Role of Radiomics Features and Machine Learning for the Histological Classification of Stage I and Stage II NSCLC at [18F]FDG PET/CT: A Comparison between Two PET/CT Scanners
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. [18F]FDG PET/CT Acquisition and Interpretation
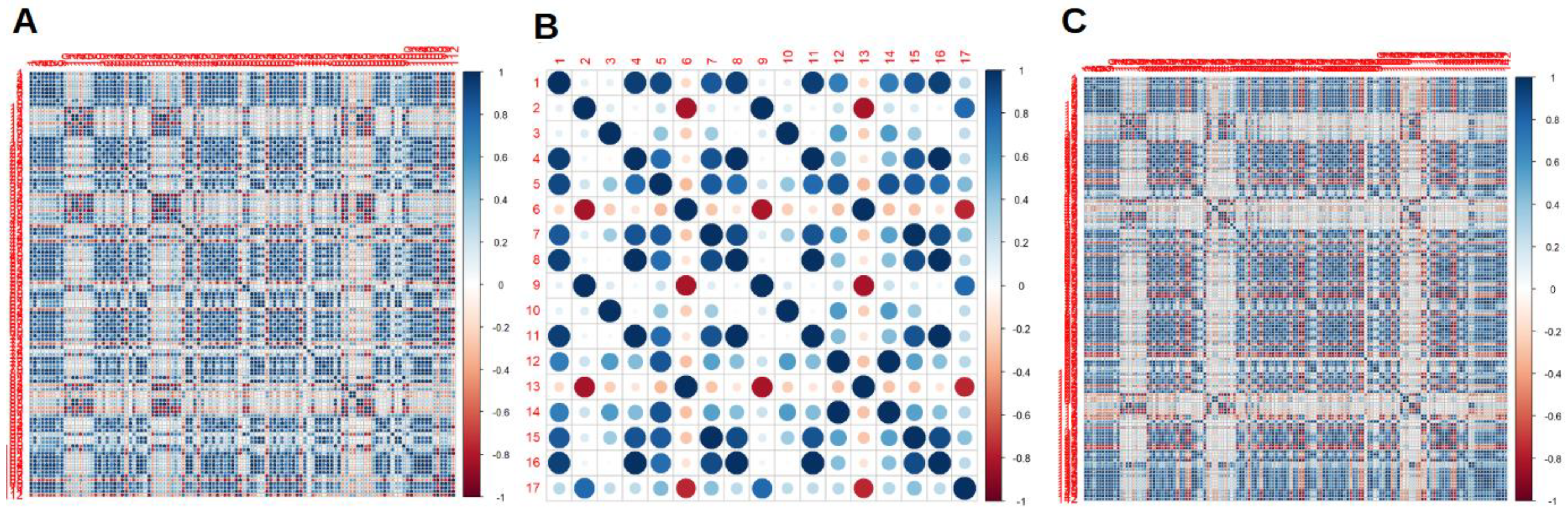
2.3. Radiomics Features Extraction
2.4. Statistical Analysis
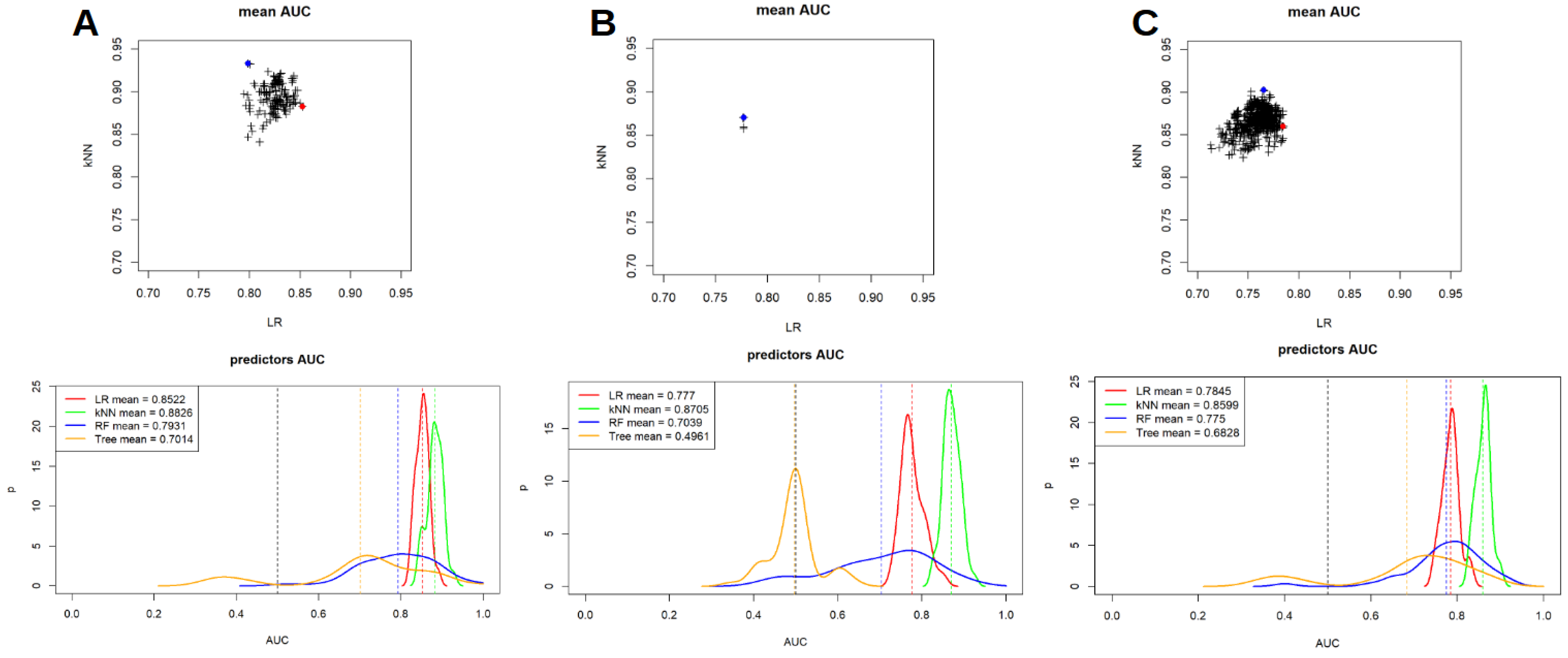
- LR: a bivariate Logistic Regressor was trained using the RaF survived at the feature selection strategy. All the possible couple of RaF with a Spearman’s correlation coefficient lower than 0.3 were tested and only the LR models were both the p-values were lower than 0.05 were considered for the testing. This bivariate analysis was conducted in order to classify these couples based on the area under the curve (AUC) value of the receiver operating characteristic (ROC) analysis. The entire process was repeated in a 50 cross-fold validation, in order to be able to measure the mean and the SD of the AUCs, for each tested couple of RaF.
- kNN: kNN was trained with a 50 cross-fold validation technique for each couple of RaF tested for LR. This was done to assess the different performances between LR and kNN on the same couple of RaF. Again, mean and SD of the AUCs were measured.
- DT and RF were tested with a 50 cross-fold validation technique on all the available RaF. In this case, for each run of the cross-fold validation, only two model were trained (one for DT and one for RF) and the mean and the SD of the AUC were measured on the base of the 50 different training runs.
