Unusual Surge of Acute Hepatitis A Cases in 2016 and 2017 in Malaga, Southern Spain: Characterization and Relationship with Other Concurrent European Outbreaks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.1.1. Description and Statistical Analyses of the Study Population Characteristics
2.1.2. Ethical Concerns
2.1.3. HAV Amplification and Sequencing
2.1.4. Molecular Genotyping of HAV Samples
2.1.5. Phylogenetic Analysis of HAV Sequence Data
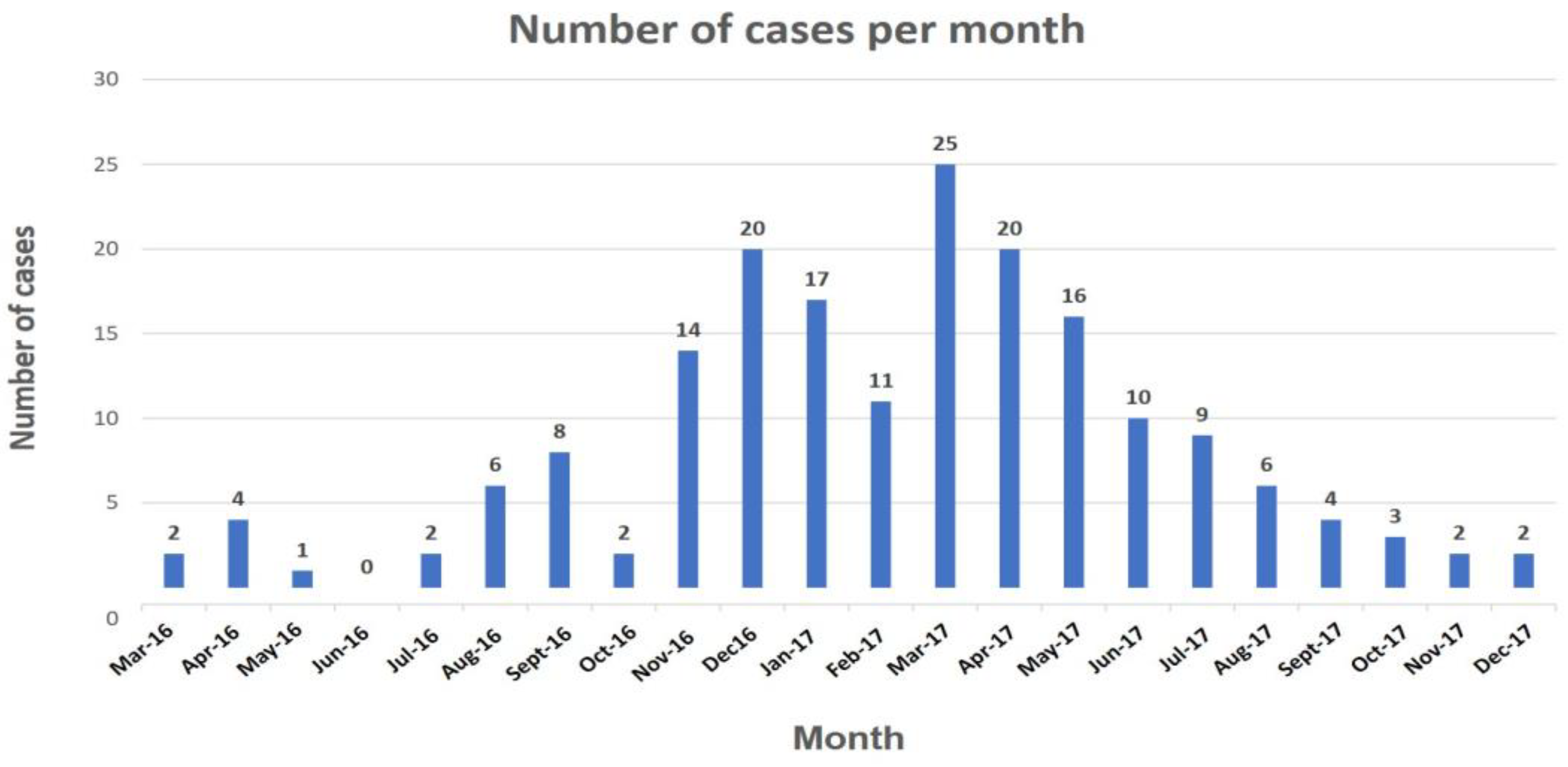
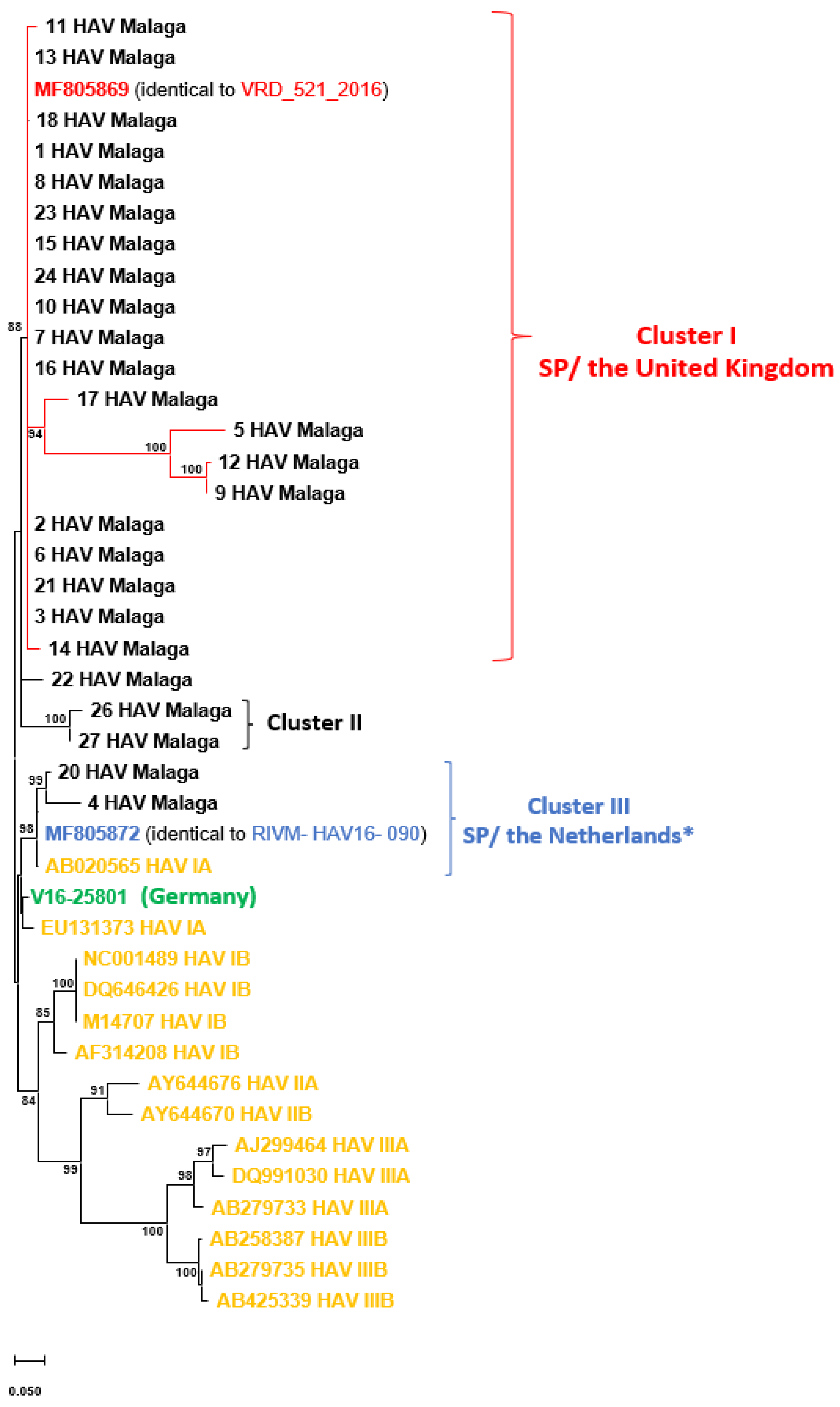
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Desbois, D.; Couturier, E.; Mackiewicz, V.; Graube, A.; Letort, M.J.; Dussaix, E.; Roque-Afonso, A.-M. Epidemiology and genetic characterization of hepatitis A virus genotype I.I.A. J. Clin. Microbiol. 2010, 48, 3306–3315. [Google Scholar] [CrossRef]
- Tong, M.J.; El-Farra, N.S.; Grew, M.I. Clinical manifestations of hepatitis A: Recent experience in a community teaching hospital. J. Infect. Dis. 1995, 171 (Suppl. S1), S15–S18. [Google Scholar] [CrossRef]
- Corey, L.; Holmes, K.K. Sexual transmission of hepatitis A in homosexual men: Incidence and mechanism. N. Engl. J. Med. 1980, 302, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Mosley, J.W. Water-borne infectious hepatitis. N. Engl. J. Med. 1959, 261, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.C.; Jeong, S.H. Natural History, Clinical Manifestations, and Pathogenesis of Hepatitis A. Cold Spring Harb. Perspect. Med. 2018, 8, a031708. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, K.H.; Koopman, J.S. The effects of socioeconomic development on worldwide hepatitis A virus seroprevalence patterns. Int. J. Epidemiol. 2005, 34, 600–609. [Google Scholar] [CrossRef]
