Combined Mutational and Clonality Analyses Support the Existence of Intra-Tumor Heterogeneity in Papillary Thyroid Cancer
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. DNA Extraction
2.3. Genotyping
2.4. HUMARA Assay
2.5. Statistical Analyses
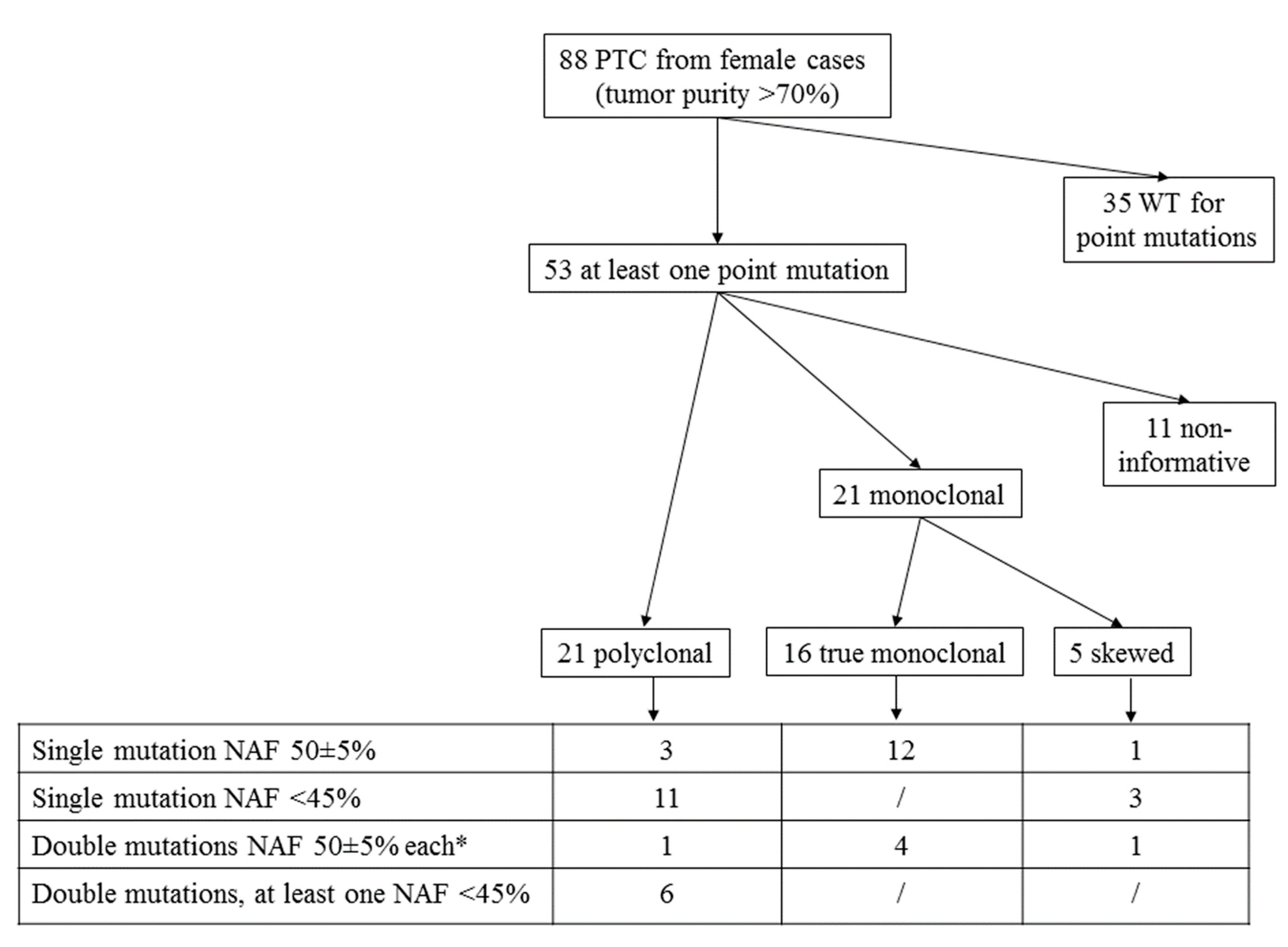
3. Results
3.1. Mutational Analysis
3.2. HUMARA Clonality Assay
3.3. Correlations between Clonal Status and Clinico-Pathological Features
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hansen, A.R.; Ratain, M.J.; Siu, L.L. Tumour heterogeneity in the clinic. Nature 2013, 501, 355–364. [Google Scholar] [CrossRef]
- Fugazzola, L.; Muzza, M.; Pogliaghi, G.; Vitale, M. Intratumoral Genetic Heterogeneity in. Papillary Thyroid Cancer: Occurrence and Clinical Significance. Cancers 2020, 7, 383. [Google Scholar] [CrossRef] [PubMed]
- Masoodi, T.; Siraj, A.K.; Siraj, S.; Azam, S.; Qadri, Z.; Parvathareddy, S.K.; Al-Sobhi, S.S.; AlDawish, M.; Alkuraya, F.S.; Al-Kuraya, K.S. Evolution and Impact of Subclonal Mutations in Papillary Thyroid Cancer. Am. J. Hum. Genet. 2019, 105, 959–973. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Wang, J.C.; Wu, Y.; Zhang, L.; Huang, C.P.; Shen, Q.; Zhu, Y.X.; Li, D.S.; Ji, Q.H. Incidentally simultaneous occurrence of RET/PTC, H4-PTEN and BRAF mutation in papillary thyroid carcinoma. Cancer Lett. 2008, 263, 44–52. [Google Scholar] [CrossRef]
- Guerra, A.; Zeppa, P.; Bifulco, M.; Vitale, M. Concomitant BRAFV600E mutation and RET/PTC rearrangement is a frequent occurrence in papillary thyroid carcinoma. Thyroid 2014, 24, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Liu, R.; Liu, X.; Murugan, A.K.; Zhu, G.; Zeiger, M.A.; Pai, S.; Bishop, J. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J. Clin. Oncol. 2014, 32, 2718–2726. [Google Scholar] [CrossRef]
- Muzza, M.; Colombo, C.; Rossi, S.; Tosi, D.; Cirello, V.; Perrino, M.; De Leo, S.; Magnani, E.; Pignatti, E.; Vigo, B.; et al. Telomerase in differentiated thyroid cancer: Promoter mutations, expression and localization. Mol. Cell. Endocrinol. 2015, 399, 288–295. [Google Scholar] [CrossRef]
- Colombo, C.; Muzza, M.; Proverbio, M.C.; Tosi, D.; Soranna, D.; Pesenti, C.; Rossi, S.; Cirello, V.; De Leo, S.; Fusco, N.; et al. Impact of mutation density and heterogeneity on papillary thyroid cancer clinical features and remission probability. Thyroid 2019, 29, 237–251. [Google Scholar] [CrossRef]
- Henderson, Y.C.; Shellenberger, T.D.; Williams, M.D.; El-Naggar, A.K.; Fredrick, M.J.; Cieply, K.M.; Clayman, G.L. High rate of BRAF and RET/PTC dual mutations associated with recurrent papillary thyroid carcinoma. Clin. Cancer Res. 2009, 15, 485–491. [Google Scholar] [CrossRef]
