Abstract
The use of radiomics and artificial intelligence applied for the diagnosis and monitoring of Alzheimer’s disease has developed in recent years. However, this approach is not yet completely applicable in clinical practice. The aim of this paper is to provide a systematic analysis of the studies that have included the use of radiomics from different imaging techniques and artificial intelligence for the diagnosis and monitoring of Alzheimer’s disease in order to improve the clinical outcomes and quality of life of older patients. A systematic review of the literature was conducted in February 2023, analyzing manuscripts and articles of the last 5 years from the PubMed, Scopus and Embase databases. All studies concerning discrimination among Alzheimer’s disease, Mild Cognitive Impairment and healthy older people performing radiomics analysis through machine and deep learning were included. A total of 15 papers were included. The results showed a very good performance of this approach in the differentiating Alzheimer’s disease patients—both at the dementia and pre-dementia phases of the disease—from healthy older people. In summary, radiomics and AI can be valuable tools for diagnosing and monitoring the progression of Alzheimer’s disease, potentially leading to earlier and more accurate diagnosis and treatment. However, the results reported by this review should be read with great caution, keeping in mind that imaging alone is not enough to identify dementia due to Alzheimer’s.
1. Introduction
Nowadays, radiomics can be considered an emergent field of research, possibly allowing for the in-depth comprehension of diseases’ etiology and evolution. For radiomics, it is intended that the mineable data are extracted by converting clinical images to quantitative features using characterization algorithms [1], with the aim of identifying prognostic and predictive biomarkers of disorders. More in detail, radiomics allows for the extraction and analysis of a large number of quantitative features from medical images, such as magnetic resonance imaging (MRI) and positron emission tomography (PET) scans, echography and computerized tomography (CT), providing detailed information from medical images using mathematical and machine learning methods to explore possible ties with biology and clinical outcomes. In this way, a radiomics approach might help clinicians provide useful insights about the prognosis and response to treatment, thus representing a potential non-invasive method of precision medicine in the era of artificial intelligence (AI) [2].
From a general point of view, radiomics, given its intrinsic capacity of obtaining and organizing a huge number of data, seems to be particularly suited for studying multifactorial and complex diseases, and, not by chance, it has been mostly investigated in the oncology field [3,4,5,6,7]. Specifically, this new approach has been widely applied mainly in solid cancers [8,9,10], since data from histopathology and immunohistochemistry as well as from genomics were largely available, proving the expected heterogeneity of diseases at both cellular and molecular levels [11].
Recently, from the oncology field, given its power in the high-dimensional data mining of radiological features and its correlation with aging progression [12,13] and clinical endpoints [14], radiomics application is expected to spread in other multifactorial diseases where new signatures are difficult to be identified, including neurological ones. In this regard, recently, radiomics has shown interesting applications in the neurology field, highlighting promising results in differential diagnosis [15] among causes of cognitive impairment and in the prediction of the conversion from mild cognitive impairment (MCI) to Alzheimer dementia. In particular, Alzheimer’s disease (AD) is known to be the most common neurodegenerative disease. It has been estimated that over 46 million people live with dementia worldwide, increasing to 131.5 million by 2050 [16]. Therefore, there is an urgent need for biomarkers to be used for screening, diagnosis, prognosis and therapy response to control the societal impact of the disease.
In order to understand a vast plethora of data for the provision of the predictive models, AI algorithms represent elective strategies to be applied for the analysis of radiomic data, especially by adopting Deep Learning (DL) approaches based on neural networks (NN) that are able to deal with a huge number of computational parameters to be processed through high-performance technologies. Even if evidence from the literature shows that data-driven deep radiomic models have a better performance than humans in understanding diseases’ pathways, the method of the adoption of radiomic biomarkers still remains a gap to be solved to ensure their clinical uptake through efforts for standardization, the harmonization of different approaches and the reproducibility of the radiomic data collection in the different clinical settings [17].
The aim of this paper is to provide a systematic analysis of the studies that have included the use of radiomics from MRI and PET and AI for the diagnosis and monitoring of AD in order to improve the clinical outcomes and quality of life of older patients.