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, S.Y.; Cho, A.; Yu, W.S.; Lee, C.Y.; Lee, J.G.; Kim, D.J.; Chung, K.Y. Prognostic value of total lesion glycolysis by 18F-FDG PET/CT in surgically resected stage IA non-small cell lung cancer. J. Nucl. Med. 2015, 56, 45–49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rami-Porta, R.; Bolejack, V.; Giroux, D.J.; Chansky, K.; Crowley, J.; Asamura, H.; Goldstraw, P.; on behalf of the International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee, Advisory Board Members and Participating Institutions. The IASLC lung cancer staging project: The new database to inform the eighth edition of the TNM classification of lung cancer. J. Thorac. Oncol. 2014, 9, 1618–1624. [Google Scholar] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Kocher, F.; Hilbe, W.; Seeber, A.; Pircher, A.; Schmid, T.; Greil, R.; Auberger, J.; Nevinny-Stickel, M.; Sterlacci, W.; Tzankov, A.; et al. Longitudinal analysis of 2293 NSCLC patients: A comprehensive study from the TYROL registry. Lung Cancer 2015, 87, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Gallamini, A.; Zwarthoed, C.; Borra, A. Positron Emission Tomography (PET) in Oncology. Cancers 2014, 6, 1821–1889. [Google Scholar] [CrossRef]
- Albano, D.; Gatta, R.; Marini, M.; Rodella, C.; Camoni, L.; Dondi, F.; Giubbini, R.; Bertagna, F. Role of 18F-FDG PET/CT Radiomics Features in the Differential Diagnosis of Solitary Pulmonary Nodules: Diagnostic Accuracy and Comparison between Two Different PET/CT Scanners. J. Clin. Med. 2021, 10, 5064. [Google Scholar] [CrossRef]
- Martini, N.; Bains, M.S.; Burt, M.E.; Zakowski, M.F.; McCormack, P.; Rusch, V.W.; Ginsberg, R.J. Incidence and local recurrence and second primary tumors in resected stage I lung cancer. J. Thorac. Cardiovasc. Surg. 1995, 109, 120–129. [Google Scholar] [CrossRef]
- Goya, T.; Asamura, H.; Yoshimura, H.; Kato, H.; Shimokata, K.; Tsuchiya, R.; Sohara, Y.; Miya, T.; Miyaoka, E. Japanese Joint Committee of Lung Cancer Registry. Prognosis of 6644 resected non-small cell lung cancers in Japan: A Japanese lung cancer registry study. Lung Cancer 2005, 50, 227–234. [Google Scholar] [CrossRef]
- Harpole, D.H., Jr.; Herndon, J.E., II; Young, W.G., Jr.; Wolfe, W.G.; Sabiston, D.C., Jr. Stage I non-small cell lung cancer: A multivariate analysis of treatment methods and patterns of recurrence. Cancer 1995, 76, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Dondi, F.; Albano, D.; Cerudelli, E.; Gazzilli, M.; Giubbini, R.; Treglia, G.; Bertagna, F. Radiolabelled PSMA PET/CT or PET/MRI in hepatocellular carcinoma (HCC): A systematic review. Clin. Transl. Imaging 2020, 8, 461–467. [Google Scholar] [CrossRef]
- Albano, D.; Dondi, F.; Gazzilli, M.; Giubbini, R.; Bertagna, F. Meta-Analysis of the Diagnostic Performance of 18F-FDG-PET/CT Imaging in Native Valve Endocarditis. JACC Cardiovasc. Imaging 2021, 14, 1063–1065. [Google Scholar] [CrossRef]
