Protein Biomarkers Shared by Multiple Neurodegenerative Diseases Are Calmodulin-Binding Proteins Offering Novel and Potentially Universal Therapeutic Targets
Abstract
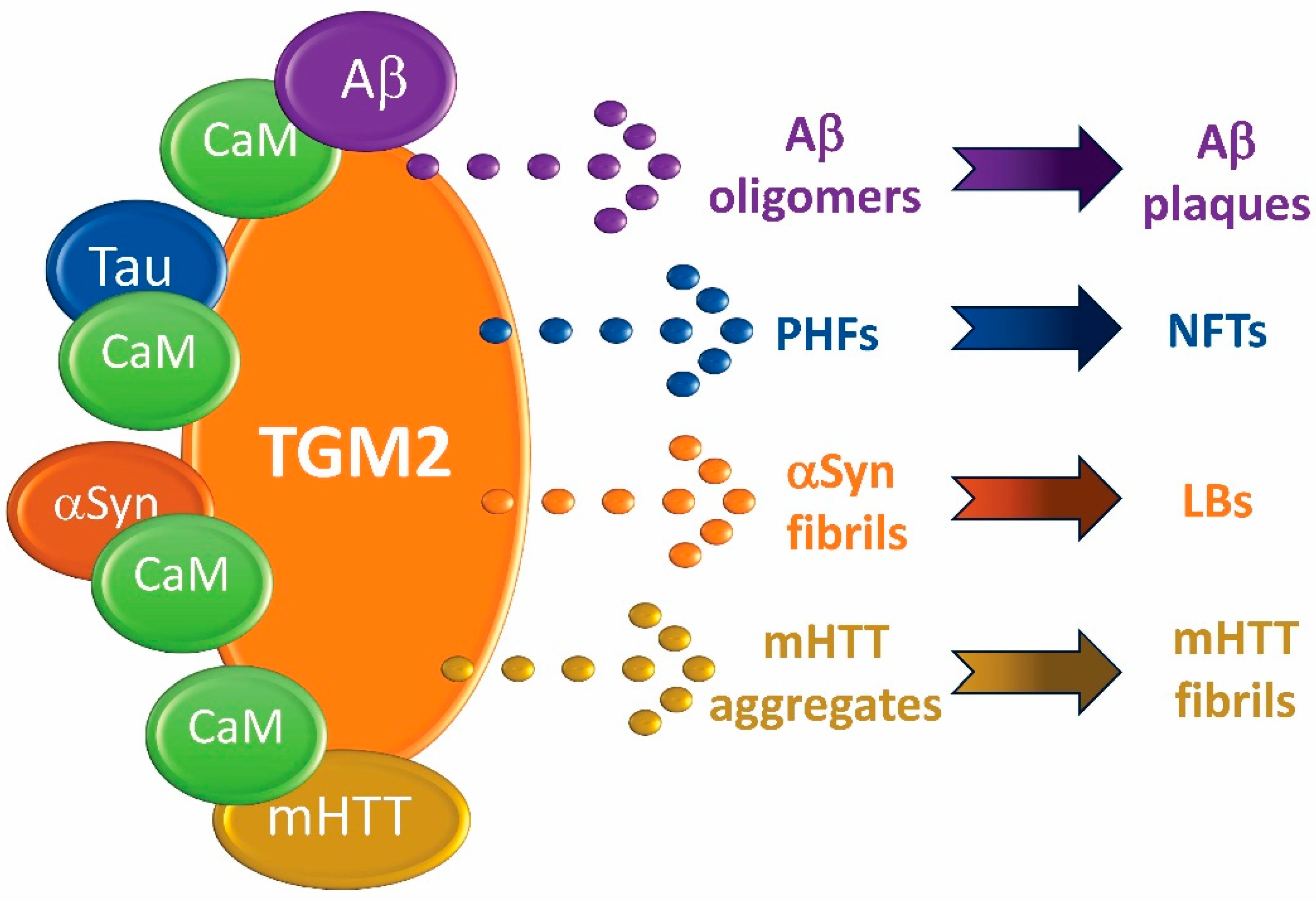
1. Introduction
2. AD
3. HD
4. PD
5. Lewy Body Diseases
6. FTD
7. ALS
8. MS
9. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCA1 | ATP-binding cassette protein A1 |
| AchR | acetylcholine receptor |
| AD | Alzheimer’s disease |
| ALS | amyotrophic lateral sclerosis |
| APOE | apolipoprotein E |
| αSyn | alpha-synuclein |
| BIN1 | bridging integrator 1 |
| C9orf72 | chromosome 9 open reading frame 72 |
| CaM | calmodulin |
| CaMBD | calmodulin-binding domain |
| CaMBP | calmodulin-binding protein |
| CaMKII | calmodulin-dependent protein kinase II |
| cdk5 | cyclin-dependent kinase 5 |
| CH3L1/YKL-40 | chitinase-3-like protein I |
| CHCHD10 | coiled-coil–helix-coiled-coil–helix domain-containing protein 10 |
| CLU | clusterin |
| CR1 | complement receptor type 1 |
| D2DR | D2-dopamine receptor |
| FTD | frontotemporal dementia |
| FUS | fused in sarcoma-RNA-binding protein |
| GBA | glucocerebrosidase |
| GRN | progranulin |
| HD | Huntington’s disease |
| LB | Lewy body |
| LBD | Lewy body disease |
| LF | lactoferrin |
| LRRK2 | leucine-rich repeat kinase 2 |
| MS | multiple sclerosis |
| NFTs | neurofibrillary tangles |
| Ng | neurogranin |
| NfL | neurofilament light chain |
| NMDAR | N-methyl-D-aspartate receptor |
| PD | Parkinson’s disease |
| PARK7 | Parkinsonism-associated deglycase 7 |
| PILRA | paired immunoglobin-like type 2 receptor alpha |
| PINK1 | PTEN induced kinase 1 |
| PP2B | calcineurin |
| ROS | reactive oxygen species |
| SNCA | α-synuclein |
| SQSTM1 | sequestosome 1 |
| TBI | traumatic brain injury |
| TGM1/2 | transglutamase ½ |
| TMEM175 | transmembrane protein 175 |
| TDP-43/TARDBP | TAR DNA-binding protein |
| TREM2 | triggering receptor expressed on myeloid cells 2 |
| VCP | valosin-containing protein |
References
- Biomarkers Definition Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Therapeut. 2001, 69, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, W.M.Y.; Kobeissym, F.; Mondello, S. Editorial: Biomarkers in neurology, vol. II. Front. Neurol. 2023, 14, 1244536. [Google Scholar] [CrossRef] [PubMed]
- Sturmey, E.; Malaspina, A. Blood biomarkers in ALS: Challenges.; applications and novel frontiers. Acta Neurol. Scand. 2022, 146, 375–388. [Google Scholar] [CrossRef]
- Azam, S.; Haque, M.E.; Balakrishnan, R.; Kim, I.-S.; Choi, D.-K. The Ageing Brain: Molecular and Cellular Basis of Neurodegeneration. Front. Cell Dev. Biol. 2021, 9, 683459. [Google Scholar] [CrossRef] [PubMed]
- Wyss-Coray, T. Ageing, neurodegeneration and brain rejuvenation. Nature 2016, 539, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Bourdenx, M.; Koulakiotis, N.S.; Sanoudou, D.; Bezard, E.; Dehay, B.; Tsarbopoulos, A. Protein aggregation and neurodegeneration in prototypical neurodegenerative diseases: Examples of amyloidopathies.; tauopathies and synucleinopathies. Prog. Neurobiol. 2017, 155, 171–193. [Google Scholar] [CrossRef]
- O’Day, D.H.; Eshak, K.; Myre, M.A. Calmodulin Binding Proteins and Alzheimer’s Disease: A Review. J. Alz. Dis. 2015, 46, 553–569. [Google Scholar] [CrossRef]
- O’Day, D.H.; Huber, R.L. Calmodulin binding proteins and neuroinflammation in multiple neurodegenerative diseases. BMC Neurosci. 2022, 23, 10. [Google Scholar] [CrossRef]
- Mark, M.D.; Schwitalla, J.C.; Groemmke, M.; Herlitze, S. Keeping our calcium in balance to maintain our balance. Biochem. Biophys. Res. Commun. 2017, 483, 1040–1050. [Google Scholar] [CrossRef]
- Ghosh, A.; Greenberg, M.E. Calcium signaling in neurons: Molecular mechanisms and cellular consequences. Science 1995, 268, 239–247. [Google Scholar] [CrossRef]