2. Materials and Methods
2.1. Literature Search and Study Selection
The methodology of this systematic review was based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [18], with the main aim of analyzing the impact of radiomic analysis and AI to diagnose and monitor AD and its progression.
We used the PICO framework (population, intervention, comparator and outcome) as follows:
P: Alzheimer patients
I: Diagnosis through artificial intelligence
C: Mild cognitive impairment and healthy subjects
O: Differential diagnosis
A systematic review of the literature was conducted in February 2023. The data were collected from PubMed, Embase and Scopus, analyzing manuscripts and articles of the last 5 years (from January 2018 to January 2023) in order to obtain the latest evidence in the field. The inclusion criteria are as follows: (1) prospective or retrospective studies; (2) Alzheimer’s disease and cognitive impairment in people aged ≥65 years; (3) use of radiomics to discriminate between Alzheimer’s and mild cognitive impairment in the older population; (4) machine/deep learning classification in terms of accuracy, specificity, sensitivity, area under the curve of the received operating characteristic (AUC-ROC), positive predictive value (PPV), negative predictive value, recall and F1-score. Systematic and narrative reviews were excluded. Based on consultation with the multidisciplinary research team, multi-modal intervention studies were searched using the following search terms, and the combination thereof: radiomic*, elderly, Alzheimer, cognitive impairment, texture analysis, neurological status, frailty and sarcopenia. The full search strings are provided in Table 1. After the preliminary search, 1679 articles resulted from PubMed, 213 resulted from Embase and 372 resulted from Scopus. The findings were analyzed and screened by four experts of the team, a bioengineer, a clinical neurologist, a psychologist and a nuclear medicine physician. In particular, the four reviewers independently analyzed the titles and abstracts retrieved from the search in order to determine if they met the predefined inclusion criteria. The full text articles were subsequently analyzed. The first screening was based on the analysis of the title and the abstract of the findings. After the first step, 45 articles resulted from PubMed, 12 resulted from Embase and 9 resulted from Scopus. A second screening was based on a deduplication analysis of the findings. After this step, 18 papers were included from PubMed, 1 was included from Embase and 1 was included from Scopus. An additional researcher, with a background in biomedical engineering, confirmed the accuracy of the papers selection and screened for any possible omission.

Table 1.
Adopted search strategy.
2.2. Data Collection
After the screening based on the inclusion/exclusion criteria conducted on the full text articles, the studies were selected as follows: 15 from PubMed, 0 from Embase and 0 from Scopus. Figure 1 shows the flowchart search strategy applied.
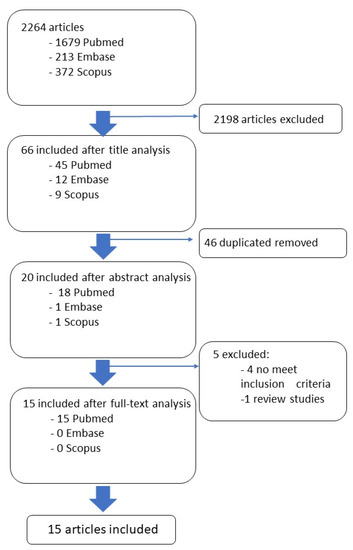
Figure 1.
Descriptive analysis of the included clinical studies.
3. Results
A total of 15 papers were included [19,20,21,22,23,24,25,26,27,28,29,30,31,32,33]. The findings reported in this section are organized under macro-concept areas of interest.
3.1. Study Quality Evaluation
The quality evaluation of 15 population-based studies was performed based on the Newcastle Ottawa Scale (NOS) scale. The NOS is designed to assess the quality of non-randomized studies [34], such as case-control and cohort studies (all the studies included in this systematic review belong to this category). The Newcastle Ottawa Scale focuses on three main areas: the selection of the study groups, the comparability of the groups and the assessment of the outcome or exposure. The final score was settled when three authors reached an agreement after repeated review and analysis. Of the fifteen studies considered, the NOS score ranged from five to a maximum of nine (Table 2).

Table 2.