- Shrestha, R.T.; Karunamurthy, A.; Amin, K.; Nikiforov, Y.E.; Caramori, M.L. Multiple mutations detected preoperatively may predict aggressive behavior of papillary thyroid cancer and guide management—A case report. Thyroid 2015, 25, 1375–1378. [Google Scholar] [CrossRef] [PubMed]
- Muzza, M. The clonal origin of multifocal papillary thyroid cancer (MPTC): Intrathyroid spread or independent tumors? Minerva Endocrinol. 2020, 46, 35–44. [Google Scholar] [CrossRef]
- Namba, H.; Matsuo, K.; Fagin, J.A. Clonal composition of benign and malignant human thyroid tumors. J. Clin. Investig. 1990, 86, 120–125. [Google Scholar] [CrossRef]
- Moniz, S.; Catarino, A.L.; Marques, A.R.; Cavaco, B.; Sobrinho, L.; Leite, V. Clonal origin of non-medullary thyroid tumours assessed by non-random X-chromosome inactivation. Eur. J. Endocrinol. 2002, 146, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef]
- Guerra, A.; Fugazzola, L.; Marotta, V.; Cirillo, M.; Rossi, S.; Cirello, V.; Forno, I.; Moccia, T.; Budillon, A.; Vitale, M. A high percentage of BRAFV600E alleles in papillary thyroid carcinoma predicts a poorer outcome. J. Clin. Endocrinol. Metab. 2012, 97, 2333–2340. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Bae, J.S.; Lim, D.J.; Lee, H.; Jeon, S.R.; Park, G.S.; Jung, C.K. Quantification of BRAF V600E alleles predicts papillary thyroid cancer progression. Endocr. Relat. Cancer 2014, 21, 891–902. [Google Scholar] [CrossRef]
- Finkel, A.; Liba, L.; Simon, E.; Bick, T.; Prinz, E.; Sabo, E.; Ben-Izhak, O.; Hershkovitz, D. Subclonality for BRAF mutation in papillary thyroid carcinoma is associated with earlier disease stage. J. Clin. Endocrinol. Metab. 2016, 101, 1407–1413. [Google Scholar] [CrossRef]
- Gandolfi, G.; Sancisi, V.; Torricelli, F.; Ragazzi, M.; Frasoldati, A.; Piana, S.; Ciarrocchi, A. Allele percentage of the BRAF V600E mutation in papillary thyroid carcinomas and corresponding lymph node metastases: No evidence for a role in tumor progression. J. Clin. Endocrinol. Metab. 2013, 98, E934–E942. [Google Scholar] [CrossRef]
- Haugen, B.R.; Alexander, E.K.; Bible, K.C.; Doherty, G.M.; Mandel, S.J.; Nikiforov, Y.E.; Pacini, F.; Randolph, G.W.; Sawka, A.M.; Schlumberger, M.; et al. 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid 2016, 26, 1–133. [Google Scholar] [CrossRef]
- Pacini, F.; Basolo, F.; Bellantone, R.; Boni, G.; Cannizzaro, M.A.; De Palma, M.; Durante, C.; Elisei, R.; Fadda, G.; Frasoldati, A.; et al. Italian consensus on diagnosis and treatment of differentiated thyroid cancer: Joint statements of six Italian societies. J. Endocrinol. Investig. 2018, 41, 849–876. [Google Scholar] [CrossRef]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer: New York, NY, USA, 2017; Volume XVII, p. 1032. [Google Scholar]