- Kolobkova, Y.A.; Vigont, V.A.; Shalygin, A.V.; Kaznacheyeva, E.V. Huntington’s disease: Calcium dyshomeostasis and pathology models. Acta Nat. 2017, 9, 33–46. [Google Scholar]
- Popugaeva, E.; Pchitskaya, E.; Bezprozvanny, I. Dysregulation of neuronal calcium homeostasis in Alzheimer’s disease—A therapeutic opportunity? Biochem. Biophys. Res. Commun. 2017, 483, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Webber, E.K.; Fivaz, M.; Stutzmann, G.E.; Griffoen, G. Cytosolic calcium: Judge jury and executioner of neurodegeneration in Alzheimer’s disease and beyond. Alz. Dement. 2023, 19, 3701–3717. [Google Scholar] [CrossRef]
- Schrank, S.; Barrington, N.; Stutzmann, G.E. Calcium-handling defects and neurodegenerative disease. Cold Spr. Harb. Perspect. Biol. 2020, 12, a035212. [Google Scholar] [CrossRef]
- Guzman-Martinez, L.; Maccioni, R.B.; Andrade, V.; Navarrete, L.P.; Pastor, M.G.; Ramos-Escobar, N. Neuroinflammation as a common feature of neurodegenerative disorders. Front. Pharmacol. 2019, 10, 1008. [Google Scholar] [CrossRef] [PubMed]
- Surmeier, D.J.; Schumacker, P.T.; Guzman, J.D.; Ilijic, E.; Yang, B.; Zampese, E. Calcium and Parkinson’s disease. Biochem. Biophys. Res. Commun. 2016, 483, 1013–1019. [Google Scholar] [CrossRef]
- Zalchick, S.V.; McGrath, K.M.; Caraveo, G. The role of Ca2+ signaling in Parkinson’s disease. Dis. Mod. Mech. 2017, 10, 519–535. [Google Scholar] [CrossRef]
- De Marco, G.; Lomartire, A.; Manera, U.; Canosa, A.; Grassano, M.; Casale, F.; Fuda, G.; Salamone, P.; Rinaudo, M.T.; Colombatto, S.; et al. Effects of intracellular calcium accumulation on proteins encoded by the major genes underlying amyotrophic lateral sclerosis. Sci. Rep. 2022, 12, 395. [Google Scholar] [CrossRef] [PubMed]
- Katzeff, J.S.; Bright, F.; Lo, K.; Kril, J.J.; Connolly, A.; Crossett, B.; Ittner, L.M.; Kassiou, M.; Loy, C.T.; Hodges, J.R.; et al. Altered serum protein levels in frontotemporal dementia and amyotrophic lateral sclerosis indicate calcium and immunity dysregulation. Sci. Rep. 2020, 10, 13741. [Google Scholar] [CrossRef]
- Enders, M.; Heider, T.; Ludwig, A.; Kuerten, S. Strategies for Neuroprotection in Multiple Sclerosis and the Role of Calcium. Int. J. Mol. Sci. 2020, 21, 1663. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K. Complicity of α-synuclein oligomer and calcium dyshomeostasis in selective neuronal vulnerability in lewy body disease. Arch. Pharm. Res. 2021, 44, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef] [PubMed]
- O’Day, D.H. Calmodulin binding domains in critical risk proteins involved in neurodegeneration. Curr. Issues Molec. Biol. 2022, 44, 5802–5814. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.R.; Friedberg, F. Sequence motifs for calmodulin recognition. FASEB J. 1997, 11, 331–340. [Google Scholar] [CrossRef]
- Yap, K.L.; Kim, J.; Truong, K.; Sherman, M.; Yuan, T.; Ikura, M. Calmodulin target database. J. Struct. Funct. Genom. 2000, 1, 8–14. [Google Scholar] [CrossRef]
- Mruk, K.; Farley, B.M.; Ritacco, A.W.; Kobertz, W.R. Calmodulation meta-analysis: Predicting calmodulin binding via canonical motif clustering. J. Gen. Physiol. 2014, 144, 105–114. [Google Scholar] [CrossRef]
- Grant, B.M.M.; Enomoto, M.; Ikura, M.; Marshall, C.B. A non-canonical calmodulin target motif comprising a polybasic region and lipidated terminal residue regulates localization. Int. J. Mol. Sci. 2020, 21, 2751. [Google Scholar] [CrossRef]
- Bohush, A.; Leśniak, W.; Weis, S.; Filipek, A. Calmodulin and its binding proteins in Parkinson’s disease. Int. J. Mol. Med. 2021, 22, 2016. [Google Scholar] [CrossRef]
- O’Day, D.H. Calmodulin binding proteins and Alzheimer’s disease: Biomarkers.; regulatory enzymes and receptors that are regulated by calmodulin. Int. J. Mol. Sci. 2020, 21, 7344. [Google Scholar] [CrossRef]