- Thuener, J. Hepatitis A and B Infections. Prim. Care 2017, 44, 621–629. [Google Scholar] [CrossRef]
- Jacobsen, K.H.; Wiersma, S.T. Hepatitis A virus seroprevalence by age and world region, 1990 and 2005. Vaccine 2010, 28, 6653–6657. [Google Scholar] [CrossRef]
- Lemon, S.M.; Ott, J.J.; van Damme, P.; Shouval, D. Type A viral hepatitis: A summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 2018, 68, 167–184. [Google Scholar] [CrossRef]
- World Health Organization (WHO). The Global Prevalence of Hepatitis A Virus Infection and Susceptibility: A Systematic Review. 2010. Available online: https://apps.who.int/iris/handle/10665/70180 (accessed on 25 May 2022).
- World Health Organization (WHO). Hepatitis A. (Last Update 27 July 2021). 2021. Available online: https://www.who.int/es/news-room/fact-sheets/detail/hepatitis-a (accessed on 25 May 2022).
- European Centre for Disease Prevention and Control (ECDC). Stockholm: 2016. Rapid Risk Assessment: Hepatitis A Outbreaks in the EU/EEA Mostly Affecting Men Who Have Sex with Men, 20 December 2016. Available online: https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-hepatitis-outbreaks-eueea-mostly-affecting-men-who-have-sex (accessed on 21 January 2022).
- European Centre for Disease Prevention and Control (ECDC). Hepatitis A. In ECDC. Annual Epidemiological Report for 2017; ECDC: Stockholm, Sweden, 2023; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/hepatits-a-annual-epidemiological-report-2017.pdf (accessed on 18 July 2023).
- Beebeejaun, K.; Degala, S.; Balogun, K.; Simms, I.; Woodhall, S.C.; Heinsbroek, E.; Crook, P.D.; Kar-Purkayastha, I.; Treacy, J.; Wedgwood, K.; et al. Outbreak of hepatitis A associated with men who have sex with men (MSM), England, July 2016 to January 2017. Euro Surveill. 2017, 22, 30454. [Google Scholar] [CrossRef]
