Assembly of Cell-Free Synthesized Ion Channel Molecules in Artificial Lipid Bilayer Observed by Atomic Force Microscopy
Abstract
:1. Introduction
2. Materials and Methods
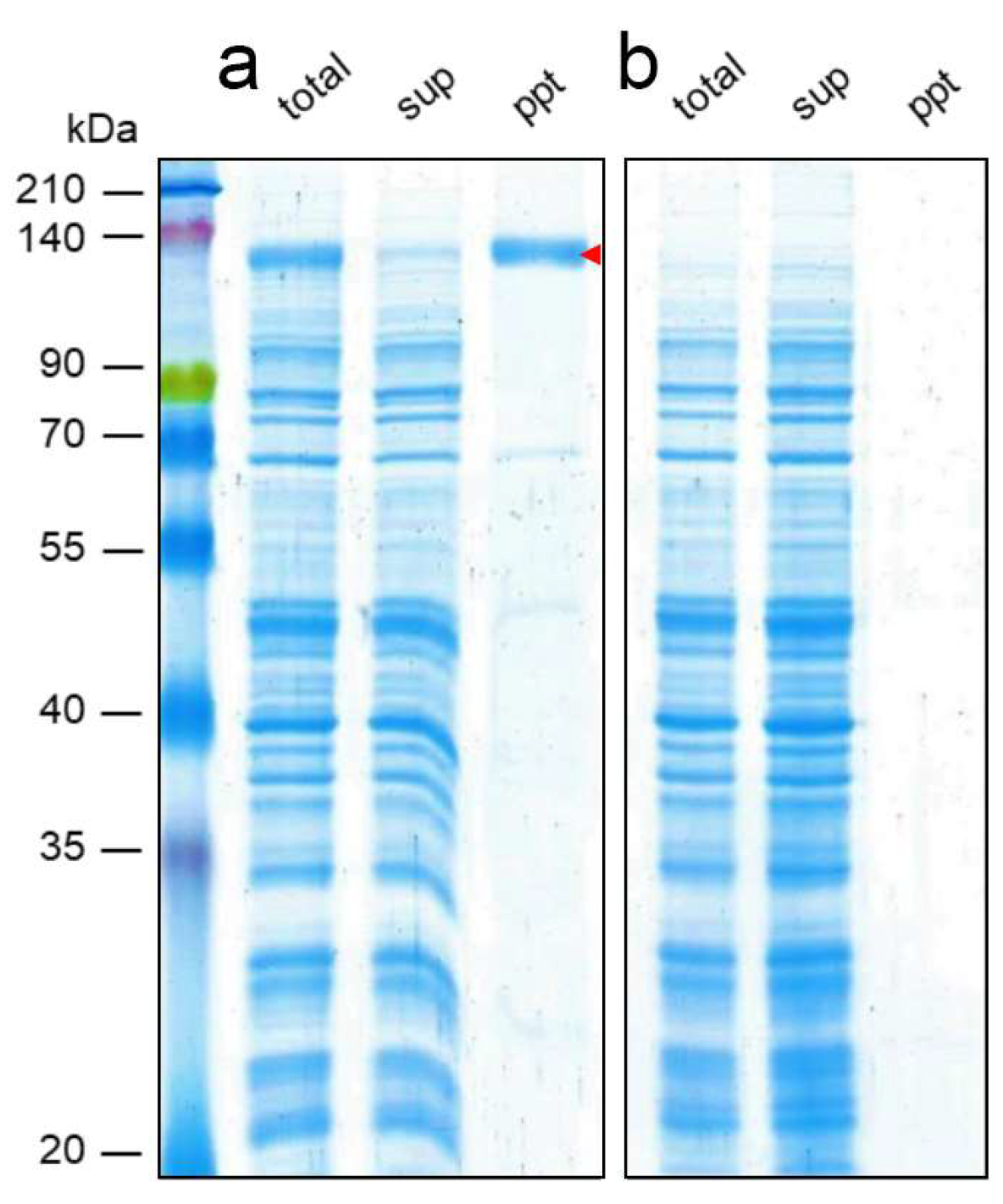
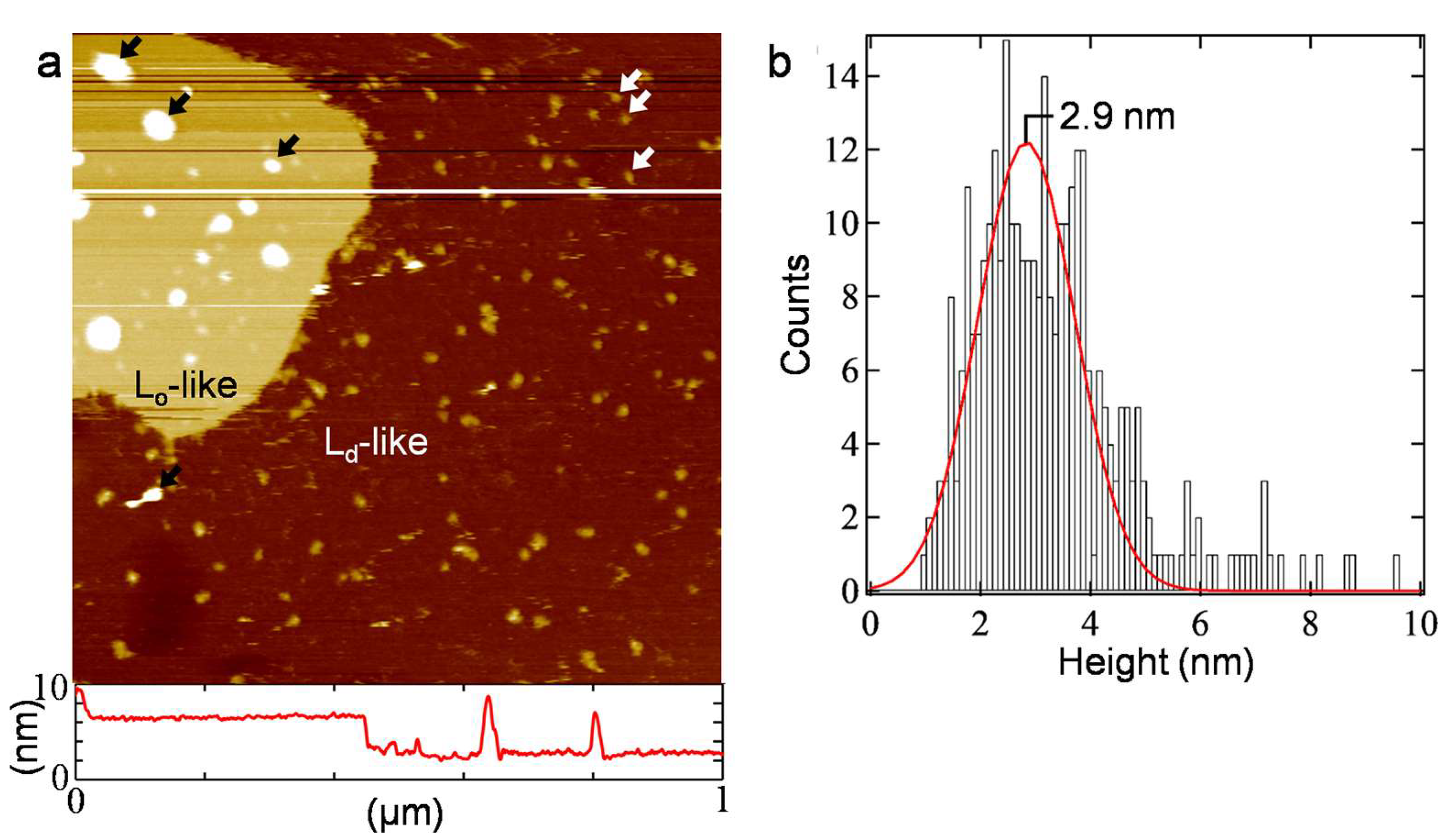
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spira, F.; Mueller, N.S.; Beck, G.; von Olshausen, P.; Beig, J.; Wedlich-Söldner, R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012, 14, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Veatch, S.L.; Rogers, N.; Decker, A.; Shelby, S.A. The plasma membrane as an adaptable fluid mosaic. Biochim. Biophys. Acta Biomembr. 2023, 1865, 184114. [Google Scholar] [CrossRef]
- Lin, Q.; London, E. Altering hydrophobic sequence lengths shows that hydrophobic mismatch controls affinity for ordered lipid domains (rafts) in the multitransmembrane strand protein perfringolysin O. J. Biol. Chem. 2013, 288, 1340–1352. [Google Scholar] [CrossRef] [PubMed]
- Lingwood, D.; Simons, K. Lipid Rafts As a Membrane-Organizing Principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Hakobyan, D.; Heuer, A. Key Molecular Requirements for Raft Formation in Lipid/Cholesterol Membranes. PLoS ONE 2014, 9, e87369. [Google Scholar] [CrossRef]
- Verkleij, A.J.; Post, J.A. Membrane phospholipid asymmetry and signal transduction. J. Membr. Biol. 2000, 178, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006, 5, 993–996. [Google Scholar] [CrossRef]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef]
- Segall, M.D.; Barber, C. Addressing toxicity risk when designing and selecting compounds in early drug discovery. Drug Discov. Today 2014, 19, 688–693. [Google Scholar] [CrossRef]