Scores of quality assessment of the included studies according to the Newcastle Ottawa Scale. * indicates that the item requirement is satisfied; 0 indicates that the item requirement is not satisfied.
3.2. General Characteristics of the Study Population
In 8 out of 15 studies [20,21,22,23,28,29,31,33], the data used were retrieved from a public repository, 6 were from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database and 2 were from the Open Access Series of Imaging Studies (OASIS) [21,22]. In the remaining seven studies [19,24,25,26,27,30,32], original data were collected from human participants hospitalized in different healthcare structures. The 15 studies had an average sample size of 396.1, ranging from the smallest sample size of 86 [18] to the largest sample size of 1650 [20]. All the studies focus on older people, with a mean age of 71.3 (±8.87) years in the AD group, 75.8 (±8.9) years in the Mild Cognitive Impairment (MCI) group and 69.5 (±7.82) in the healthy control (HC). There were 1137 males and 1106 females in the AD group, 1188 males and 874 females in the MCI group and 1187 males and 1337 females in the HC group. The Mini-Mental State Examination and Clinical Dementia Rating are the most common neuropsychological assessment tests used to evaluate the cognitive and functional abilities of patients. Mini-Mental State Examination values are reported in 14 out of 15 studies [19,20,22,23,24,25,26,27,28,29,30,31,32,33], and a Clinical Dementia Rating is calculated in 7 out of 15 [21,22,23,24,27,28,30]. Other tests like the Montreal Cognitive Assessment, Auditory Verbal Learning Test, Alzheimer’s Disease Assessment Scale, Activities of Daily Living Scale and Geriatric Depression Scale are adopted. Table 3 summarizes the imaging method adopted in each study, the type of imaging evaluation, the AI algorithm adopted and the performance metrics used to check the classification/prediction performance of each ML/DL model, and Table 4 shows the characteristics of the included studies.

Table 3.
Studies grouped by macro-categories. For each article, the following is reported: the state or condition of the disease, the method and evaluation of each imaging technique, the AI algorithm adopted for the classification and prediction and the metrics for evaluating the model’s performance.

Table 4.
Descriptive analysis.
3.3. Radiomic Features
Radiomic features are extracted directly after segmentation from the area of interest. All studies used automatic feature extraction methods. The number of features calculated varies from a minimum of 35 [21] to 3360 [25]. Radiomic features can be broadly categorized into five classes: first-order features, shape-based features, texture features, wavelet-based features, and deep learning-based features. First-order features, or statistical features, were used in [19,20,21,23,24,27] to describe the basic statistics of the voxel intensity distribution within a region of interest, such as the mean, median, variance, skewness, and kurtosis. Shape-based features describe the geometric properties of the region of interest (ROI), such as the volume, surface area, and compactness. Shape features are calculated in all papers. Texture features capture the spatial distribution of voxel intensities within the ROI and provide information on the heterogeneity, coarseness, and complexity of the tissue. Texture features are extracted using different methods, such as the gray-level co-occurrence matrix (GLCM), gray-level run-length matrix (GLRLM), gray-level size zone matrix (GLSZM), gray-level dependence matrix (GLDM) and neighboring gray-tone difference matrix (NGTDM), as accomplished in all reported studies. Wavelet-based features use wavelet transforms to decompose the image into multiple frequency bands and extract features from each band. Wavelet features are calculated in all the studies except for [19,20,31].
Finally, deep learning-based features are extracted by training a neural network on a large dataset of images to identify patterns and features that are relevant for the task at hand. These features can then be used to extract radiomic features from new images, as accomplished in [22,33].
Radiomics features are then reduced in dimensionality with different techniques: Least Absolute Shrinkage and Selection Operator (LASSO) was used in six studies [19,23,24,25,26,27], t-test statistical correlation analysis was adopted in three studies [23,28,29], Cox regression was used in two studies [26,28] and Fisher’s score was adopted in one [30].
