Liver X Receptor Expression and Pentraxin 3 Production in Chronic Rhinosinusitis and Sinonasal Mucosal Fibroblast Cells
Abstract
1. Introduction
2. Experimental Section
3. Results
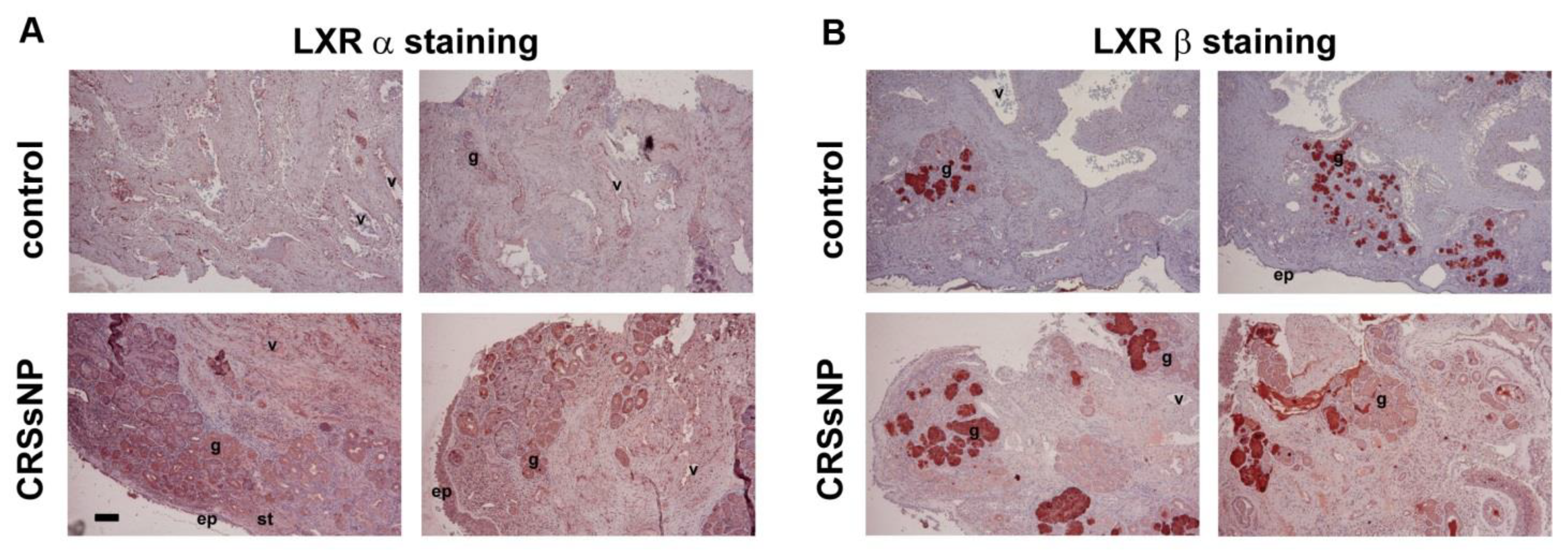
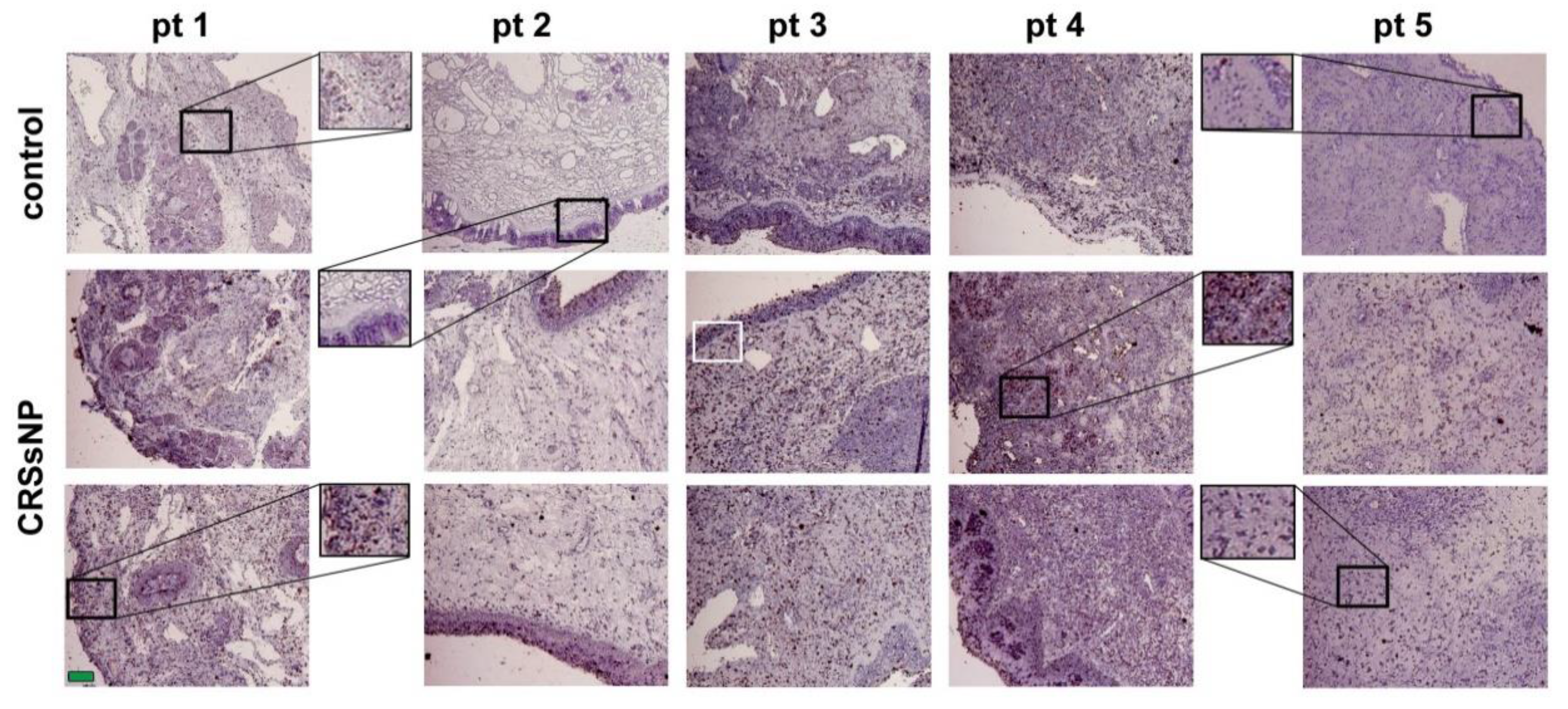
3.1. LXRα and β Expression and Lipid/Fat Deposition in CRSsNP Nasal Mucosae
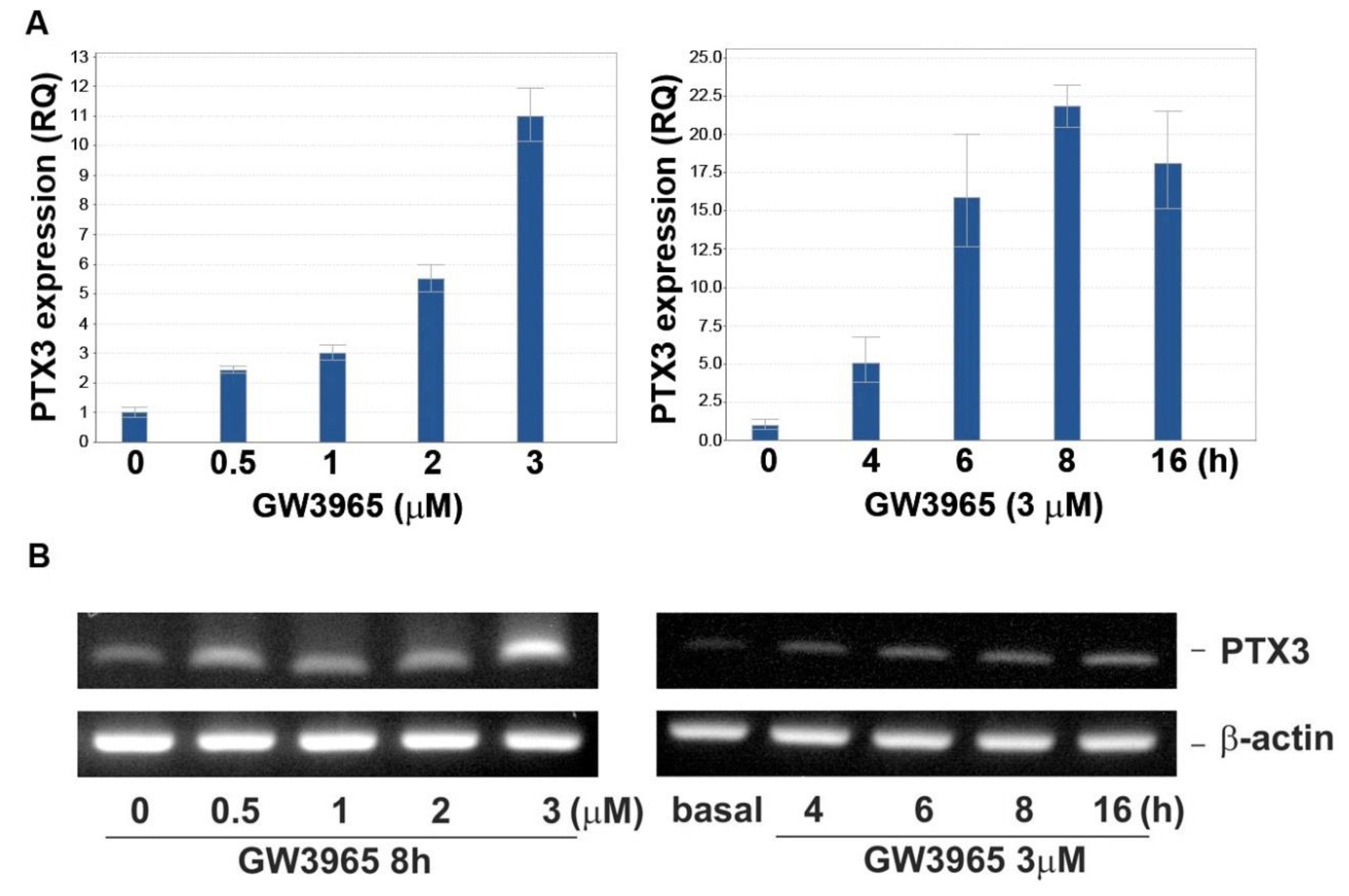
3.2. GW3965 Enhances PTX3 Protein and mRNA Expression
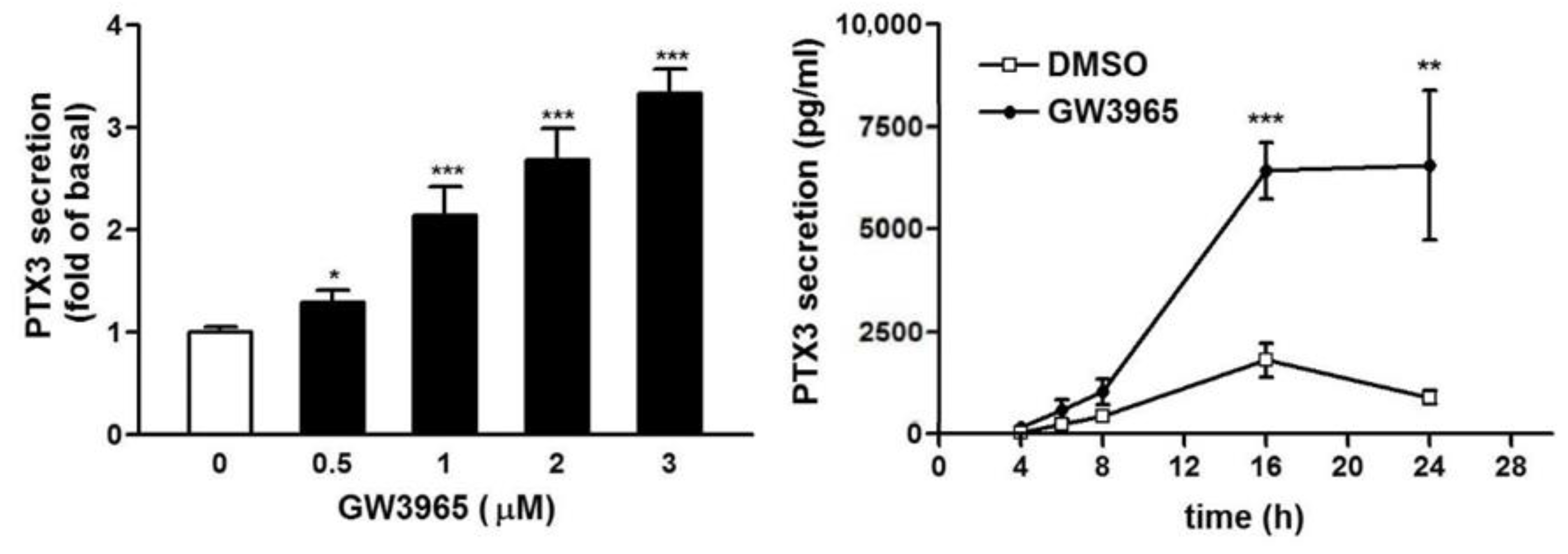
3.3. GW3965 Enhances PTX3 Protein Release
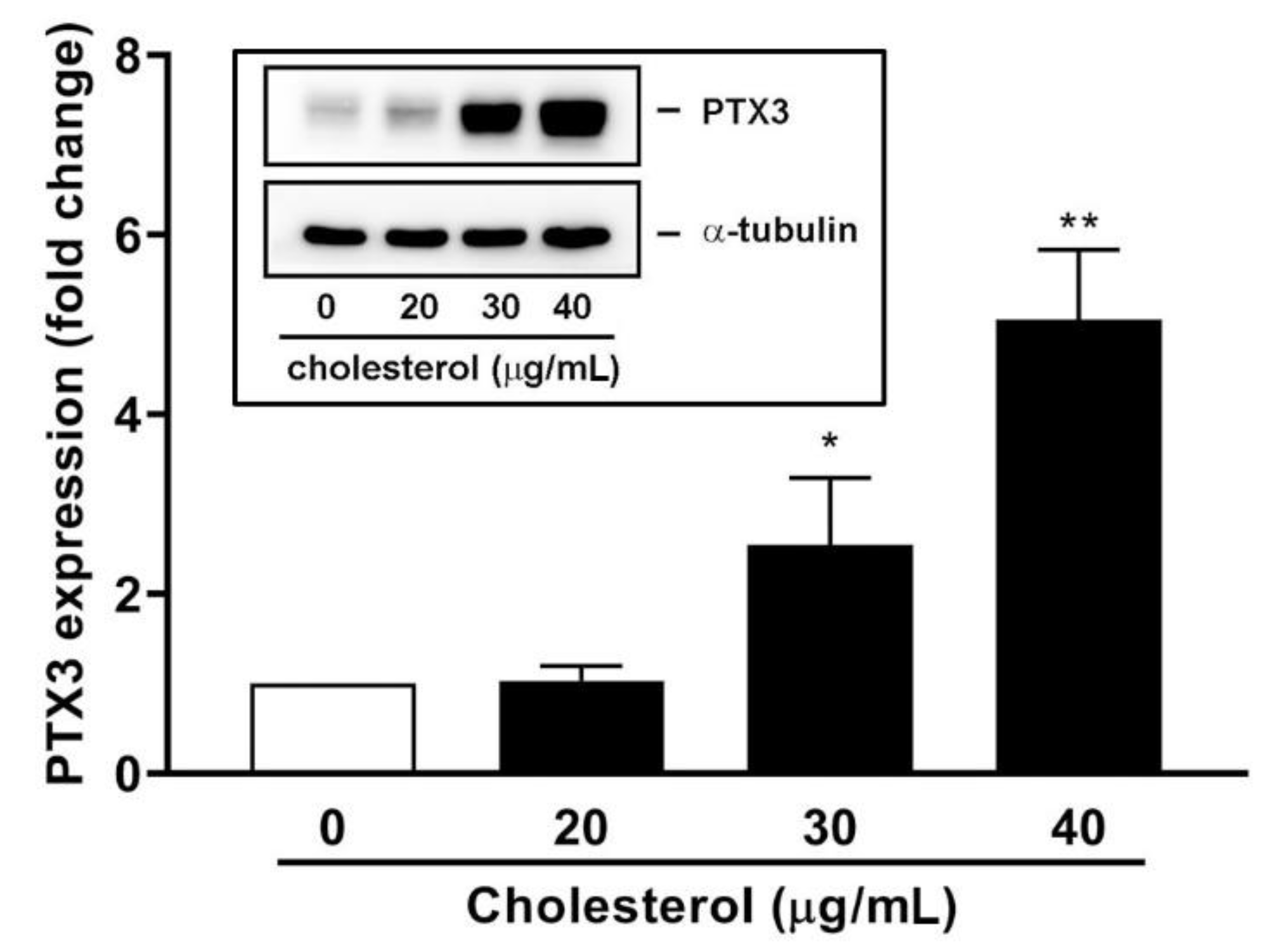
3.4. An Endogenous LXR Agonist-Cholesterol Induces PTX3 Expression
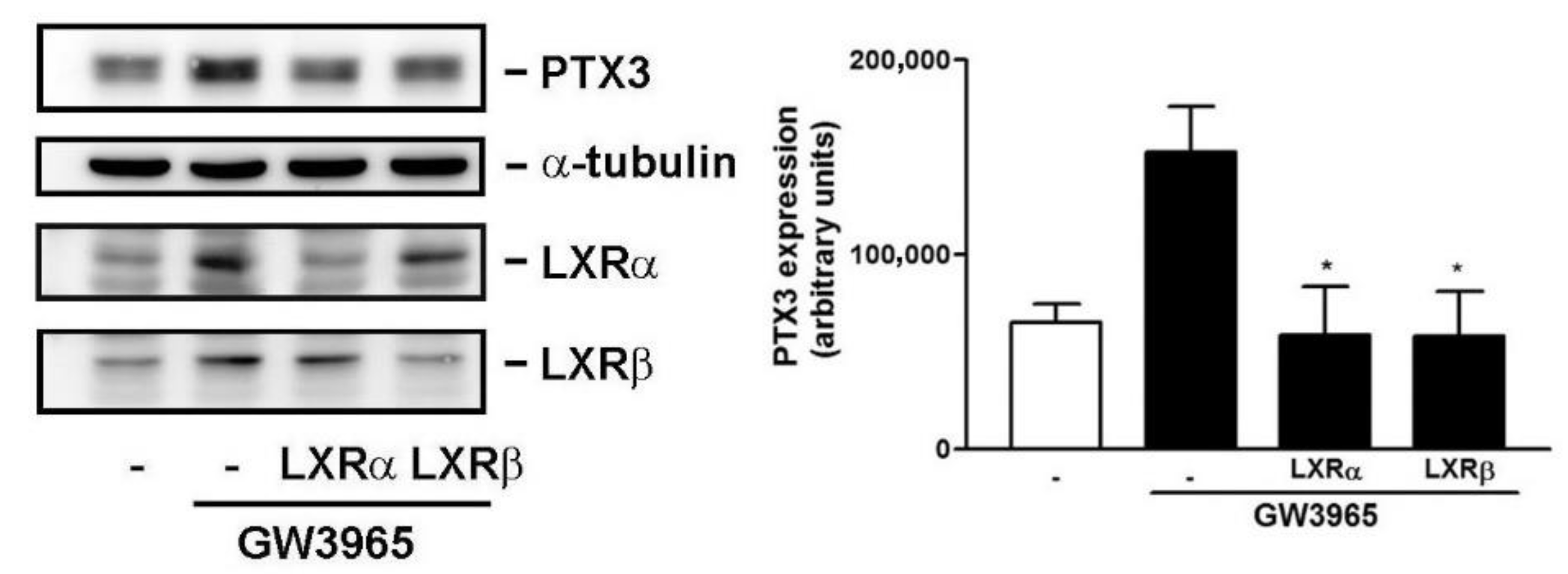
3.5. Knockdown of LXRα and β Expression Compromises GW3965-Induced PTX3 Expression
3.6. A Collaboration of PI3K/Akt and LXR Activation in PTX3 Induction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Im, S.S.; Osborne, T.F. Liver x receptors in atherosclerosis and inflammation. Circ. Res. 2011, 108, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Kliewer, S.A.; Lenhard, J.M.; Willson, T.M.; Patel, I.; Morris, D.C.; Lehmann, J.M. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor γ and promotes adipocyte differentiation. Cell 1995, 83, 813–819. [Google Scholar] [CrossRef]
- Janowski, B.A.; Willy, P.J.; Devi, T.R.; Falck, J.R.; Mangelsdorf, D.J. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 1996, 383, 728–731. [Google Scholar] [CrossRef]
- Repa, J.J.; Liang, G.; Ou, J.; Bashmakov, Y.; Lobaccaro, J.M.; Shimomura, I.; Shan, B.; Brown, M.S.; Goldstein, J.L.; Mangelsdorf, D.J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000, 14, 2819–2830. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Steffensen, K.R.; Gustafsson, J.-Å. Structural characterisation of the mouse nuclear oxysterol receptor genes LXRα and LXRβ. Gene 2000, 243, 93–103. [Google Scholar] [CrossRef]