- Hayashi, Y. Molecular mechanism of hippocampal long-term potentiation—Towards multiscale understanding of learning and memory. Neurosci. Res. 2022, 175, 3–15. [Google Scholar] [CrossRef]
- Robinson, A.J. Emerging role of CaMKII in neuropsychiatric disorders. Trends Neurosci. 2014, 37, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. Alzheimer’s Association Report: 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 16, 391–460. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Benoist, C.; Weidner, W. World Alzheimer Report 2023: Reducing Dementia Risk: Never Too Early, Never Too Late; Alzheimer’s Disease International: London, UK, 2023. [Google Scholar]
- Gunes, S.; Aizawa, Y.; Sugashi, T.; Sugimoto, M.; Rodrigues, P.P. Biomarkers for Alzheimer’s Disease in the Current State: A Narrative Review. Int. J. Mol. Sci. 2022, 23, 4962. [Google Scholar] [CrossRef]
- Yang, Y.; Bagyinszky, E.; An, S.S.A. Presenilin-1 (PSEN1) Mutations: Clinical Phenotypes beyond Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 8417. [Google Scholar] [CrossRef]
- Fernández-Calle, R.; Konings, S.C.; Frontiñán-Rubio, J.; García-Revilla, J.; Camprubí-Ferrer, L.; Svensson, M.; Martinson, I.; Boza-Serrano, A.; Venero, J.L.; Nielsen, H.M.; et al. APOE in the bullseye of neurodegenerative diseases: Impact of the APOE genotype in Alzheimer’s disease pathology and brain diseases. Mol. Neurodegener. 2022, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Canobbio, I.; Catricalà, S.; Balduini, C.; Torti, M. Calmodulin regulates the non-amyloidogenic metabolism of amyloid precursor protein in platelets. Biochem. Biophys. Acta 2011, 1813, 500–506. [Google Scholar] [CrossRef]
- Chavez, S.E.; O’Day, D.H. Calmodulin binds to and regulates the activity of beta-secretase (BACE1). Curr. Res. Alz. Dis. 2007, 1, 37–47. [Google Scholar]
- Michno, K.; Knight, D.; Campusano, J.M.; van de Hoef, D.; Boulianne, G.L. Intracellular calcium deficits in Drosophila cholinergic neurons expressing wild type or FAD-mutant presenilin. PLoS ONE 2009, 4, e6904. [Google Scholar] [CrossRef]
- Corbacho, I.; Berrocal, M.; Torok, K.; Mata, A.M.; Gutierrez-Merino, C. High affinity binding of amyloid-peptide to calmodulin: Structural and functional implications. Biochem. Biophys. Res. Commun. 2017, 486, 992–997. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Q.; Mobley, W.C. Alzheimer Disease Pathogenesis: Insights From Molecular and Cellular Biology Studies of Oligomeric Aβ and Tau Species. Front. Neurosci. 2019, 13, 659–680. [Google Scholar] [CrossRef]
- Penke, B.; Szűcs, M.; Bogár, F. Oligomerization and Conformational Change Turn Monomeric β-Amyloid and Tau Proteins Toxic: Their Role in Alzheimer’s Pathogenesis. Molecules 2020, 25, 1659. [Google Scholar] [CrossRef] [PubMed]
- Chong, F.P.; Ng, K.Y.; Koh, R.Y.; Chye, S.M. Tau Proteins and Tauopathies in Alzheimer’s Disease. Cell. Mol. Neurobiol. 2018, 38, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Padilla, R.; Maccioni, R.B.; Avila, J. Calmodulin binds to a tubulin binding site of the microtubule associated protein tau. Mol. Cell. Biochem. 1990, 97, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Min, B.; Chung, K.C. New insight into transglutamase 2 and link to neurodegenerative diseases. BMB Rep. 2018, 51, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Curro, M.; Ferlazzo, N.; Condello, S.; Caccamo, D.; Lentile, R. Transglutaminase 2 silencing reduced the beta-amyloid-effects on the activation of human THP-1 cells. Amino Acids 2010, 39, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Zemaitaitis, M.O.; Lee, J.M.; Troncoso, J.C.; Muma, N.A. Transglutaminase-induced cross-linking of tau proteins in progressive supranuclear palsy. J. Neuropathol. Exp. Neurol. 2000, 59, 983–989. [Google Scholar] [CrossRef]