- Sofińska, K.; Lupa, D.; Chachaj-Brekiesz, A.; Czaja, M.; Kobierski, J.; Seweryn, S.; Skirlińska-Nosek, K.; Szymonski, M.; Wilkosz, N.; Wnętrzak, A.; et al. Revealing local molecular distribution, orientation, phase separation, and formation of domains in artificial lipid layers: Towards comprehensive characterization of biological membranes. Adv. Colloid Interface Sci. 2022, 301, 102614. [Google Scholar] [CrossRef]
- Hirano-Iwata, A.; Niwano, M.; Sugawara, M. The design of molecular sensing interfaces with lipid-bilayer assemblies. TrAC Trends Anal. Chem. 2008, 27, 512–520. [Google Scholar] [CrossRef]
- Trudeau, M.C.; Warmke, J.W.; Ganetzky, B.; Robertson, G.A. HERG, a Human Inward Rectifier in the Voltage-Gated Potassium Channel Family. Science 1995, 269, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Danker, T.; Möller, C. Early identification of hERG liability in drug discovery programs by automated patch clamp. Front. Pharmacol. 2014, 5, 203. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, J.I.; Perry, M.D.; Perrin, M.J.; Mann, S.A.; Ke, Y.; Hill, A.P. hERG K(+) channels: Structure, function, and clinical significance. Physiol. Rev. 2012, 92, 1393–1478. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.C.; Tristani-Firouzi, M. hERG potassium channels and cardiac arrhythmia. Nature 2006, 440, 463–469. [Google Scholar] [CrossRef]
- Warmke, J.W.; Ganetzky, B.; Hamilton, B.; Meyerowitz, E. A family of potassium channel genes related to eag in Drosophila and mammals. Proc. Natl. Acad. Sci. USA 1994, 91, 3438–3442. [Google Scholar] [CrossRef]