- Willy, P.J.; Mangelsdorf, D.J. Unique requirements for retinoid-dependent transcriptional activation by the orphan receptor LXR. Genes Dev. 1997, 11, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Willy, P.J.; Umesono, K.; Ong, E.S.; Evans, R.M.; Heyman, R.A.; Mangelsdorf, D.J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995, 9, 1033–1045. [Google Scholar] [CrossRef]
- Svensson, S.; Ostberg, T.; Jacobsson, M.; Norstrom, C.; Stefansson, K.; Hallen, D.; Johansson, I.C.; Zachrisson, K.; Ogg, D.; Jendeberg, L. Crystal structure of the heterodimeric complex of LXR [alpha] and RXR [beta] ligand-binding domains in a fully agonistic conformation. EMBO J. 2003, 22, 4625–4633. [Google Scholar] [CrossRef]
- Wang, Y.; Rogers, P.M.; Stayrook, K.R.; Su, C.; Varga, G.; Shen, Q.; Nagpal, S.; Burris, T.P. The selective Alzheimer’s disease indicator-1 gene (Seladin-1/DHCR24) is a liver X receptor target gene. Mol. Pharm. 2008, 74, 1716–1721. [Google Scholar] [CrossRef]
- Wang, Y.; Rogers, P.M.; Su, C.; Varga, G.; Stayrook, K.R.; Burris, T.P. Regulation of cholesterologenesis by the oxysterol receptor, LXRalpha. J. Biol. Chem. 2008, 283, 26332–26339. [Google Scholar] [CrossRef]
- Wang, B.; Tontonoz, P. Liver X receptors in lipid signalling and membrane homeostasis. Nat. Rev. Endocrinol. 2018, 14, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Repa, J.J.; Mangelsdorf, D.J. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu. Rev. Cell Dev. Biol. 2000, 16, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Barbier, O.; Trottier, J.; Kaeding, J.; Caron, P.; Verreault, M. Lipid-activated transcription factors control bile acid glucuronidation. Mol. Cell. Biochem. 