- Zainelli, G.M.; Ross, C.A.; Troncoso, J.C.; Fitzgerald, J.K.; Muma, N.A. Calmodulin regulates transglutamase 2 cross-linking of huntingtin. J. Neurosci. 2004, 24, 1954–1961. [Google Scholar] [CrossRef]
- Agnello, L.; Gambino, C.M.; Lo Sasso, B.; Bivona, G.; Milano, S.; Ciaccio, A.M.; Piccoli, T.; La Bella, V.; Ciaccio, M. Neurogranin as a Novel Biomarker in Alzheimer’s Disease. Lab Med. 2021, 52, 188–196. [Google Scholar] [CrossRef]
- Klyucherev, T.O.; Olszewski, P.; Shalimova, A.A.; Cubarev, V.N.; Tarasov, V.V.; Attwood, M.M.; Syvänen, S.; Schiöth, H.B. Advances in the development of new biomarkers for Alzheimer’s disease. Transl. Neurodegen. 2022, 11, 25. [Google Scholar] [CrossRef]
- Zhong, L.; Cherry, T.; Bies, C.E.; Florence, M.A.; Gerges, N.Z. Neurogranin enhances synaptic strength through its interaction with calmodulin. EMBO J. 2009, 28, 3027–3039. [Google Scholar] [CrossRef]
- Xiang, Y.; Xin, J.; Le, W.; Yang, Y. Neurogranin: A potential biomarker of neurological and mental diseases. Front. Aging Neurosci. 2020, 12, 58743. [Google Scholar] [CrossRef]
- Li, L.; Massimo, L.; Cole, S.; Novere, N.L.; Edelstein, S.J. Neurogranin stimulates Ca2+/calmodulin-dependent kinase II by suppressing calcineurin activity at specific calcium spike frequencies. PLoS ONE 2020, 16, e1006991. [Google Scholar] [CrossRef] [PubMed]
- Jacobo-Albavera, L.; Domínguez-Pérez, M.; Medina-Leyte, D.J.; González-Garrido, A.; Villarreal-Molina, T. The Role of the ATPBinding Cassette A1 (ABCA1) in Human Disease. Int. J. Mol. Sci. 2021, 22, 1593. [Google Scholar] [CrossRef]
- Pineda-Torra, I.; Siddique, S.; Waddington, K.E.; Farrell, R.; Jury, E.C. Disrupted Lipid Metabolism in Multiple Sclerosis: A Role for Liver X Receptors? Front Endocrinol. 2021, 12, 639757. [Google Scholar] [CrossRef]
- Lewandowski, C.T.; Laham, M.S.; Thatcher, G.R.J. Remembering your A.; B.; C’s: Alzheimer’s disease and ABCA1. Acta Pharm. Sin B 2022, 12, 995–1018. [Google Scholar] [CrossRef]
- Iwamoto, N.; Lu, R.; Abe-Dohmae, S.; Yokoyama, S. Calmodulin interacts with ATP binding cassette trans- porter A1 to protect from calpain-mediated degradation and upregulates high-density lipoprotein generation. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1446–1452. [Google Scholar] [CrossRef] [PubMed]
- Foster, E.M.; Dangla-Valls, A.; Lovestone, S.; Ribe, E.M.; Buckley, N.J. Clusterin in Alzheimer’s Disease: Mechanisms, Genetics and Lessons From Other Pathologies. Front. Neurosci. 2019, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, C.; Ramazzina, I.; Russo, I.; Filippini, A.; Betuzzi, S.; Rizzi, F. The down-regulation of cluserin expression enhances the αsynuclein aggregation process. Int. J. Mol. Sci. 2020, 21, 7181. [Google Scholar] [CrossRef]
- Jay, T.R.; von Saucken, V.E.; Landreth, G.E. TREM2 in Neurodegenerative Diseases. Mol. Neurodegener. 2017, 12, 56. [Google Scholar] [CrossRef]
- Hampel, H.; Caraci, F.; Cuello, A.C.; Caruso, G.; Nisticò, R.; Corbo, M.; Baldacci, F.; Toschi, N.; Garaci, F.; Chiesa, P.A.; et al. A path toward precision medicine for neuroinflammatory mechanisms in Alzheimer’s disease. Front. Immunol. 2020, 11, 456. [Google Scholar] [CrossRef]
- González-Sánchez, M.; Bartolome, F.; Antequera, D.; Puertas-Martín, V.; González, P.; Gómez-Grande, A.; Llamas-Velasco, S.; Herrero-San Martín, A.; Pérez-Martínez, D.; Villarejo-Galende, A.; et al. Decreased salivary lactoferrin levels are specific to Alzheimer’s disease. EBioMedicine 2020, 57, 102834. [Google Scholar] [CrossRef]
- Li, Y.-Q.; Guo, C. A Review on Lactoferrin and Central Nervous System Diseases. Cells 2021, 10, 1810. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Ishida, H.; Vogel, H.J. Structural Characterization of the Interaction of Human Lactoferrin with Calmodulin. PLoS ONE 2012, 7, e51026. [Google Scholar] [CrossRef]