- Pesenti, C.; Muzza, M.; Colombo, C.; Proverbio, M.C.; Farè, C.; Ferrero, S.; Miozzo, M.; Fugazzola, L.; Tabano, S. MassARRAY-based simultaneous detection of hotspot somatic mutations and recurrent fusion genes in papillary thyroid carcinoma: The PTC-MA assay. Endocrine 2018, 61, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.C.; Zoghbi, H.Y.; Moseley, A.B.; Rosenblatt, H.M.; Belmont, J.W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 1992, 51, 1229–1239. [Google Scholar]
- Jovanovic, L.; Delahunt, B.; McIver, B.; Eberhardt, N.L.; Grebe, S.K. Thyroid gland clonality revisited: The embryonal patch size of the normal human thyroid gland is very large, suggesting X-chromosome inactivation tumor clonality studies of thyroid tumors have to be interpreted with caution. J. Clin. Endocrinol. Metab. 2003, 88, 3284–3291. [Google Scholar] [CrossRef]
- Lin, J.; Takata, M.; Murata, H.; Goto, Y.; Kido, K.; Ferrone, S.; Saida, T. Polyclonality of BRAF mutations in acquired melanocytic nevi. J. Natl. Cancer. Inst. 2009, 101, 1423–1427. [Google Scholar] [CrossRef]
- Masoodi, T.; Siraj, A.K.; Siraj, S.; Azam, S.; Qadri, Z.; Albalawy, W.N.; Parvathareddy, S.K.; Al-Sobhi, S.S.; Al-Dayel, F.; Alkuraya, F.S.; et al. Whole-Exome Sequencing of Matched Primary and Metastatic Papillary Thyroid Cancer. Thyroid 2020, 30, 42–56. [Google Scholar] [CrossRef]
- Bozic, I.; Reiter, J.G.; Allen, B.; Antal, T.; Chatterjee, K.; Shah, P.; Moon, Y.S.; Yaqubie, A.; Kelly, N.; Le, D.T.; et al. Evolutionary dynamics of cancer in response to targeted combination therapy. eLife 2013, 2, e00747. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.G.; Riaz, N.; Desrichard, A.; Şenbabaoğlu, Y.; Hakimi, A.A.; Makarov, V.; Reis-Filho, J.S.; Chan, T.A. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 2016, 7, 10051–10063. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.Y.; Shin, H.T.; Yun, J.W.; Kim, K.T.; Kim, J.; Bae, J.S.; Cho, Y.B.; Lee, W.Y.; Yun, S.H.; Park, Y.A.; et al. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci. Rep. 2019, 9, 4542. [Google Scholar] [CrossRef]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, J.; Mayor, R.; Mancuso, F.M.; Iglesias, C.; Caratú, G.; Matos, I.; Zafón, C.; Hernando, J.; Petit, A.; Nuciforo, P.; et al. Early evolutionary divergence between papillary and anaplastic thyroid cancers. Ann. Oncol. 2019, 30, 1843. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Hu, J.Q.; Wei, W.J.; Ma, B.; Lu, Z.W.; Wang, Y.L.; Wang, Y.; Ji, Q.H. Dedifferentiation patterns in DTC: Is PDTC an intermediate state between DTC and ATC? Clin. Cancer Res. 2018, 24, 3059–3068. [Google Scholar]
