, E.; Alibeu, O.; Nitschke, P.; Colleaux, L. Role of miR-146a in neural stem cell differentiation and neural lineage determination: Relevance for neurodevelopmental disorders. Mol. Autism 2018, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.D.; Ignacio, C.; Gentile, K.; Middleton, F.A. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016, 16, 52. [Google Scholar] [CrossRef]
- Vasu, M.; Anitha, A.; Thanseem, I.; Suzuki, K.; Yamada, K.; Takahashi, T.; Wakuda, T.; Iwata, K.; Tsujii, M.; Sugiyama, T.; et al. Serum microRNA profiles in children with autism. Mol. Autism 2014, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Cirnigliaro, M.; Barbagallo, C.; Gulisano, M.; Domini, C.N.; Barone, R.; Barbagallo, D.; Ragusa, M.; Di Pietro, C.; Rizzo, R.; Purrello, M. Expression and regulatory network analysis of miR-140-3p, a new potential serum biomarker for autism spectrum disorder. Front. Mol. Neurosci. 2017, 10, 250. [Google Scholar] [CrossRef]
- Yu, D.; Jiao, X.; Cao, T.; Huang, F. Serum miRNA expression profiling reveals miR-486-3p may play a significant role in the development of autism by targeting ARID1B. NeuroReport 2018, 29, 1431–1436. [Google Scholar] [CrossRef]
- Hosokawa, R.; Yoshino, Y.; Funahashi, Y.; Horiuchi, F.; Iga, J.-I.; Ueno, S.-I. MiR-15b-5p Expression in the Peripheral Blood: A Potential Diagnostic Biomarker of Autism Spectrum Disorder. Brain Sci. 2023, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Caracci, M.O.; Avila, M.E.; Espinoza-Cavieres, F.A.; Lopez, H.R.; Ugarte, G.D.; De Ferrari, G.V. Wnt/beta-Catenin-Dependent Transcription in Autism Spectrum Disorders. Front. Mol. Neurosci. 2021, 14, 764756. [Google Scholar] [CrossRef]
- Hicks, S.D.; Carpenter, R.L.; Wagner, K.E.; Pauley, R.; Barros, M.; Tierney-Aves, C.; Barns, S.; Greene, C.D.; Middleton, F.A. Saliva microRNA differentiates children with autism from peers with typical and atypical development. J. Am. Acad. Child. Adolesc. Psychiatry 2020, 59, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.E.; Parikshak, N.N.; Belgard, T.G.; Geschwind, D.H. Genome- wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat. Neurosci. 2016, 19, 1463–1476. [Google Scholar] [CrossRef]
- Courchesne, E.; Carper, R.; Akshoomoff, N. Evidence of brain overgrowth in the first year of life in autism. JAMA 2003, 290, 337–344. [Google Scholar] [CrossRef]
- Dementieva, Y.A.; Vance, D.D.; Donnelly, S.L.; Elston, L.A.; Wolpert, C.M.; Ravan, S.A.; DeLong, G.R.; Abramson, R.K.; Wright, H.H.; Cuccaro, M.L. Accelerated Head Growth in Early Development of Individuals with Autism. Pediatr. Neurol. 2005, 32, 102–108. [Google Scholar] [CrossRef]
- Muratori, F.; Calderoni, S.; Apicella, F.; Filippi, T.; Santocchi, E.; Calugi, S.; Cosenza, A.; Tancredi, R.; Narzisi, A. Tracing back to the onset of abnormal head circumference growth in Italian children with autism spectrum disorder. Res. Autism Spect. Disord. 2012, 6, 442–449. [Google Scholar] [CrossRef]
- Wheeler, G.; Ntounia-Fousara, S.; Granda, B.; Rathjen, T.; Dalmay, T. Identification of new central nervous system specific mouse microRNAs. FEBS Fed. Eur. Biochem. Soc. Lett. 2006, 580, 2195–2200. [Google Scholar] [CrossRef]
- Bai, X.; Bian, Z. MicroRNA-21 Is a Versatile Regulator and Potential Treatment Target in Central Nervous System Disorders. Front. Mol. Neurosci. 2022, 15, 842288. [Google Scholar] [CrossRef] [PubMed]
- Behrouzi, A.; Ashrafian, F.; Mazaheri, H.; Lari, A.; Nouri, M.; Riazi Rad, F.; Hoseini Tavassol, Z.; Siadat, S.D. The importance of interaction between microRNAs and gut microbiota in several pathways. Microb. Pathog. 2020, 144, 104200. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant derived exosomal microRNAs shape the gut microbiota. Cell Host. Microbe. 2018, 24, 637–652. [Google Scholar] [CrossRef]