- Ferrari, C.; Sorbi, S. The complexity of Alzheimer’s disease: An evolving puzzle. Physiol. Rev. 2021, 101, 1047–1081. [Google Scholar] [CrossRef]
- Bates, G.P.; Dorsey, R.; Gusella, J.F.; Hayden, M.R.; Chris, K. Huntington disease. Nat. Rev. Dis. Primers 2015, 1, 15005. [Google Scholar] [CrossRef] [PubMed]
- Martí-Martínez, S.; Valor, L.M. A Glimpse of Molecular Biomarkers in Huntington’s Disease. Int. J. Mol. Sci. 2022, 23, 5411. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Flower, M.D.; Ross, C.A.; Wild, E.J. Huntington disease: New insights into molecular pathogenesis and therapeutic opportunities. Nat. Rev. Neurol. 2020, 16, 529–546. [Google Scholar] [CrossRef]
- Bao, J.; Sharp, A.H.; Wagster, M.V.; Becher, M.; Schilling, G.; Ross, C.A.; Dawson, V.L.; Dawson, T.M. Expansion of polyglutamine repeat in huntingtin leads to abnormal protein interactions involving calmodulin. Proc. Natl. Acad. Sci. USA 1996, 93, 5037–5042. [Google Scholar] [CrossRef]
- Puszkin, E.G.; Raghuraman, V. Catalytic properties of a calmodulin-regulated transglutaminase from human platelet and chicken gizzard. J. Biol. Chem. 1985, 260, 16012–16020. [Google Scholar] [CrossRef]
- Zainelli, G.M.; Dudek, N.L.; Ross, C.A.; Kim, S.-Y.; Muma, N.A. Mutant Huntingtin protein A substrate for transglutaminase 1.; 2.; and 3. J. Neuropathol. Exp. Neurol. 2005, 64, 58–65. [Google Scholar]
- Lesort, M.; Chun, W.; Johnson, G.V.; Ferrante, R.J. Tissue transglutamase is increased in Huntington’s disease brain. J. Neurochem. 1999, 73, 2018–2027. [Google Scholar]
- Tomás-Zapico, C.; Díez-Zaera, M.; Ferrer, I.; Gómez-Ramos, P.; Morán, M.A.; Miras-Portugal, M.T.; Díaz-Hernández, M.; Lucas, J.J. α-Synuclein accumulates in huntingtin inclusions but forms independent filaments and its deficiency attenuates early phenotype in a mouse model of Huntington’s disease. Hum. Mol. Genet. 2012, 21, 495–510. [Google Scholar] [CrossRef]
- Samii, A.; Nutt, J.G.; Ransom, B.R. Parkinson’s disease. Lancet 2004, 363, 1783–1793. [Google Scholar] [CrossRef] [PubMed]
- Vijiaratnam, N.; Simuni, T.; Bandmann, O.; Morris, H.R.; Foltynie, T. Progress towards therapies for disease modification in Parkinson’s disease. Lancet Neurol. 2021, 20, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Lill, C.M. Genetics of Parkinson’s disease. Mol. Cell. Probes 2016, 30, 386–396. [Google Scholar] [CrossRef]
- Kwon, E.H.; Tennagels, S.; Gold, R.; Gerwert, K.; Beyer, L.; Tönges, L. Update on CSF Biomarkers in Parkinson’s Disease. Biomolecules 2022, 12, 329. [Google Scholar] [CrossRef] [PubMed]
- Bofill-Cardona, E.; Kudlacek, O.; Yang, Q.; Ahorn, H.; Freissmuth, M.; Nanoff, C. Binding of calmodulin to the D2-Dopamine receptor reduces receptor signaling by arresting the G protein activation switch. J. Biol. Chem. 2020, 275, 32672–32680. [Google Scholar] [CrossRef]
- Lange, J.; Lunde, K.A.; Sletten, C.; Moller, S.G.; Tysnes, O.-B.; Alves, G.; Larsen, J.P.; Maple-Grodem, J. Association of a BACE1 gene polymorphism with Parkinson’s disease in a Norwegian population. Parkinson’s Dis. 2015, 2015, 973290. [Google Scholar]
- Sieber, B.A.; Landis, S.; Koroshetz, W.; Bateman, R.; Siderowf, A.; Galpern, W.R. Prioritized research recommendations from the National Institute of Neurological Disorders and Stroke Parkinson’s Disease 2014 conference. Ann. Neurol. 2014, 76, 469–472. [Google Scholar] [CrossRef]
- Fereshtehnejad, S.M.; Romenets, S.R.; Anang, J.B.; Latreille, V.; Gagnon, J.F.; Postuma, R.B. New clinical subtypes of Parkinson disease and their longitudinal progression. JAMA Neurol. 2015, 72, 863. [Google Scholar] [CrossRef]