- Freidl, G.S.; Sonder, G.J.; Bovée, L.P.; Friesema, I.H.; van Rijckevorsel, G.G.; Ruijs, W.L.; van Schie, F.; Siedenburg, E.C.; Yang, J.-Y.; Vennema, H. Hepatitis A outbreak among men who have sex with men (MSM) predominantly linked with the EuroPride, The Netherlands, July 2016 to February 2017. Euro Surveill. 2017, 22, 30468. [Google Scholar] [CrossRef]
- Werber, D.; Michaelis, K.; Hausner, M.; Sissolak, D.; Wenzel, J.; Bitzegeio, J.; Belting, A.; Sagebiel, D.; Faber, M. Ongoing outbreaks of hepatitis A among men who have sex with men (MSM), Berlin, November 2016 to January 2017—Linked to other German cities and European countries. Euro Surveill. 2017, 22, 30457. [Google Scholar] [CrossRef]
- Rodríguez-Tajes, S.; Perpiñán, E.; Caballol, B.; Lens, S.; Mariño, Z.; Costa, J.; Vilella, A.; Pérez-del-Pulgar, S.; Forns, X.; Koutsoudakis, G. Hepatitis A outbreak in Barcelona among men who have sex with men (MSM), January–June 2017: A hospital perspective. Liver Int. 2018, 38, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Minosse, C.; Messina, F.; Garbuglia, A.R.; Meschi, S.; Scognamiglio, P.; Capobianchi, M.R.; Ippolito, G.; Lanini, S. Origin of HAV strains responsible for 2016–2017 outbreak among MSM: Viral phylodynamics in Lazio region. PLoS ONE 2020, 15, e0234010. [Google Scholar] [CrossRef] [PubMed]
- Spanish National Epidemiology Center; Institute of Health Carlos III (ISCIII). Epidemiological Surveillance of Communicable Diseases. Annual Report. Years 2017–2018. Madrid. 2020. Available online: https://www.isciii.es/QueHacemos/Servicios/VigilanciaSaludPublicaRENAVE/EnfermedadesTransmisibles/Documents/INFORMES/INFORMES%20RENAVE/RENAVE_Informe_anual__2017-2018.pdf (accessed on 25 May 2022).