2009, 326, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Volle, D.H.; Repa, J.J.; Mazur, A.; Cummins, C.L.; Val, P.; Henry-Berger, J.; Caira, F.; Veyssiere, G.; Mangelsdorf, D.J.; Lobaccaro, J.M. Regulation of the aldo-keto reductase gene akr1b7 by the nuclear oxysterol receptor LXRalpha (liver X receptor-alpha) in the mouse intestine: Putative role of LXRs in lipid detoxification processes. Mol. Endocrinol. 2004, 18, 888–898. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cha, J.Y.; Repa, J.J. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 2007, 282, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Zelcer, N.; Tontonoz, P. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Investig. 2006, 116, 607–614. [Google Scholar] [CrossRef]
- Demerjian, M.; Choi, E.H.; Man, M.Q.; Chang, S.; Elias, P.M.; Feingold, K.R. Activators of PPARs and LXR decrease the adverse effects of exogenous glucocorticoids on the epidermis. Exp. Dermatol. 2009, 18, 643–649. [Google Scholar] [CrossRef]
- Koldamova, R.; Lefterov, I. Role of LXR and ABCA1 in the pathogenesis of Alzheimer’s disease-implications for a new therapeutic approach. Curr. Alzheimer Res. 2007, 4, 171–178. [Google Scholar] [CrossRef]
- Fokkens, W.J.; Lund, V.J.; Mullol, J.; Bachert, C.; Alobid, I.; Baroody, F.; Cohen, N.; Cervin, A.; Douglas, R.; Gevaert, P.; et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol. Suppl. 2012, 50, 1–298. [Google Scholar] [CrossRef]
- Tomassen, P.; Van Zele, T.; Zhang, N.; Perez-Novo, C.; Van Bruaene, N.; Gevaert, P.; Bachert, C. Pathophysiology of chronic rhinosinusitis. Proc. Am. Thorac. Soc. 2011, 8, 115–120. [Google Scholar] [CrossRef]