- Menšíková, K.; Matěj, R.; Colosimo, C.; Rosales, R.; Tučková, L.; Ehrmann, J.; Hraboš, D.; Kolaříková, K.; Vodička, R.; Vrtěl, R.; et al. Lewy body disease or diseases with Lewy bodies? NPJ Park. Dis. 2022, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Compta, Y.; Parkkinen, L.; O’Sullivan, S.S.; Vandrovcova, J.; Holton, J.L.; Collins, C.; Lashley, T.; Kallis, C.; Williams, D.R.; de Silva, R.; et al. Lewy- and Alzheimer-type pathologies in Parkinson’s disease dementia: Which is more important? Brain 2011, 134, 1493–1505. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Walker, D.E.; Goldstein, J.M.; de Laat, R.; Banducci, K.; Caccavello, R.J.; Barbour, R.; Huang, J.; Kling, K.; Lee, M.; et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 2006, 281, 29739–29752. [Google Scholar] [CrossRef] [PubMed]
- Kurata, T.; Karaabayashi, T.; Murakami, T.; Miyazaki, K.; Morimoto, N.; Ohta, Y.; Takehisa, Y.; Nagai, M.; Matsubara, E.; Westaway, D.; et al. Enhanced accumulation of phosphorylated α-synuclein in double transgenic mice expressing mutant b-amyloid precursor protein and presenilin-1. J. Neuro. Res. 2007, 85, 2246–2252. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Moeller, I.; Erdjument-Bromagel, H.; Tempst, P.; Lauring, B. Parkinson’s disease-associated α-synuclein is a calmodulin substrate. J. Biol. Chem. 2003, 278, 17379–17387. [Google Scholar] [CrossRef]
- Yap, T.L.; Gruschus, J.M.; Velayati, A.; Westboek, W.; Goldin, E.; Moaven, N.; Sidransky, E.; Lee, J.C. α-synuclein interacts with glucocerebrocidase providing a molecular link between Parkinson and Gaucher diseases. J. Biol. Chem. 2011, 286, 28080–28088. [Google Scholar] [CrossRef]
- Lim, E.W.; Aarsland, D.; Ffytche, D.; Taddei, R.N.; van Wamelen, D.J.; Wan, Y.-M.; Tan, E.T.; Chaudhuri, K.R. Kings Parcog groupMDS Nonmotor study group. Amyloid-β and Parkinson’s disease. Neurology 2019, 266, 2605–2619. [Google Scholar] [CrossRef]
- Clinton, L.K.; Blurton-Jones, M.; Myczek, K.; Trojanowski, J.Q.; LaFerla, F.M. Synergistic Interactions between Aβ, Tau, and α-Synuclein: Acceleration of Neuropathology and Cognitive Decline. J. Neurosci. 2010, 30, 7281–7289. [Google Scholar] [CrossRef]
- McKeith, I.G.; Boeve, B.F.; Dickson, D.W.; Halliday, G.; Taylor, J.P.; Weintraub, D.; Aarsland, D.; Galvin, J.; Attems, J.; Ballard, C.G.; et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology 2017, 89, 88–100. [Google Scholar] [CrossRef]
- Bousiges, O.; Blanc, F. Biomarkers of Dementia with Lewy Bodies: Differential Diagnostic with Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 6371. [Google Scholar] [CrossRef]
- Scott, G.D.; Arnold, M.R.; Beach, T.G.; Gibbons, C.H.; Kanthasamy, A.G.; Lebovitz, R.M.; Lemstra, A.W.; Shaw, L.M.; Teunissen, C.E.; Zetterberg, H.; et al. Fluid and Tissue Biomarkers of Lewy Body Dementia: Report of an LBDA Symposium. Front. Neurol. 2022, 12, 805135. [Google Scholar] [CrossRef] [PubMed]
- Orme, T.; Guerreiro, R.; Bras, J. The Genetics of dementia with Lewy Bodies: Current understanding and future directions. Curr. Neurol. Neurosci. Rep. 2018, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Katseff, J.S.; Bright, F.; Phan, K.; Kril, J.J.; Ittner, L.M.; Kassiou, M.; Hodges, J.R.; Piquet, O.; Kiernan, M.C.; Halliday, G.M.; et al. Biomarker discovery and development for frontemporal dementia and amyotrophic lateral sclerosis. Brain 2022, 145, 1598–1609. [Google Scholar] [CrossRef] [PubMed]
- Ntymenou, S.; Tsantzali, I.; Kalamatianos, T.; Voumvourakis, K.I.; Kapaki, E.; Tsivgoulis, G.; Stranjalis, G.; Paraskevas, G.P. Blood Biomarkers in Frontotemporal Dementia: Review and Meta-Analysis. Brain Sci. 2021, 11, 244. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Falcon, B.; Zhang, W.; Newell, K.L.; Hasegawa, M.; Scheres, S.H.W.; Ghetti, B. Tau Protein and Frontotemporal Dementias. Adv. Exp. Med. Biol. 2021, 1281, 177–199. [Google Scholar]
- Che, X.; Song, N.; Gao, Y.; Ren, R.; Wang, G. Precision medicine of frontotemporal dementia: From genotype to phenotype. Front. Biosci. Landmark 2018, 23, 1144–1165. [Google Scholar]