3.4. Machine and Deep Learning Methods Applied to the Diagnosis of AD
Machine learning models can be pooled into two categories: support vector machine (SVM) and logistic regression (LR). In seven studies, an SVM classification model was built. In particular, the radial basis function was chosen as the SVM algorithm kernel [20,23,29,32,33], while a multi-function kernel (linear, polynomial and sigmoid) was involved in two studies [28,30] in order to evaluate the performance of each SVM model. On the other side, in three studies, an LR algorithm was used to build a diagnostic model [19,25,26]. Convolutional neural networks (CNN) were used as deep learning algorithms to perform AD diagnosis in three studies [21,22].
3.5. Performance Metrics
Different metrics were used to assess the performance of ML and DL models. All studies report metrics referring both to the test set and validation set. Accuracy is adopted in 12 out of 15 studies [20,23,24,25,26,27,28,29,30,31,32,33]. It was used individually only in one study [28], while in all the other studies, it was calculated in combination with other metrics. The most common metric is the area under the curve (AUC), used in 13 out of 15 studies [19,20,21,22,23,24,25,26,27,29,31,32,33], while sensitivity and specificity were used in 9 out of 15 studies [19,23,24,25,26,27,30,31,32,33]. The least used metrics for evaluating the model classification performance are precision (3 out of 15) [19,26,27], positive predictive value (PPV) and negative predictive value (NPV) (2 out of 15) [19,26], recall and F1 score (2 out of 15) [19,27].
3.6. Differential Diagnosis
In 9 out of 15 studies [20,21,22,25,26,27,29,31,32], radiomics analysis is used to perform a differential diagnosis between AD and HC. In detail, among these 10 articles, 4 of them [21,22,26,27] focus exclusively on the discrimination between AD and HC, while in the other research [20,25,29,31,32], the authors focused in parallel on radiomic analysis aimed at the discrimination between MCI and AD, AD and HC.
In [19,30,33], the main core is the differential diagnosis between Subjective cognitive decline (SCD) and HC. It is noteworthy that SCD is often considered a potential early indicator of cognitive impairment or dementia. However, SCD is not a diagnostic criterion for dementia or other cognitive disorders, as many individuals with SCD do not progress to develop dementia or cognitive impairment.
In addition, in [28,29], relevance is given to progression from MCI to AD, while in [23], early-onset AD and late-onset AD against HC are classified. Indeed, a recent study has suggested considerable differences between early-onset and late-onset AD from a clinical point of view [35]; thus, we found it interesting to add this article within this review. For instance, early-onset AD patients show a faster and more severe disease progression compared to late-onset AD patients. They also present an unusual pattern of preserved memory function alongside cortical symptoms that affect language, visuospatial skills, and executive function, as reported by Cacace et al. [36]. Additionally, early-onset AD patients have less damage in the hippocampus but more severe damage in the neocortex. Furthermore, they display functional disruption between the hippocampus and middle frontal cortex, which distinguishes them from late-onset AD patients. On the other hand, in [24], the authors made a radiomic analysis to predict amyloid β peptide positivity and negativity. The amyloid β peptide status is crucial not only for diagnostic purposes but also for predicting the clinical course of patients in the early stage of AD. In particular, the Aβ status is associated with clinical deterioration and the transition to dementia in patients with mild cognitive impairment (MCI).
3.7. Brain Regions and Classification Results
In 14 out of 15 studies [19,20,21,22,23,24,25,26,27,28,29,30,32,33], regions of interest were extracted from MR images, while only in [31], Aβ PET was used to detect the accumulation of the beta-amyloid protein in the brain. For the identification of AD from HCs with the radiomics features of Aβ PET images, the authors obtained an AUC = 0.93 with the standard machine learning SVM method, while in all other studies using MRI, the AUC values ranged from 0.72 to 0.93. The segmentation of the MRI/PET image of the brain is aimed at extracting one or more anatomical areas of interest from which radiomic features are then calculated.