- Liu, N.; Feng, G.; Zhang, X.; Hu, Q.; Sun, S.; Sun, J.; Sun, Y.; Wang, R.; Zhang, Y.; Wang, P.; et al. The Functional Role of Lactoferrin in Intestine Mucosal Immune System and Inflammatory Bowel Disease. Front. Nutr. 2021, 8, 759507. [Google Scholar] [CrossRef]
- Wang, H.; Chao, K.; Ng, S.C.; Bai, A.H.; Yu, Q.; Yu, J.; Li, M.; Cui, Y.; Chen, M.; Hu, J.F.; et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Lonnerdal, B.; Iyer, S. Lactoferrin: Molecular structure and biological function. Annu. Rev. Nutr. 1995, 15, 93–110. [Google Scholar] [CrossRef]
- Sanchez, L.; Calvo, M.; Brock, J.H. Biological role of lactoferrin. Arch. Dis. Child. 1992, 67, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, P.; Decembrino, L.; Stolfi, I.; Pugni, L.; Rinaldi, M.; Cattani, S.; Romeo, M.G.; Messner, H.; Laforgia, N.; Vagnarelli, F.; et al. Lactoferrin and prevention of late-onset sepsis in the pre-term neonates. Early Hum. Dev. 2010, 86, 59–61. [Google Scholar] [CrossRef]
- Baveye, S.; Elass, E.; Mazurier, J.; Spik, G.; Legrand, D. Lactoferrin: A multifunctional glycoprotein involved in the modulation of the inflammatory process. Clin. Chem. Lab. Med. 1999, 37, 281–286. [Google Scholar] [CrossRef]
- Asbjornsdottir, B.; Snorradottir, H.; Andresdottir, E.; Fasano, A.; Lauth, B.; Gudmundsson, L.S.; Gottfredsson, M.; Halldorsson, T.I.; Birgisdottir, B.E. Zonulin-Dependent Intestinal Permeability in Children Diagnosed with Mental Disorders: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 1982. [Google Scholar] [CrossRef]
- Esnafoglu, E.; Cırrık, S.; Ayyıldız, S.N.; Erdil, A.; Ertürk, E.Y.; Daglı, A.; Noyan, T. Increased serum zonulin levels as an intestinal permeability marker in autistic subjects. J. Pediatr. 2017, 188, 240–244. [Google Scholar] [CrossRef]
- Karagözlü, S.; Dalgıç, B.; İşeri, E. The Relationship of Severity of Autism with Gastrointestinal Symptoms and Serum Zonulin Levels in Autistic Children. J. Autism Dev. Disord. 2021, 52, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Jyonouchi, H.; Geng, L.; Streck, D.L.; Dermody, J.J.; Toruner, G.A. MicroRNA expression changes in association with changes in interleukin-1./interleukin10 ratios produced by monocytes in autism spectrum disorders: Their association with neuropsychiatric symptoms and comorbid conditions (observational study). J. Neuroinflamm. 2017, 14, 229. [Google Scholar] [CrossRef]
- Kichukova, T.M.; Popov, N.T.; Ivanov, I.S.; Vachev, T.I. Profiling of circulating serum microRNAs in children with autism spectrum disorder using stem-loop qRT-PCR assay. Folia Med. 2017, 59, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Mor, M.; Nardone, S.; Sams, D.S.; Elliott, E. Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol. Autism 2015, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Santagati, M.; Mirabella, F.; Lauretta, G.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; Domini, C.N.; Gulisano, M.; Barone, R.; et al. Potential associations among alteration of salivary miRNAs, saliva microbiome structure, and cognitive impairments in autistic children. Int. J. Mol. Sci. 2020, 21, 6203. [Google Scholar] [CrossRef]
- Abu-Elneel, K.; Liu, T.; Gazzaniga, F.S.; Nishimura, Y.; Wall, D.P.; Geschwind, D.H.; Lao, K.; Kosik, K.S. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics 2008, 9, 153–161. [Google Scholar] [CrossRef]