- Owen, D.H.; Konda, B.; Sipos, J.; Liu, T.; Webb, A.; Ringel, M.D.; Timmers, C.D.; Shah, M.H. KRAS G12V mutation in acquired resistance to combined BRAF and MEK inhibition in papillary thyroid cancer. JNCCN J. Natl. Compr. Cancer Netw. 2019, 17, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, M.E.; Dadu, R.; Iyer, P.; Wanland, K.B.; Busaidy, N.L.; Ying, A.; Gule-Monroe, M.; Wang, J.R.; Zafereo, M.; Hofmann, M.C. Acquired Secondary RAS Mutation in BRAF(V600E)-Mutated Thyroid Cancer Patients Treated with BRAF Inhibitors. Thyroid 2020, 30, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Pesenti, C.; Paganini, L.; Fontana, L.; Veniani, E.; Runza, L.; Ferrero, S.; Bosari, S.; Menghi, M.; Marfia, G.; Caroli, M.; et al. Mass spectrometry-based assay for the molecular diagnosis of glioma: Concomitant detection of chromosome 1p/19q codeletion, and IDH1, IDH2, and TERT mutation status. Oncotarget 2017, 8, 57134–57148. [Google Scholar] [CrossRef][Green Version]
- Parsons, B.L. Many different tumor types have polyclonal tumor origin: Evidence and implications. Mutat. Res. 2008, 659, 232–247. [Google Scholar] [CrossRef]

| # | Normalized Allelic Frequency | Clonality (Corrected Ratio) | ||
|---|---|---|---|---|
| BRAF V600E | TERT c.-124C>T | N-RAS Q61K/R/ H-RAS Q61R | ||
| 1 | 50 | monoclonal (0.3) | ||
| 2 | 50 | monoclonal (0.08) | ||
| 3 | 50 | monoclonal (0.13) | ||
| 4 | 55 | monoclonal (0.24) | ||
| 5 | 45 | monoclonal (16.14) | ||
| 6 | 48 | monoclonal (5) | ||
| 7 | 45 | monoclonal (0.29) | ||
| 8 | 49 | monoclonal (9.13) | ||
| 9 | 49 | monoclonal (8.16) | ||
| 10 | 52 | monoclonal (3.1) | ||
| 11 | 46 | monoclonal (4.31) | ||
| 12 | 53 | monoclonal (0.08) | ||
| 13 | 47 | monoclonal (4.2), normal tissue skewed (3.5) | ||
| 14 | 47 | polyclonal (0.68) | ||
| 15 | 53 | polyclonal (0.6) | ||
| 16 | 48 | polyclonal (1.27) | ||
| 17 | 15 | monoclonal (12.3), normal tissue skewed (3.2) | ||
| 18 | 13 | monoclonal (5.18), normal tissue skewed (18.8) | ||
| 19 | 31 | monoclonal (8.33), normal tissue skewed (3) | ||
| 20 | 30 | polyclonal (1.33) | ||
| 21 | 44 | polyclonal (0.73) | ||
| 22 | 43 | polyclonal (1.15) | ||
| 23 | 12 | polyclonal (0.8) | ||
| 24 | 17 | polyclonal (1.6) | ||
| 25 | 36 | polyclonal (0.91) | ||
| 26 | 36 | polyclonal (0.62) | ||
| 27 | 35 | polyclonal (1.66) | ||
| 28 | 29 | polyclonal (2.73) | ||
| 29 | 40 | polyclonal (1.18) | ||
| 30 | 37 | polyclonal (0.91) | ||
| 31 | 49 | 50 | monoclonal (8.11) | |
| 32 | 53 | 50 | monoclonal (0.16) | |
| 33 | 49 | 49 | monoclonal (0.16) | |
| 34 | 46 | 55 | monoclonal (3.29) | |
| 35 | 45 | 53 | monoclonal (18.8), normal tissue skewed (66.7) | |
| 36 | 50 | 80 | polyclonal (2.38) | |
| 37 | 14 | 40 | polyclonal (1.32) | |
| 38 | 23 | 39 | polyclonal (0.76) | |
| 39 | 35 | 20 | polyclonal (1.17) | |