- Van Crombruggen, K.; Zhang, N.; Gevaert, P.; Tomassen, P.; Bachert, C. Pathogenesis of chronic rhinosinusitis: Inflammation. J. Allergy Clin. Immunol. 2011, 128, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.J.; Douglas, R.G. Pathogenesis and treatment of chronic rhinosinusitis. Postgrad. Med. J. 2010, 86, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Brook, I. Microbiology of chronic rhinosinusitis. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Frank, D.N.; Ramakrishnan, V. Microbiome of the paranasal sinuses: Update and literature review. Am. J. Rhinol. Allergy 2016, 30, 3–16. [Google Scholar] [CrossRef]
- Su, W.; Jiang, Y. [Bacterial culture analysis for patients with chronic rhinosinusitis with or without polyps]. Zhong nan da xue xue bao. Yi xue ban J. Cent. South Univ. Med. Sci. 2015, 40, 1253–1257. [Google Scholar]
- Zhang, P.; Liu, X.; Cao, X. Extracellular pattern recognition molecules in health and diseases. Cell. Mol. Immunol. 2015, 12, 255–257. [Google Scholar] [CrossRef]
- Mantovani, A.; Valentino, S.; Gentile, S.; Inforzato, A.; Bottazzi, B.; Garlanda, C. The long pentraxin PTX3: A paradigm for humoral pattern recognition molecules. Ann. N. Y. Acad. Sci. 2013, 1285, 1–14. [Google Scholar] [CrossRef]
- Tsai, Y.J.; Hao, C.Y.; Chen, C.L.; Wu, P.H.; Wu, W.B. Expression of long pentraxin 3 in human nasal mucosa fibroblasts, tissues, and secretions of chronic rhinosinusitis without nasal polyps. J. Mol. Med. (Berl. Ger.) 2020, 98, 673–689. [Google Scholar] [CrossRef]
- Inforzato, A.; Baldock, C.; Jowitt, T.A.; Holmes, D.F.; Lindstedt, R.; Marcellini, M.; Rivieccio, V.; Briggs, D.C.; Kadler, K.E.; Verdoliva, A.; et al. The Angiogenic Inhibitor Long Pentraxin PTX3 Forms an Asymmetric Octamer with Two Binding Sites for FGF2. J. Biol. Chem. 2010, 285, 17681–17692. [Google Scholar] [CrossRef]