- Greaves, C.V.; Rohrer, J.D. An update on genetic frontotemporal dementia. J. Neurol. 2019, 266, 2075–2086. [Google Scholar] [CrossRef]
- Wagner, M.; Lorenz, G.; Volk, A.E.; Brunet, T.; Edbauer, D.; Berutti, R.; Zhao, C.; Anderl-Straub, S.; Danek, A.; Bertram, L.; et al. Clinico-genetic findings in 509 frontotemporal disease patients. Mol. Psych. 2021, 26, 5824–5832. [Google Scholar] [CrossRef]
- Rossi, G.; Salvi, E.; Mehmeti, E.; Ricci, M.; Villa, C.; Prioni, S.; Moda, F.; Di Fede, G.; Tiraboschi, P.; Redaelli, V.; et al. Semantic and right temporal variant of FTD: Next generation sequencing genetic analysis on a single-center cohort. Front. Aging Neurosci. 2022, 14, 1085406. [Google Scholar] [CrossRef]
- Gifford, A.; Praschan, N.; Newhouse, A.; Chemali, Z. Biomarkers in frontotemporal dementia: Current landscape and future directions. Biomark. Neuropsychiatry 2023, 8, 100065. [Google Scholar] [CrossRef]
- Tan, R.H.; Kril, J.J.; Yang, Y.; Tom, N.; Hodges, J.R.; Villemagne, V.L.; Rowe, C.C.; Leyton, C.E.; Kwok, J.B.J.; Ittner, L.M.; et al. Assessment of amyloid β in pathologically confirmed frontotemporal dementia syndromes. Alz. Dement. 2017, 9, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Aoki, N.; Boyer, P.J.; Lund, C.; Lin, W.L.; Koga, S.; Ross, O.A.; Weiner, M.; Lipton, A.; Powers, J.M.; White, C.L., 3rd; et al. Atypical multiple system atrophy is a new subtype of frontotemporal lobar degeneration: Frontotemporal lobar degeneration associated with α-synuclein. Acta Neuropathol. 2015, 130, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Masrori, P.; Van Damme, P. Amyotrophic lateral sclerosis: A clinical review. Eur. J. Neurol. 2020, 27, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Abramzon, Y.A.; Fratta, P.; Traynor, B.J.; Chia, R. The overlapping genetics of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Front. Neurosci. 2020, 14, 42. [Google Scholar] [CrossRef]
- Oono, M.; Okado-Matsumoto, A.; Shodai, A.; Ido, A.; Ohta, Y.; Abe, K.; Ayaki, T.; Ito, H.; Takahashi, R.; Taniguchi, N.; et al. Transglutaminase 2 accelerates neuroinflammation in amyotrophic lateral sclerosis through interaction with misfolded superoxide dismutase 1. J Neurochem. 2014, 128, 403–418. [Google Scholar] [CrossRef] [PubMed]
- Calingasan, N.Y.; Chen, J.; Kiaei, M.; Beal, M.F. Beta-amyloid 42 accumulation in the lumbar spinal cord motor neurons of amyotrophic lateral sclerosis patients. Neurobiol Dis. 2005, 19, 340–347. [Google Scholar] [CrossRef]
- Bryson, J.B.; Hobbs, C.; Parsons, M.J.; Bosch, K.D.; Pandraud, A.; Walsh, F.S.; Doherty, P.; Greensmith, L. Amyloid precursor protein (APP) contributes to pathology of SOD1G93A mouse model of amyotrophic lateral sclerosis. Human Mol. Genet. 2012, 21, 3871–3882. [Google Scholar] [CrossRef][Green Version]
- Roberts, B.; Theunissen, F.; Mastaglia, F.L.; Akkari, P.A.; Flynn, L.L. Synucleinopathy in Amyotrophic Lateral Sclerosis: A Potential Avenue for Antisense Therapeutics? Int. J. Mol. Sci. 2022, 23, 9364. [Google Scholar] [CrossRef]
- Ziemssen, T.; Akgün, K.; Brück, W. Molecular biomarkers in multiple sclerosis. J. Neuroinflam. 2019, 16, 272. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Tejerina, D.; Llaurado, A.; Sotoca, J.; Lopez-Diego, V.; Vidal Taboada, J.M.; Salvado, M.; Juntas-Morales, R. Biofluid Biomarkers in the Prognosis of Amyotrophic Lateral Sclerosis: Recent Developments. Therapeut. Applic. Cells 2023, 12, 1180. [Google Scholar]
- Yang, J.; Hamade, M.; Wu, Q.; Wang, Q.; Axtell, R.; Giri, S.; Mao-Draayer, Y. Current and Future Biomarkers in Multiple Sclerosis. Int. J. Mol. Sci. 2022, 23, 5877. [Google Scholar] [CrossRef] [PubMed]