| 40 | 30 | 34 | polyclonal (1.14) | |
| 41 | 53 | 21 | polyclonal (0.78) | |
| 42 | 15 | 50 | polyclonal (1.34) | |
| Features (%) | Monoclonal PTCs (n = 16) | Polyclonal PTCs (n = 21) | p Value |
|---|---|---|---|
| BRAFV600E | 12/4 (75/25%) | 20/1 (95/5%) | 0.074 |
| H-/N-RAS codon 61 | 3/13 (19/81%) | 1/20 (5/95%) | 0.174 |
| TERT c.-124C>T | 5/11 (31/69%) | 7/14 (33/67%) | 0.893 |
| BRAFV600E + TERT c.-124C>T | 4/12 (25/75%) | 4/17 (19/81%) | 0.663 |
| Median age at diagnosis, years (range) | 45.5 (15–77) | 56 (24–87) | 0.747 |
| Pre-surgical diagnosis, yes/indeterminate/no | 14/1/1 (88/6/6) | 21/0/0 (100/0/0%) | 0.249 |
| Size, mm (mean) | 25.4 (8–55) | 23.7 (8–44) | 0.268 |
| Extrathyroidal extension, yes/no | 4/12 (25/75) | 12/9 (57/43%) | 0.053 |
| Multifocality, yes/no | 7/9 (44/56) | 8/13 (38/62) | 0.732 |
| Histological variants of PTC, classical/follicular/other | 12/3/1 (75/19/6) | 18/0/3 (86/0/14%) | 0.099 |
| T1/T2/T3/T4 | 9/2/4/1 (56/12/25/7%) | 9/6/3/3 (43/29/14/14%) | 0.472 |
| N1/N0/NX | 3/7/6 (18/44/38%) 3/7 * (30/70%) | 11/5/5 (52/24/24%) 11/5 * (69/31%) | 0.110 0.053 |
| M1/M0 | 1/15 (6/94%) | 0/21 (0/100%) | 0.251 |
| AJCC Stage, I/II/III/IV | 14/0/2/0 (88/0/12/0%) | 14/4/2/1 (67/19/10/4%) | 0.220 |
| Radioiodine Ablation, yes/no | 6/10 (38/42%) | 16/5 (76/24%) | 0.019 |
| Disease outcome, persistence/remission | 3/13 (19/81%) | 5/16 (24/76%) | 0.714 |
| Disease-specific mortality, yes/no | 1/15 (6/94%) | 0/21 (0/100%) | 0.251 |
| Follow-up, months (mean, range) | 62 (6–217) | 54 (6–225) | 0.145 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muzza, M.; Pogliaghi, G.; Persani, L.; Fugazzola, L.; Colombo, C. Combined Mutational and Clonality Analyses Support the Existence of Intra-Tumor Heterogeneity in Papillary Thyroid Cancer. J. Clin. Med. 2021, 10, 2645. https://doi.org/10.3390/jcm10122645
Muzza M, Pogliaghi G, Persani L, Fugazzola L, Colombo C. Combined Mutational and Clonality Analyses Support the Existence of Intra-Tumor Heterogeneity in Papillary Thyroid Cancer. Journal of Clinical Medicine. 2021; 10(12):2645. https://doi.org/10.3390/jcm10122645
Chicago/Turabian StyleMuzza, Marina, Gabriele Pogliaghi, Luca Persani, Laura Fugazzola, and Carla Colombo. 2021. "Combined Mutational and Clonality Analyses Support the Existence of Intra-Tumor Heterogeneity in Papillary Thyroid Cancer" Journal of Clinical Medicine 10, no. 12: 2645. https://doi.org/10.3390/jcm10122645
APA StyleMuzza, M., Pogliaghi, G., Persani, L., Fugazzola, L., & Colombo, C. (2021). Combined Mutational and Clonality Analyses Support the Existence of Intra-Tumor Heterogeneity in Papillary Thyroid Cancer. Journal of Clinical Medicine, 10(12), 2645. https://doi.org/10.3390/jcm10122645