- Williams, D.E.; Pairolero, P.C.; Davis, C.S.; Bernatz, P.E.; Payne, W.S.; Taylor, W.F.; Uhlenhopp, M.A.; Fontana, R.S. Survival of patients surgically treated for stage I lung cancer. J. Thorac. Cardiovasc. Surg. 1981, 82, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Wisnivesky, J.P.; Henschke, C.; McGinn, T.; Iannuzzi, M.C. Prognosis of stage II non small cell lung cancer according to tumor and nodal status at diagnosis. Lung Cancer 2005, 49, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Al-Sarraf, N.; Gately, K.; Lucey, J.; Aziz, R.; Doddakula, K.; Wilson, L.; McGovern, E.; Young, V. Clinical implication and prognostic significance of standardised uptake value of primary non-small cell lung cancer on positron emission tomography: Analysis of 176 cases. Eur. J. Cardiothorac. Surg. 2008, 34, 892–897. [Google Scholar] [CrossRef]
- Dondi, F.; Pasinetti, N.; Gatta, R.; Albano, D.; Giubbini, R.; Bertagna, F. Comparison between Two Different Scanners for the Evaluation of the Role of 18F-FDG PET/CT Semiquantitative Parameters and Radiomics Features in the Prediction of Final Diagnosis of Thyroid Incidentalomas. J. Clin. Med. 2022, 11, 615. [Google Scholar] [CrossRef]
- Sollini, M.; Cozzi, L.; Antunovic, L.; Chiti, A.; Kirienko, M. PET Radiomics in NSCLC: State of the art and a proposal for harmonization of methodology. Sci. Rep. 2017, 7, 358. [Google Scholar] [CrossRef]
- Bianconi, F.; Palumbo, I.; Fravolini, M.L.; Chiari, R.; Minestrini, M.; Brunese, L.; Palumbo, B. Texture Analysis on [18F]FDG PET/CT in Non-Small-Cell Lung Cancer: Correlations Between PET Features, CT Features, and Histological Types. Mol. Imaging Biol. 2019, 21, 1200–1209. [Google Scholar] [CrossRef]
- Ma, Y.; Feng, W.; Wu, Z.; Liu, M.; Zhang, F.; Liang, Z.; Cui, C.; Huang, J.; Li, X.; Guo, X. Intra-tumoural heterogeneity characterization through texture and colour analysis for differentiation of non-small cell lung carcinoma subtypes. Phys. Med. Biol. 2018, 63, 165018. [Google Scholar] [CrossRef]
- Aydos, U.; Ünal, E.R.; Özçelik, M.; Akdemir, D.; Ekinci, Ö.; Taştepe, A.İ.; Memiş, L.; Atay, L.Ö.; Akdemir, Ü.Ö. Texture features of primary tumor on 18F-FDG PET images in non-small cell lung cancer: The relationship between imaging and histopathological parameters. Rev. Esp. Med. Nucl. Imagen Mol. 2021, 40, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Orlhac, F.M.; Soussan, M.; Chouahnia, K.; Martinod, E.; Buvat, I. 18F-FDG PET-derived textural indices reflect tissue-specific uptake pattern in non-small cell lung cancer. PLoS ONE 2015, 10, e0145063. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Choi, H.; Cheon, G.J.; Kang, K.W.; Chung, J.K.; Kim, E.E.; Lee, D.S. Autoclustering of Non-small Cell Lung Carcinoma Subtypes on 18F-FDG PET Using Texture Analysis: A Preliminary Result. Nucl. Med. Mol. Imaging 2014, 48, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Jung, J.H.; Son, S.H.; Kim, C.Y.; Hong, C.M.; Oh, J.R.