- Garlanda, C.; Bottazzi, B.; Magrini, E.; Inforzato, A.; Mantovani, A. PTX3, a Humoral Pattern Recognition Molecule, in Innate Immunity, Tissue Repair, and Cancer. Physiol. Rev. 2018, 98, 623–639. [Google Scholar] [CrossRef]
- Doni, A.; Musso, T.; Morone, D.; Bastone, A.; Zambelli, V.; Sironi, M.; Castagnoli, C.; Cambieri, I.; Stravalaci, M.; Pasqualini, F.; et al. An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J. Exp. Med. 2015, 212, 905–925. [Google Scholar] [CrossRef] [PubMed]
- Rua, R.; McGavern, D.B. Pentraxin 3 innately preps damaged tissue for wound healing. J. Exp. Med. 2015, 212, 829. [Google Scholar] [CrossRef] [PubMed]
- Bonavita, E.; Gentile, S.; Rubino, M.; Maina, V.; Papait, R.; Kunderfranco, P.; Greco, C.; Feruglio, F.; Molgora, M.; Laface, I.; et al. PTX3 Is an Extrinsic Oncosuppressor Regulating Complement-Dependent Inflammation in Cancer. Cell 2015, 160, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Niho, M. [Cholesterol deposits in the paranasal sinus (author′s transl)]. Nihon Jibiinkoka Gakkai Kaiho 1980, 83, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.J.; Hao, S.P.; Chen, C.L.; Lin, B.J.; Wu, W.B. Involvement of B2 receptor in bradykinin-induced proliferation and proinflammatory effects in human nasal mucosa-derived fibroblasts isolated from chronic rhinosinusitis patients. PLoS ONE 2015, 10, e0126853. [Google Scholar]
- Lai, T.-H.; Shieh, J.-M.; Tsou, C.-J.; Wu, W.-B. Gold nanoparticles induce heme oxygenase-1 expression through Nrf2 activation and Bach1 export in human vascular endothelial cells. Int. J. Nanomed. 2015, 10, 5925–5939. [Google Scholar]