The hippocampus is the most interesting research area, with 6 out of 15 studies focusing on it [19,20,23,28,29,32]. In all of these studies, the hippocampus was taken singularly. For studies that take the hippocampus as the anatomical reference, AUC values ranging from 0.88 to 0.94 are obtained for studies that focus on the differentiation between AD patients and healthy subjects. Among these, the ADNI database was used in four papers [20,23,28,29], while in the remaining two studies [19,32], the sample was collected inside the healthcare structure. Individually, the amygdala [25] and corpus callosum [26] were also analyzed. For the study that refers to radiomic analysis of the amygdala, a satisfactory diagnostic performance in terms of accuracy (90%) in discriminating between AD and healthy subjects was reported, while slightly lower values were obtained with respect to the differentiation between patients with AD dementia and MCI. In this case, a database of patients internal to the referring hospital was used. As for the corpus callosum, the study investigating it provides a classification accuracy level between AD and healthy people of 79.2%. In this study, the referral patients were selected from within the healthcare center too. In two studies, gray matter, white matter and cerebrospinal fluid were segmented together [30,33]. For these two studies, the accuracy in differentiating between SCD or pre-clinical AD subjects on one hand and healthy subjects on the other hand reached over 89% in both [30,33].
In this case, in [33], the sample is retrieved from the ADNI repository, while the other study examines an in-house repository at the health institution. In the remaining four studies [21,22,27,31], researchers do not involve the segmentation of a specific anatomical region; rather, they perform training of their classifiers by whole-brain imaging. In [21], the authors achieved 93.9% accuracy in the differential analysis between AD and healthy subjects from the OASIS repository. In [22], an accuracy of 92.6% was achieved, in [27], one of 96.2% was achieved and in [31], the AUC was 0.93 for distinguishing AD from healthy subjects and 0.83 for the prediction of MCI conversion to AD. In this case, only in [27] was a local repository used, while the others adopted the OASIS database for the sample acquisition.
4. Discussion
In the current AD diagnostic criteria, cerebrospinal fluid (CSF), namely, CSF amyloid β 42 (Aβ 42) or Aβ 42/40, phosphorylated tau and total tau levels and/or PET biomarkers, i.e., amyloid PET and tau PET, were included [37,38]. However, these biomarkers are affected by some limitations. In particular, CSF biomarkers can be assessed only after having performed a lumbar puncture, which is considered a quite invasive procedure, and amyloid- PET and tau PET are expensive and not always available. Therefore, growing efforts are being made by researchers in order to identify reliable, easily available and non-invasive AD biomarkers. In this regard, radiomics might be an interesting option.
The present systematic review included all studies focused on the use of radiomics in the AD field, of which 14 [19,20,21,22,23,24,25,26,27,28,29,30,32,33] considered data from MRI, and only 1 [31] was based on data analysis from amyloid PET imaging. Globally considered, the results showed a very good performance of this approach in the differentiating AD patients—both at the dementia and pre-dementia phases of the disease—from HC patients. Indeed, the majority of studies reported a very high accuracy, with an AUC ranging from 80 to 95%, approximately. It is noteworthy that, according to current AD diagnostic criteria, MRI is considered a progression biomarker, rather than a diagnostic one, because of the suboptimal diagnostic performance, mainly in the view of early AD diagnosis, in terms of sensitivity and specificity, compared to the CSF and tau/amyloid PET biomarkers. Interestingly, some radiomics studies, based on MRI and focused on early AD diagnosis, reported such impressive results in terms of the accuracy of the prediction of MCI conversion to AD. In [24], T1 and T2 FLAIR radiomics data, together with demographic features, were able to distinguish between amyloid-positive and amyloid-negative patients, as classified by means of amyloid PET, with a quite satisfactory accuracy (AUC for test = 0.79; AUC for validation = 0.76). Despite this diagnostic, the performance is still clearly inferior compared to that of the CSF and PET biomarkers. These preliminary results could be considered encouraging, but they need to be validated by further investigations.