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. Prognostic Significance of Intratumoral Metabolic Heterogeneity on 18F-FDG PET/CT in Pathological N0 Non-Small Cell Lung Cancer. Clin. Nucl. Med. 2015, 40, 708–714. [Google Scholar] [CrossRef] [PubMed]
- van Gómez López, O.; García Vicente, A.M.; Honguero Martínez, A.F.; Soriano Castrejón, A.M.; Jiménez Londoño, G.A.; Udias, J.M.; León Atance, P. Heterogeneity in [18F]fluorodeoxyglucose positron emission tomography/computed tomography of non-small cell lung carcinoma and its relationship to metabolic parameters and pathologic staging. Mol. Imaging 2014, 13. [Google Scholar] [CrossRef]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuzé, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterization of Tumor Heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef]
- Dinapoli, N.; Alitto, A.R.; Vallati, M.; Gatta, R.; Autorino, R.; Boldrini, L.; Damiani, A.; Valentini, V. Moddicom: A complete and easily accessible library for prognostic evaluations relying on image features. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2015, 2015, 771–774. [Google Scholar] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]

| Characteristic | n (%) |
|---|---|
| Sex | |
| Male | 147 (64.8%) |
| Female | 80 (35.2%) |
| Age (mean ± SD, range) | 70 ± 8, 38–87 |
| Histology | |
| Adenocarcinoma | 169 (74.4%) |
| Squamous cell carcinoma | 58 (25.6%) |
| Size (mean ± SD, range) (mm) | 32 ± 15, 7–69 |
| Grading * | |
| G1 | 1 (1.0%) |
| G2 | 42 (42.0%) |
| G3 | 57 (57.0%) |
| Lobe | |
| LSL | 54 (23.8%) |
| LIL | 39 (17.2%) |
| RSL | 82 (36.1%) |
| ML | 7 (3.1%) |
| RIL | 45 (19.8%) |
| TNM stage | |
| T1mi | 1 (0.4%) |
| T1a | 26 (11.5%) |
| T1b | 31 (13.7%) |
| T1c | 11 (4.8%) |
| T1aN1 | 2 (0.9%) |
| T1bN1 | 3 (1.3%) |
| T1cN1 | 1 (0.4%) |
| T2a | 59 (26.0%) |
| T2b | 23 (10.1%) |
| T2aN1 | 23 (10.1%) |
| T2bN1 | 9 (4.0%) |
| T3 | 38 (16.7%) |
| AJCC stage | |
| I | |
| IA1 | 27 (11.9%) |
| IA2 | 31 (13.7%) |
| IA3 | 11 (4.8%) |
| IB | 59 (26.0%) |
| II | |
| IIA | 23 (10.1%) |
| IIB | 76 (33.5%) |
| Nodal metastasis | |
| Yes | 38 (16.7%) |
| No | 189 (83.3%) |
| PET/CT scanner | |
| Scanner 1 (Discovery 690) | 142 (62.6%) |
| Scanner 2 (Discovery STE) | 85 (37.4%) |
| Covariate 1 | Covariate 2 | Mean AUC | SD AUC | Mean p-Value 1 | Mean p-Value 2 |
|---|---|---|---|---|---|
| Scanner 1 | |||||
| L_least | F_cm.2.5Dmerged.diff.entr | 0.852 | 0.015 | <0.001 | 0.020 |
| L_least | F_cm_2.5D.diff.entr | 0.850 | 0.017 | <0.001 | 0.020 |
| L_major | F_cm_2.5D.diff.entr | 0.847 | 0.018 | <0.001 | <0.001 |
| L_major | F_cm.2.5Dmerged.diff.entr | 0.846 | 0.019 | <0.001 | <0.001 |