- Tsai, Y.-J.; Chi, J.C.-Y.; Hao, C.-Y.; Wu, W.-B. Peptidoglycan induces bradykinin receptor 1 expression through Toll-like receptor 2 and NF-κB signaling pathway in human nasal mucosa-derived fibroblasts of chronic rhinosinusitis patients. J. Cell. Physiol. 2018, 233, 7226–7238. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, Y.-T.; Chen, S.-Y.; He, H.; Tseng, S.C.G. Constitutive Expression of Pentraxin 3 (PTX3) Protein by Human Amniotic Membrane Cells Leads to Formation of the Heavy Chain (HC)-Hyaluronan (HA)-PTX3 Complex. J. Biol. Chem. 2014, 289, 13531–13542. [Google Scholar] [CrossRef]
- Rong, J.X.; Shapiro, M.; Trogan, E.; Fisher, E.A. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc. Natl. Acad. Sci. USA 2003, 100, 13531–13536. [Google Scholar] [CrossRef]
- Fontaine, C.; Rigamonti, E.; Nohara, A.; Gervois, P.; Teissier, E.; Fruchart, J.-C.; Staels, B.; Chinetti-Gbaguidi, G. Liver X Receptor Activation Potentiates the Lipopolysaccharide Response in Human Macrophages. Circ. Res. 2007, 101, 40–49. [Google Scholar] [CrossRef]
- Dykewicz, M.S.; Hamilos, D.L. Rhinitis and sinusitis. J. Allergy Clin. Immunol. 2010, 125, S103–S115. [Google Scholar] [CrossRef] [PubMed]
- Shieh, J.M.; Tsai, Y.J.; Chi, J.C.; Wu, W.B. TGFβ mediates collagen production in human CRSsNP nasal mucosa-derived fibroblasts through Smad 2/3-dependent pathway and CTGF induction and secretion. J. Cell. Physiol. 2019, 234, 10489–10499. [Google Scholar] [CrossRef] [PubMed]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef]
- Higo, R.; Iwamori, M. Significantly High Synthetic Activities of Cholesterol Sulfate in the Nasal, Oral and Tracheal Mucosae of Guinea Pigs. ORL 1995, 57, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Do, T.Q.; Moshkani, S.; Castillo, P.; Anunta, S.; Pogosyan, A.; Cheung, A.; Marbois, B.; Faull, K.F.; Ernst, W.; Chiang, S.M.; et al. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J. Immunol. Baltim. Md. 1950 2008, 181, 4177–4187. [Google Scholar] [CrossRef]