Interesting findings also come from studies exploring the ability of radiomics in differentiating patients with subjective cognitive decline (SCD) and/or pre-clinical AD from controls [30,33]. Subjective Cognitive Decline may represent a very early phase of AD, which precedes MCI; however, not all SCD subjects will develop AD during follow-up. In one of those two studies, SCD subjects were included, but they were not well characterized due to the lack of clinical follow-up data and/or assessment by means of AD-validated biomarkers at baseline [30]. Considering these limitations, no conclusive statement about the findings from this study with respect to early AD diagnosis will be made. The second investigation in this field, by Jiang and colleagues [33], included a large cohort of well-characterized, cognitively unimpaired subjects, who were divided in two groups (amyloid-positive and amyloid-negative), according to the amyloid PET status. In this investigation, a Deep Learning Radiomics method was used, obtaining a very high diagnostic accuracy (89.85 ± 1.12%) in differentiating amyloid-positive cognitively normal subjects (i.e., pre-clinical AD patients according to the current diagnostic criteria) from amyloid-negative healthy subjects (i.e., persons without any clinical and/or biological evidence of a neurodegenerative disorder). If confirmed by future investigations, those results might open a new era for the non-invasive assessment of aged healthy people in the view of early AD diagnosis, making it crucial to enroll subjects in similar clinical trials in order to test new disease-modifying therapies.
Also, the study on radiomics applied to amyloid-PET imaging [31] showed a very satisfactory accuracy (AUC = 0.93) in differentiating patients with AD from patients from normal controls and in predicting MCI conversion to AD dementia. However, in our opinion, the application of radiomics to amyloid-PET cannot be easily implemented in clinical practice in the future, due to the different availability of PET machines in hospitals and the absence of a standardization of a radiomics approach in this field. Despite the promising findings reported in papers included in this review, radiomics studies in the AD field are still affected by a great heterogeneity in terms of patients’ enrolment criteria, region of interest and other technical issues. Those aspects have impeded us from performing a rigorous meta-analysis; thus, we conducted a qualitative systemic review of the studies on this topic. Since radiomics is nowadays a quite complicated and time-consuming approach to be applicable in clinical practice for AD, larger prospective studies are needed to clarify its potential usefulness in the field.
5. Conclusions
Radiomics and artificial intelligence have the potential to help in the early detection of and diagnosis of Alzheimer’s by analyzing medical images and other data to identify patterns that contribute to developing the disease. In summary, radiomics and AI can be valuable tools for diagnosing and monitoring the progression of Alzheimer’s disease, potentially leading to earlier and more accurate diagnosis and treatment. However, the most critical limitation of this review is that all included articles do not have pathological confirmation of the clinical diagnosis. Although all patients included in the databases were meticulously screened by clinical experts in defining the pathology (MCI or Alzheimer’s disease), no further biological references were taken in the reviewed articles, and only imaging information was considered. In this regard, the results reported by this review should be read with great caution, keeping in mind that imaging alone is not enough to identify dementia due to Alzheimer’s disease. It is necessary to conduct a multi-omics analysis that considers a combination of biomarkers to increase the degree of accuracy with which an early diagnosis of the pathology can be made. It is for this reason that, for clinical practice, it is still difficult to rely solely on the use of imaging to diagnose pathology. However, the review presents interesting results for research purposes that pave the way for future developments in artificial intelligence related to Alzheimer’s identification.