| F_cm_2.5D.diff.entr | F_cm_2.5D.inv.diff.mom.norm | 0.845 | 0.019 | <0.001 | <0.001 |
| F_cm_2.5D.diff.entr | F_cm.2.5Dmerged.inv.diff.mom.norm | 0.845 | 0.019 | <0.001 | <0.001 |
| F_cm_2.5D.diff.entr | F_szm_2.5D.zsnu | 0.845 | 0.018 | 0.012 | <0.001 |
| F_cm.2.5Dmerged.diff.entr | F_cm.2.5Dmerged.inv.diff.mom.norm | 0.844 | 0.019 | <0.001 | <0.001 |
| F_cm.diff.entr | F_cm_2.5D.inv.diff.mom.norm | 0.844 | 0.017 | <0.001 | <0.001 |
| F_cm.diff.entr | F_cm.2.5Dmerged.inv.diff.mom.norm | 0.844 | 0.016 | <0.001 | <0.001 |
| Scanner 2 | |||||
| F_cm.clust.shade | F_cm_merged.inv.var | 0.777 | 0.027 | 0.029 | 0.021 |
| F_cm.inv.var | F_cm.clust.shade | 0.777 | 0.028 | 0.019 | 0.030 |
| F_cm.inv.var | F_cm_merged.clust.shade | 0.777 | 0.028 | 0.019 | 0.030 |
| F_cm_merged.inv.var | F_cm_merged.clust.shade | 0.777 | 0.027 | 0.021 | 0.029 |
| Scanner 1 + 2 | |||||
| L_major | F_rlm.2.5Dmerged.sre | 0.784 | 0.020 | <0.001 | <0.001 |
| F_stat.entropy | F_rlm.2.5Dmerged.sre | 0.784 | 0.020 | <0.001 | <0.001 |
| L_least | F_rlm.2.5Dmerged.sre | 0.784 | 0.021 | <0.001 | <0.001 |
| F_stat.entropy | F_rlm_2.5D.sre | 0.784 | 0.020 | <0.001 | <0.001 |
| L_major | F_rlm_2.5D.sre | 0.784 | 0.020 | <0.001 | <0.001 |
| L_least | F_rlm_2.5D.sre | 0.784 | 0.021 | <0.001 | <0.001 |
| L_minor | F_rlm.2.5Dmerged.sre | 0.782 | 0.020 | <0.001 | <0.001 |
| L_minor | F_rlm_2.5D.sre | 0.782 | 0.020 | <0.001 | <0.001 |
| F_morph.surface | F_rlm.2.5Dmerged.sre | 0.782 | 0.021 | <0.001 | <0.001 |
| F_szm_2.5D.zsnu | F_rlm.2.5Dmerged.sre | 0.782 | 0.020 | <0.001 | <0.001 |
| Covariate 1 | Covariate 2 | Mean AUC | SD AUC | Mean p-Value 1 | Mean p-Value 2 |
|---|---|---|---|---|---|
| Scanner 1 | |||||
| F_stat.entropy | F_cm_2.5D.clust.shade | 0.938 | 0.012 | <0.001 | 0.271 |
| F_stat.entropy | F_cm.2.5Dmerged.clust.shade | 0.938 | 0.012 | <0.001 | 0.272 |
| F_morph.surface | F_cm_merged.clust.prom | 0.936 | 0.016 | <0.001 | 0.613 |
| F_morph.surface | F_cm.clust.prom | 0.936 | 0.016 | <0.001 | 0.613 |
| F_stat.entropy | F_cm_merged.clust.prom | 0.935 | 0.013 | <0.001 | 0.610 |
| F_stat.entropy | F_cm.clust.prom | 0.935 | 0.013 | <0.001 | 0.611 |
| F_cm.energy | F_cm.2.5Dmerged.inv.diff.mom.norm | 0.933 | 0.015 | 0.046 | 0.008 |
| F_stat.entropy | F_cm_2.5D.clust.prom | 0.933 | 0.013 | <0.001 | 0.515 |
| F_stat.entropy | F_cm.2.5Dmerged.clust.prom | 0.933 | 0.013 | <0.001 | 0.516 |
| F_cm.energy | F_cm_2.5D.inv.diff.mom.norm | 0.932 | 0.014 | 0.044 | 0.007 |
| Scanner 2 | |||||
| F_stat.median | F_cm.clust.shade | 0.903 | 0.014 | 0.065 | 0.026 |
| F_stat.median | F_cm_merged.clust.shade | 0.909 | 0.014 | 0.063 | 0.026 |
| F_cm.joint.max | F_cm.clust.shade | 0.899 | 0.025 | 0.014 | 0.140 |
| F_cm.joint.max | F_cm_merged.clust.sade | 0.897 | 0.024 | 0.014 | 0.139 |
| F_cm.clust.shade | F_cm_2.5D.joint.max | 0.893 | 0.016 | 0.111 | 0.028 |