- Lee, J.T.; Jansen, M.; Yilma, A.N.; Nguyen, A.; Desharnais, R.; Porter, E. Antimicrobial lipids: Novel innate defense molecules are elevated in sinus secretions of patients with chronic rhinosinusitis. Am. J. Rhinol. Allergy 2010, 24, 99–104. [Google Scholar] [CrossRef]
- Lee, J.T.; Escobar, O.H.; Anouseyan, R.; Janisiewicz, A.; Eivers, E.; Blackwell, K.E.; Keschner, D.B.; Garg, R.; Porter, E. Assessment of epithelial innate antimicrobial factors in sinus tissue from patients with and without chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2014, 4, 893–900. [Google Scholar] [CrossRef]
- Du Clos, T.W. Pentraxins: Structure, function, and role in inflammation. ISRN Inflamm. 2013, 2013, 379040. [Google Scholar] [CrossRef]
- He, X.; Han, B.; Liu, M. Long pentraxin 3 in pulmonary infection and acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L1039–L1049. [Google Scholar] [CrossRef]
- Huwait, E.A.; Greenow, K.R.; Singh, N.N.; Ramji, D.P. A novel role for c-Jun N-terminal kinase and phosphoinositide 3-kinase in the liver X receptor-mediated induction of macrophage gene expression. Cell Signal. 2011, 23, 542–549. [Google Scholar] [CrossRef]
- Sun, T.; Li, Y.J.; Tian, Q.Q.; Wu, Q.; Feng, D.; Xue, Z.; Guo, Y.Y.; Yang, L.; Zhang, K.; Zhao, M.G.; et al. Activation of liver X receptor β-enhancing neurogenesis ameliorates cognitive impairment induced by chronic cerebral hypoperfusion. Exp. Neurol. 2018, 304, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, N.; Silveyra, P. Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135–170. [Google Scholar] [PubMed]