- Ekawidyani, K.R.; Abdullah, M. Diet, nutrition and intestinal permeability: A mini review. Asia Pac. J. Clin. Nutr. 2023, 32, 8–12. [Google Scholar] [CrossRef]
- Veres-Székely, A.; Szász, C.; Pap, D.; Szebeni, B.; Bokrossy, P.; Vannay, Á. Zonulin as a Potential Therapeutic Target in Microbiota-Gut-Brain Axis Disorders: Encouraging Results and Emerging Questions. Int. J. Mol. Sci. 2023, 24, 7548. [Google Scholar] [CrossRef]
| ASD Subjects (n = 31) | TD Subjects (n = 10) | p-Value | |
|---|---|---|---|
| Males (%) | 77.4 | 77.7 | n.s. |
| AGE (years) | 4.28 ± 0.22 | 5.00 ± 0.61 | n.s. |
| WEIGHT (kg) | 17.62 ± 0.67 | 15.18 ± 1.45 | n.s. |
| HEIGHT (cm) | 104.68 ± 1.62 | 100.85 ± 5.53 | n.s. |
| BMI (kg/m2) | 15.93 ± 0.35 | 14.73 ± 0,11 | n.s. |
| BMI Z-SCORE | 0.03 ± 0.26 | −0.93 ± 0.29 | n.s. |
| Gene | Forward Primer Sequence (5′-3′) | Genbank Accession Number | Location | Ta, °C |
|---|---|---|---|---|
| hsa-miR-197-5p | CGGGTAGAGAGGGCAGTGGGAGG | NR_029583.1 | chr 1p13.3 | 55 |
| hsa-miR-424-5p | CAGCAGCAATTCATGTTTTGAA | NR_029946.1 | chr Xq26.3 | 55 |
| hsa-miR-664a-3p | TATTCATTTATCCCCAGCCTACA | NR_031705.1 | chr 1q41 | 55 |
| hsa-miR-500a-5p | TAATCCTTGCTACCTGGGTGAGA | NR_030224.1 | chr Xp11.23 | 55 |
| hsa-miR-21-5p | TAGCTTATCAGACTGATGTTGA | NR_029493.1 | chr 17q23.1 | 55 |
| hsa-miR-320a-5p | GCCTTCTCTTCCCGGTTCTTCC | NR_029714.2 | chr 8p21.3 | 55 |
| hsa-miR-31-5p | AGGCAAGATGCTGGCATAGCT | NR_029505.1 | chr 9p21.3 | 55 |
| hsa-miR-223-5p | CGTGTATTTGACAAGCTGAGTT | LM608368 | chr Xq12 | 55 |
| miR-197-5p | miR-21-5p | miR-320a-5p | miR-424-5p | miR-500a-5p | miR-664a-5p | |
|---|---|---|---|---|---|---|
| miR-197-5p | - | R = 0.36; p = 0.003 | R = 0.35; p = 0.004 | R = 0.33; p = 0.007 | R = 0.60; p < 0.0001 | R = 0.55; p < 0.0001 |
| miR-21-5p | R= 0.36; p = 0.003 | - | - | R= 0.29; p = 0.02 | R = 0.74; p < 0.0001 | R = 0.64; p < 0.0001 |
| miR-320a-5p | R= 0.35; p = 0.004 | - | - | R= 0.45; p = 0.002 | - | - |
| miR-500a-5p | R= 0.60; p < 0.0001 | R = 0.74; p < 0.0001 | - | R = 0.25; p = 0.05 | - | R = 0.71; p < 0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guiducci, L.; Cabiati, M.; Santocchi, E.; Prosperi, M.; Morales, M.A.; Muratori, F.; Randazzo, E.; Federico, G.; Calderoni, S.; Del Ry, S. Expression of miRNAs in Pre-Schoolers with Autism Spectrum Disorders Compared with Typically Developing Peers and Its Effects after Probiotic Supplementation. J. Clin. Med. 2023, 12, 7162. https://doi.org/10.3390/jcm12227162
Guiducci L, Cabiati M, Santocchi E, Prosperi M, Morales MA, Muratori F, Randazzo E, Federico G, Calderoni S, Del Ry S. Expression of miRNAs in Pre-Schoolers with Autism Spectrum Disorders Compared with Typically Developing Peers and Its Effects after Probiotic Supplementation. Journal of Clinical Medicine. 2023; 12(22):7162. https://doi.org/10.3390/jcm12227162
Chicago/Turabian StyleGuiducci, Letizia, Manuela Cabiati, Elisa Santocchi, Margherita Prosperi, Maria Aurora Morales, Filippo Muratori, Emioli Randazzo, Giovanni Federico, Sara Calderoni, and Silvia Del Ry. 2023. "Expression of miRNAs in Pre-Schoolers with Autism Spectrum Disorders Compared with Typically Developing Peers and Its Effects after Probiotic Supplementation" Journal of Clinical Medicine 12, no. 22: 7162. https://doi.org/10.3390/jcm12227162
APA StyleGuiducci, L., Cabiati, M., Santocchi, E., Prosperi, M., Morales, M. A., Muratori, F., Randazzo, E., Federico, G., Calderoni, S., & Del Ry, S. (2023). Expression of miRNAs in Pre-Schoolers with Autism Spectrum Disorders Compared with Typically Developing Peers and Its Effects after Probiotic Supplementation. Journal of Clinical Medicine, 12(22), 7162. https://doi.org/10.3390/jcm12227162