Author Contributions
Study concept and design: L.B., E.M. and R.B.; Acquisition of data (literature search and study selection): F.B., L.B. and L.F.; Analysis and interpretation of data (literature): L.B., E.M., R.B. and L.F.; Writing—original draft preparation: L.B., F.B., E.M. and R.B.; Critical revision of the manuscript for important intellectual content: D.F., L.P., E.P. and F.L.; Supervision: G.P., G.R.R. and E.P.; Writing—review and editing: F.B., C.G. and S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated, used and analyzed during the trial and its preceding pilot trial are or will be available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021, 83, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, Y.; Wang, Y.; Jiang, S.; Fan, R.; Zhang, H.; Jiang, W. Application of radiomics and machine learning in head and neck cancers. Int. J. Biol. Sci. 2021, 17, 475–486. [Google Scholar] [CrossRef]
- Chetan, M.R.; Gleeson, F.V. Radiomics in predicting treatment response in non-small-cell lung cancer: Current status, challenges and future perspectives. Eur. Radiol. 2021, 31, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics 2019, 9, 1303–1322. [Google Scholar] [CrossRef] [PubMed]
- Kohavi, R. Wrappers for Performance Enhancement and Obvious Decision Graphs. Ph.D. Thesis, Stanford University, Computer Science Department, Stanford, CA, USA, 1995. [Google Scholar]
- Harding-Theobald, E.; Louissaint, J.; Maraj, B.; Cuaresma, E.; Townsend, W.; Mendiratta-Lala, M.; Singal, A.G.; Su, G.L.; Lok, A.S.; Parikh, N.D. Systematic review: Radiomics for the diagnosis and prognosis of hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2021, 54, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Ligero, M.; Garcia-Ruiz, A.; Viaplana, C.; Villacampa, G.; Raciti, M.V.; Landa, J.; Matos, I.; Martin-Liberal, J.; Ochoa-de-Olza, M.; Hierro, C.; et al. A CT-based Radiomics Signature Is Associated with Response to Immune Checkpoint Inhibitors in Advanced Solid Tumors. Radiology 2021, 299, 109–119. [Google Scholar] [CrossRef]
- Wang, T.; She, Y.; Yang, Y.; Liu, X.; Chen, S.; Zhong, Y.; Deng, J.; Zhao, M.; Sun, X.; Xie, D.; et al. Radiomics for Survival Risk Stratification of Clinical and Pathologic Stage IA Pure-Solid Non-Small Cell Lung Cancer. Radiology 2022, 302, 425–434. [Google Scholar] [CrossRef]
- Yip, S.S.; Kim, J.; Coroller, T.P.; Parmar, C.; Velazquez, E.R.; Huynh, E.; Mak, R.H.; Aerts, H.J. Associations Between Somatic Mutations and Metabolic Imaging Phenotypes in Non-Small Cell Lung Cancer. J. Nucl. Med. 2017, 58, 569–576. [Google Scholar] [CrossRef]
- Salvatore, C.; Castiglioni, I.; Cerasa, A. Radiomics Approach in the Neurodegenerative Brain. Aging Clin. Exp. Res. 2021, 33, 1709–1711. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Ding, Z. MRI Radiomics Classification and Prediction in Alzheimer’s Disease and Mild Cognitive Impairment: A Review. Curr. Alzheimer Res. 2020, 17, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Reginelli, A.; Grassi, R.; Boldrini, L.; Vacca, G.; D’Ippolito, E.; Annunziata, S.; Farchione, A.; Belfiore, M.P.; Desideri, I.; et al. Delta radiomics: A systematic review. Radiol. Med. 2021, 126, 1571–1583. [Google Scholar] [CrossRef]
- Sotoudeh, H.; Sarrami, A.H.; Roberson, G.H.; Shafaat, O.; Sadaatpour, Z.; Rezaei, A.; Choudhary, G.; Singhal, A.; Sotoudeh, E.; Tanwar, M. Emerging Applications of Radiomics in Neurological Disorders: A Review. Cureus 2021, 13, e20080. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 5 February 2023).
- Wu, G.; Jochems, A.; Refaee, T.; Ibrahim, A.; Yan, C.; Sanduleanu, S.; Woodruff, H.C.; Lambin, P. Structural and functional radiomics for lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 3961–3974. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Song, Q.; Wang, M.; Pang, P.; Liao, Z.; Jiang, H.; Ding, Z. Hippocampus radiomic biomarkers for the diagnosis of amnestic mild cognitive impairment: A machine learning method. Front. Aging Neurosci. 2019, 11, 323. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhang, Y.; Li, H.; Tong, X.; Ouyang, M. How segmentation methods affect hippocampal radiomic feature accuracy in Alzheimer’s disease analysis? Eur. Radiol. 2022, 32, 6965–6976. [Google Scholar] [CrossRef]