- Ferreira, A.J.G.; Alonso, A.O.; Rodríguez, J.C.; de Tena, P.B.; Cid, M.C.; García, A.M.G.; Escaño, M.D.G.; Benitez, R.A.; Frutos, E.C.; García-Cortés, M.; et al. Outbreak of acute hepatitis A in the health area served by the Hospital Universitario Virgen de la Victoria (HUVV): A change in epidemiology. Rev. Esp. Enferm. Dig. 2018, 110, 380–385. [Google Scholar]
- Michaelis, K.; Wenzel, J.J.; Stark, K.; Faber, M. Hepatitis A virus infections and outbreaks in asylum seekers arriving to Germany, September 2015 to March 2016. Emerg. Microbes Infect. 2017, 6, e26. [Google Scholar] [CrossRef]
- HAV Net; Dutch National Institute of Public Health and the Environment (RIVM). PROTOCOL: Molecular Detection and Typing of VP1 Region of Hepatitis A Virus, HAVnet. Available online: https://www.rivm.nl/sites/default/files/2018-11/Typing%20protocol%20HAVNET%20VP1P2A%20a1a.pdf (accessed on 5 August 2021).
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Gallian, P.; Barlet, V.; Mouna, L.; Gross, S.; Lecam, S.; Ricard, C.; Wind, F.; Pouchol, E.; Fabra, C.; Flan, B.; et al. Hepatitis A: An epidemiological survey in blood donors, France 2015 to 2017. Euro Surveill. 2018, 23, 1800237. [Google Scholar] [CrossRef] [PubMed]
- Hernández, E.; de Castro, V.; Avellón, A.; González, I.; Muniozguren, N.; Vázquez, S.; Muñoz-Chimeno, M. Brote de hepatitis A asociado a un manipulador de alimentos en Bizkaia, 2017. Enferm. Infecc. Microbiol. Clín. 2019, 37, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Marosevic, D.; Belting, A.; Schönberger, K.; Carl, A.; Wenzel, J.J.; Brey, R. Hepatitis A Outbreak in the General Population due to a MSM-Associated HAV Genotype Linked to a Food Handler, November 2017–February 2018, Germany. Food Environ. Virol. 2019, 11, 149–156. [Google Scholar] [CrossRef] [PubMed]
| Variables | SP/UK Cluster | p-Value | |
|---|---|---|---|
| Number of patients | 20 | ||
| Age (years) | 32.5 (27.8–49.8) | ||
| Sex | Male | 16 (80) | <0.001 |
| Female | 4 (20) | ||
| Origin | Spanish | 15 (75) | <0.001 |
| Immigrants | |||
| South American | 1 (5) | ||
| European countries * | 3 (15) | ||
| Unknown | 1 (5) | ||
| Risk behaviour | MSM | 5 (25) | <0.001 |
| HTX | 1 (5) | ||
| Piercing, tattoos | 0 (0) | ||
| Trip to endemic country | 0 (0) | ||
| Family contact with HAV+ | 1 (5) | ||
| Coinfection | No | 16 (80) | <0.001 |
| Yes | 4 (20) | ||
| HIV | 3 (15) | ||
| HIV + Syphilis | 1 (5) | ||
| Assistance department | Primary care | 4 (20) | |
| Emergency department | 8 (40) | ||
| Out-Patient Consultation | 1 (5) | ||
| In-patient stay | 5 (25) | ||
| Other hospital department | 2 (10) | ||
| Clinical symptoms | No | 2 (10) | <0.001 |
| Yes | 18 (90) | ||
| Diarrhea | 3 (15) | ||
| Free intraperitoneal fluid | 1 (5) | ||
| Choluria (bile in urine) | 16 (80) | ||
| Acholia (white stools) | 4 (20) | ||
| Jaundice | 11 (55) | ||
| Fever | 13 (65) | ||
| Sickness | 12 (60) | ||