- Witzel, S.; Mayer, K.; Oeckl, P. Biomarkers for amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2022, 35, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Razia, R.; Majeed, F.; Amin, R.; Mukhtar, S.; Mahmood, K.; Abualait, T.; Bashir, S.; Baig, D.N. Predictive value of α-synuclein expression in peripheral blood of multiple sclerosis patients: A two-dimensional assessment of a selected biomarker. PLoS ONE 2023, 18, e0285022. [Google Scholar] [CrossRef] [PubMed]
- Robey, T.; Panagyres, P.K. Cerebrospinal fluid biomarkers in neurodegenerative diseases. Future Neurol. 2019, 14, FNL6. [Google Scholar] [CrossRef]
- O’Day, D.H. Calmodulin and amyloid beta as coregulators of critical events during the onset and progression of Alzheimer’s disease. Int. J. Mol. Sci. 2023, 24, 1393–1401. [Google Scholar] [CrossRef]
- Yuan, K.; Yong, S.; Xu, F.; Zhou, T.; McDonald, J.M.; Chen, Y. Calmodulin antagonists promote TRA-8 therapy resistant pancreatic cancer. Oncotarget 2015, 6, 25308–25319. [Google Scholar] [CrossRef] [PubMed]
- Taglialatella, G.; Rastellini, C.; Cicalese, L. Reduced incidence of dementia in solid organ transplant patients treated with calcineurin inhibitors. J. Alz. Dis. 2015, 47, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Rozkalne, A.; Hyman, B.T.; Spires-Jones, T.L. Calcineurin inhibition with FK506 ameliorates dendritic spine density deficits in plaque-bearing Alzheimer model mice. Neurobiol. Dis. 2011, 41, 650–654. [Google Scholar] [CrossRef]
- Nassal, D.; Gratz, D.; Hund, T.J. Challenges and opportunities for therapeutic targeting of calmodulin kinase in heart. Front. Pharmacol. 2020, 11, 35. [Google Scholar] [CrossRef] [PubMed]
| I. Experimentally Validated CaMBP Biomarkers | |||||||
| Biomarker | AD | ALS | FTD | HD | LBD | MS | PD |
| αSyn | ✓* | ✓ | ✓* | ✓ | ✓ | ✓ | ✓ |
| Aβ | ✓ | ✓ | ✓* | - | ✓* | ✓ | ✓ |
| ABCA1 | ✓ | - | ✓ | - | ✓ | ✓ | ✓ |
| AβPP | ✓ | ✓ | - | - | - | - | - |
| APOE4 | ✓ | ✓ | ✓ | - | ✓ | ✓ | ✓ |
| BACE1 | ✓ | - | - | - | - | - | ✓ |
| HTT | - | - | - | ✓ | - | - | ✓ |
| LF | ✓ | ✓ | - | - | - | - | ✓ |
| Ng | ✓ | - | - | ✓ | ✓ | - | ✓ |
| PSEN1 | ✓ | - | ✓ | - | ✓ | - | ✓ |
| Tau | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| TGM2 | ✓ | ✓ | - | ✓ | ✓ | - | ✓ |
| II. Biomarkers with CaM-Binding Domains | |||||||
| Biomarker | AD | ALS | FTD | HD | LBD | MS | PD |
| ABCA7 | ✓ | - | ✓ | - | - | - | ✓ |
| BIN1 | ✓ | - | - | - | ✓ | - | - |
| C9orf72 | - | ✓ | ✓ | - | - | - | - |
| CLU | ✓ | - | - | - | - | - | ✓ |
| FUS | - | ✓ | ✓ | - | - | - | - |
| GBA | - | ✓ | ✓ | - | ✓ | - | ✓ |
| LRRK2 | - | - | - | - | ✓ | - | ✓ |
| PARK7 | - | - | - | - | - | - | ✓ |
| PINK1 | - | - | - | - | ✓ | - | ✓ |
| SQSTM1 | ✓* | ✓ | ✓ | - | - | - | - |
| TBK1 | - | ✓ | ✓ | - | - | - | - |
| TDP-43 | - | ✓ | ✓ | ✓ | ✓ | - | ✓ |
| TMEM175 | - | ✓ | ✓ | - | - | - | ✓ |
| TREM2 | ✓ | ✓ | ✓ | - | - | - | ✓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Day, D.H. Protein Biomarkers Shared by Multiple Neurodegenerative Diseases Are Calmodulin-Binding Proteins Offering Novel and Potentially Universal Therapeutic Targets. J. Clin. Med. 2023, 12, 7045. https://doi.org/10.3390/jcm12227045
O’Day DH. Protein Biomarkers Shared by Multiple Neurodegenerative Diseases Are Calmodulin-Binding Proteins Offering Novel and Potentially Universal Therapeutic Targets. Journal of Clinical Medicine. 2023; 12(22):7045. https://doi.org/10.3390/jcm12227045
Chicago/Turabian StyleO’Day, Danton H. 2023. "Protein Biomarkers Shared by Multiple Neurodegenerative Diseases Are Calmodulin-Binding Proteins Offering Novel and Potentially Universal Therapeutic Targets" Journal of Clinical Medicine 12, no. 22: 7045. https://doi.org/10.3390/jcm12227045
APA StyleO’Day, D. H. (2023). Protein Biomarkers Shared by Multiple Neurodegenerative Diseases Are Calmodulin-Binding Proteins Offering Novel and Potentially Universal Therapeutic Targets. Journal of Clinical Medicine, 12(22), 7045. https://doi.org/10.3390/jcm12227045