- Chaddad, A.; Toews, M.; Desrosiers, C.; Niazi, T. Deep radiomic analysis based on modeling information flow in convolutional neural networks. IEEE Access 2019, 7, 97242–97252. [Google Scholar] [CrossRef]
- Chaddad, A.; Desrosiers, C.; Niazi, T. Deep radiomic analysis of MRI related to Alzheimer’s disease. IEEE Access 2018, 6, 58213–58221. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, S.; Fang, Y.; Qiu, Q.; Zhao, L.; Wei, W.; Tang, Y.; Li, X. Radiomic Features of the Hippocampus for Diagnosing Early-Onset and Late-Onset Alzheimer’s Disease. Front. Aging Neurosci. 2022, 13, 1014. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.P.; Kim, J.; Jang, H.; Kim, J.; Kang, S.H.; Kim, J.S.; Lee, J.; Na, D.L.; Kim, H.J.; Seo, S.W. Predicting amyloid positivity in patients with mild cognitive impairment using a radiomics approach. Sci. Rep. 2021, 11, 6954. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Niu, J.; Wang, L.; Pang, P.; Wang, M.; Liao, Z.; Song, Q.; Jiang, H.; Ding, Z. Comprehensive classification models based on amygdala radiomic features for Alzheimer’s disease and mild cognitive impairment. Brain Imaging Behav. 2021, 15, 2377–2386. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Chen, Y.; Liao, Z.; Jiang, H.; Mao, D.; Wang, M.; Yu, E.; Ding, Z. Corpus callosum radiomics-based classification model in Alzheimer’s disease: A case-control study. Front. Neurol. 2018, 9, 618. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jie, C.; Zheng, W.; Cui, J.; Wang, Z. Investigation of underlying association between whole brain regions and alzheimer’s disease: A research based on an artificial intelligence model. Front. Aging Neurosci. 2022, 14, 872530. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, J.; Shen, T.; Wu, P.; Zuo, C. Radiomics features as predictors to distinguish fast and slow progression of Mild Cognitive Impairment to Alzheimer’s disease. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; IEEE: Piscataway, NJ, USA; 2018; pp. 127–130. [Google Scholar]
- Zhao, K.; Ding, Y.; Han, Y.; Fan, Y.; Alexander-Bloch, A.F.; Han, T.; Jin, D.; Liu, B.; Lu, J.; Song, C.; et al. Independent and reproducible hippocampal radiomic biomarkers for multisite Alzheimer’s disease: Diagnosis, longitudinal progress and biological basis. Sci. Bull. 2020, 65, 1103–1113. [Google Scholar] [CrossRef]
- Wu, Y.; Li, T.; Han, Y.; Jiang, J. Use of radiomic features and support vector machine to discriminate subjective cognitive decline and healthy controls. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; IEEE: Piscataway, NJ, USA; 2020; pp. 1762–1765. [Google Scholar]
- Ding, Y.; Zhao, K.; Che, T.; Du, K.; Sun, H.; Liu, S.; Zheng, Y.; Li, S.; Liu, B.; Liu, Y.; et al. Quantitative radiomic features as new biomarkers for Alzheimer’s disease: An amyloid PET study. Cereb. Cortex 2021, 31, 3950–3961. [Google Scholar] [CrossRef]
- Feng, F.; Wang, P.; Zhao, K.; Zhou, B.; Yao, H.; Meng, Q.; Wang, L.; Zhang, Z.; Ding, Y.; Wang, L. Radiomic features of hippocampal subregions in Alzheimer’s disease and amnestic mild cognitive impairment. Front. Aging Neurosci. 2018, 10, 290. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, J.; Li, Z.; Li, L.; Huang, B.; Alzheimer’s Disease Neuroimaging Initiative. Using Deep Learning Radiomics to Distinguish Cognitively Normal Adults at Risk of Alzheimer’s Disease from Normal Control: An Exploratory Study Based on Structural MRI. Front. Med. 2022, 9, 894726. [Google Scholar] [CrossRef]
- Stang, A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010, 25, 603–605. [Google Scholar] [CrossRef]
- Ayodele, T.; Rogaeva, E.; Kurup, J.T.; Beecham, G.; Reitz, C. Earlyonset Alzheimer’s disease: What is missing in research? Curr. Neurol. Neurosci. Rep. 2021, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Cacace, R.; Sleegers, K.; Van Broeckhoven, C. Molecular geneticsof early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016, 12, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; Feldman, H.H.; Jacova, C.; Hampel, H.; Molinuevo, J.L.; Blennow, K.; DeKosky, S.T.; Gauthier, S.; Selkoe, D.; Bateman, R.; et al. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).