| Vomiting | 9 (45) | ||
| Outcome | Hospitalization | 7 (35) | |
| Hospitalization in ICU | 0 (0) | ||
| Variables | SP/UK Cluster | Reference Ranges |
|---|---|---|
| Mean ± SD | ||
| Total of patients | 20 | |
| Biochemical parameters | ||
| Albumin (g/dL) | 3.4 ± 0.6 | 3.5–5.5 |
| Alkaline phosphatase (IU/L) | 214.8 ± 106.9 | 20–125 |
| C-reactive protein (mg/dL) | 13.1 ± 9.0 | <0.5 |
| Direct bilirubin (mg/dL) | 5.3 ± 2.7 | 0–0.3 |
| Direct bilirubin zenith (mg/dL) | 6.1 ± 2.1 | |
| Total bilirubin (mg/dL) | 5.9 ± 3.8 | 0.3–1.2 |
| Total bilirubin zenith (mg/dL) | 6.6 ± 3.3 | |
| SGGT (IU/L) | 347.1 ± 186.9 | 8–78 |
| SGOT (IU/L) | 1382.4 ± 1000.0 | 0–35 |
| SGOT zenith (IU/L) | 1732.4 ± 1178.2 | |
| SGPT (IU/L) | 1812.3 ± 1236.8 | 0–35 |
| SGPT zenith (IU/L) | 2068.3 ± 1710.0 | |
| Lactate dehydrogenase (IU/L) | 495.5 ± 246.8 | 60–160 |
| Total cholesterol (mg/dL) | 135.5 ± 65.9 | 140–200 |
| Serum creatinine (mg/dL) | 0.9 ± 0.2 | 0.7–1.3 |
| Urea (mg/dL) | 25.2 ± 12.5 | 20–50 |
| Hemogram | ||
| Prothrombin time (%) | 74.0 ± 18.2 | 80–120 |
| International Normalized Ratio (INR) | 1.2 ± 0.2 | 0.8–1.2 |
| Red blood cell count (×106/mL) | 5.0 ± 0.5 | 3.6–6.0 |
| Hematocrit (%) | 42.5 ± 3.8 | 35–46 |
| Leukocyte count (×103/mL) | 5.5 ± 1.6 | 4–11.5 |
| Platelet count (×103/mL) | 205.9 ± 42.0 | 140–450 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bardón De Tena, P.; Tapia Paniagua, S.T.; Vico Sevilla, J.A.; Clavijo, E.; Martínez Manzanares, E.; Gonzalez-Domenech, C.M. Unusual Surge of Acute Hepatitis A Cases in 2016 and 2017 in Malaga, Southern Spain: Characterization and Relationship with Other Concurrent European Outbreaks. J. Clin. Med. 2023, 12, 6613. https://doi.org/10.3390/jcm12206613
Bardón De Tena P, Tapia Paniagua ST, Vico Sevilla JA, Clavijo E, Martínez Manzanares E, Gonzalez-Domenech CM. Unusual Surge of Acute Hepatitis A Cases in 2016 and 2017 in Malaga, Southern Spain: Characterization and Relationship with Other Concurrent European Outbreaks. Journal of Clinical Medicine. 2023; 12(20):6613. https://doi.org/10.3390/jcm12206613
Chicago/Turabian StyleBardón De Tena, Paula, Silvana Teresa Tapia Paniagua, José Alberto Vico Sevilla, Encarnación Clavijo, Eduardo Martínez Manzanares, and Carmen Maria Gonzalez-Domenech. 2023. "Unusual Surge of Acute Hepatitis A Cases in 2016 and 2017 in Malaga, Southern Spain: Characterization and Relationship with Other Concurrent European Outbreaks" Journal of Clinical Medicine 12, no. 20: 6613. https://doi.org/10.3390/jcm12206613
APA StyleBardón De Tena, P., Tapia Paniagua, S. T., Vico Sevilla, J. A., Clavijo, E., Martínez Manzanares, E., & Gonzalez-Domenech, C. M. (2023). Unusual Surge of Acute Hepatitis A Cases in 2016 and 2017 in Malaga, Southern Spain: Characterization and Relationship with Other Concurrent European Outbreaks. Journal of Clinical Medicine, 12(20), 6613. https://doi.org/10.3390/jcm12206613