| F_cm.2.5Dmerged.energy | F_cm.2.5Dmerged.clust.shade | 0.892 | 0.021 | 0.013 | 0.791 |
| F_cm_2.5D.joint.max | F_cm_merged.clust.shade | 0.892 | 0.016 | 0.028 | 0.111 |
| F_cm_2.5D.clust.shade | F_cm.2.5Dmerged.energy | 0.892 | 0.022 | 0.790 | 0.013 |
| F_cm.clust.shade | F_cm.2.5Dmerged.joint.max | 0.886 | 0.020 | 0.108 | 0.031 |
| F_cm_merged.clust.shade | F_cm.2.5Dmerged.joint.max | 0.885 | 0.020 | 0.107 | 0.031 |
| Scanner 1 + 2 | |||||
| F_stat.uniformity | F_cm.clust.shade | 0.912 | 0.011 | 0.008 | 0.110 |
| F_stat.uniformity | F_cm_merged.clust.shade | 0.911 | 0.011 | 0.008 | 0.111 |
| F_rlm.lre | F_cm.2.5Dmerged.sum.entr | 0.910 | 0.010 | 0.857 | <0.001 |
| F_stat.uniformity | F_cm_2.5D.clust.shade | 0.907 | 0.014 | 0.008 | 0.158 |
| F_stat.uniformity | F_cm.2.5Dmerged.clust.shade | 0.907 | 0.015 | 0.008 | 0.158 |
| F_morph.volume | F_rlm_2.5D.gl.var | 0.903 | 0.013 | 0.117 | 0.030 |
| F_rlm.lre | F_cm_2.5D.sum.entr | 0.903 | 0.013 | 0.838 | <0.001 |
| F_morph.volume | F_rlm.2.5Dmerged.gl.var | 0.903 | 0.014 | 0.117 | 0.031 |
| F_stat.uniformity | F_cm.diff.avg | 0.903 | 0.014 | 0.008 | 0.005 |
| F_stat.uniformity | F_cm.dissimilarity | 0.903 | 0.014 | 0.008 | 0.005 |
| ML Model | Scanner 1 | Scanner 2 | Scanner 1 + 2 |
|---|---|---|---|
| LR | 0.852 | 0.777 | 0.784 |
| kNN | 0.882 | 0.870 | 0.860 |
| RF | 0.793 | 0.704 | 0.775 |
| DT | 0.701 | 0.496 | 0.682 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dondi, F.; Gatta, R.; Albano, D.; Bellini, P.; Camoni, L.; Treglia, G.; Bertagna, F. Role of Radiomics Features and Machine Learning for the Histological Classification of Stage I and Stage II NSCLC at [18F]FDG PET/CT: A Comparison between Two PET/CT Scanners. J. Clin. Med. 2023, 12, 255. https://doi.org/10.3390/jcm12010255
Dondi F, Gatta R, Albano D, Bellini P, Camoni L, Treglia G, Bertagna F. Role of Radiomics Features and Machine Learning for the Histological Classification of Stage I and Stage II NSCLC at [18F]FDG PET/CT: A Comparison between Two PET/CT Scanners. Journal of Clinical Medicine. 2023; 12(1):255. https://doi.org/10.3390/jcm12010255
Chicago/Turabian StyleDondi, Francesco, Roberto Gatta, Domenico Albano, Pietro Bellini, Luca Camoni, Giorgio Treglia, and Francesco Bertagna. 2023. "Role of Radiomics Features and Machine Learning for the Histological Classification of Stage I and Stage II NSCLC at [18F]FDG PET/CT: A Comparison between Two PET/CT Scanners" Journal of Clinical Medicine 12, no. 1: 255. https://doi.org/10.3390/jcm12010255
APA StyleDondi, F., Gatta, R., Albano, D., Bellini, P., Camoni, L., Treglia, G., & Bertagna, F. (2023). Role of Radiomics Features and Machine Learning for the Histological Classification of Stage I and Stage II NSCLC at [18F]FDG PET/CT: A Comparison between Two PET/CT Scanners. Journal of Clinical Medicine, 12(1), 255. https://doi.org/10.3390/jcm12010255