- Montenegro, K.R.; Cruzat, V.; Carlessi, R.; Newsholme, P. Mechanisms of vitamin D action in skeletal muscle. Nutr. Res. Rev. 2019, 32, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Dalmolin, R.J.S.; Zanotto-filho, A.; De oliveira, R.B.; Duarte, R.F.; Pasquali, M.A.B.; Moreira, J.C.F. Retinol and retinoic acid increase MMP-2 activity by different pathways in cultured Sertoli cells. Free. Radic. Res. 2007, 41, 1338–1347. [Google Scholar] [CrossRef]


| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Product Size (bp) |
|---|---|---|---|
| ptx3 | GCTCTCTGGTCTGCAGTGTT | CTTGTCCCATTCCGAGTGCT | 147 |
| β-actin | ATCATGTTTGAGACCTTCAA | CATCTCTTGCTCGAAGTCCA | 314 |
| Cyc-A | TATCTGCACTGCCAAGACTGAGTG | CTTCTTGCTGGTCTTGCCATTCC | 127 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, Y.-J.; Shen, P.-H.; Luo, S.-D.; Wu, W.-B. Liver X Receptor Expression and Pentraxin 3 Production in Chronic Rhinosinusitis and Sinonasal Mucosal Fibroblast Cells. J. Clin. Med. 2021, 10, 452. https://doi.org/10.3390/jcm10030452
Tsai Y-J, Shen P-H, Luo S-D, Wu W-B. Liver X Receptor Expression and Pentraxin 3 Production in Chronic Rhinosinusitis and Sinonasal Mucosal Fibroblast Cells. Journal of Clinical Medicine. 2021; 10(3):452. https://doi.org/10.3390/jcm10030452
Chicago/Turabian StyleTsai, Yih-Jeng, Ping-Hung Shen, Sheng-Dean Luo, and Wen-Bin Wu. 2021. "Liver X Receptor Expression and Pentraxin 3 Production in Chronic Rhinosinusitis and Sinonasal Mucosal Fibroblast Cells" Journal of Clinical Medicine 10, no. 3: 452. https://doi.org/10.3390/jcm10030452
APA StyleTsai, Y.-J., Shen, P.-H., Luo, S.-D., & Wu, W.-B. (2021). Liver X Receptor Expression and Pentraxin 3 Production in Chronic Rhinosinusitis and Sinonasal Mucosal Fibroblast Cells. Journal of Clinical Medicine, 10(3), 452. https://doi.org/10.3390/jcm10030452