- Stevens, J.L.; Baker, T.K. The future of drug safety testing: Expanding the view and narrowing the focus. Drug Discov. Today 2009, 14, 162–167. [Google Scholar] [CrossRef]
- Wang, W.; MacKinnon, R. Cryo-EM Structure of the Open Human Ether-à-go-go-Related K+Channel hERG. Cell 2017, 169, 422–430. [Google Scholar] [CrossRef]
- Manzer, Z.A.; Selivanovitch, E.; Ostwalt, A.R.; Daniel, S. Membrane protein synthesis: No cells required. Trends Biochem. Sci. 2023, 48, 642–654. [Google Scholar] [CrossRef]
- Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A user’s guide to cell-free protein synthesis. Methods Protoc. 2019, 2, 24. [Google Scholar] [CrossRef]
- Bernhard, F.; Tozawa, Y. Cell-free expression-making a mark. Curr. Opin. Struct. Biol. 2013, 23, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Kuruma, Y.; Ueda, T. The PURE system for the cell-free synthesis of membrane proteins. Nat. Protoc. 2015, 10, 1328–1344. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.M.; Swartz, J.R. Efficient Production of a Bioactive, Multiple Disulfide-Bonded Protein Using Modified Extracts of Escherichia coli. Biotechnol. Bioeng. 2004, 85, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Madin, K.; Sawasaki, T.; Ogasawara, T.; Endo, Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: Plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. USA 2000, 97, 559–564. [Google Scholar] [CrossRef]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef]
- Tadaki, D.; Yamaura, D.; Araki, S.; Yoshida, M.; Arata, K.; Ohori, T.; Ishibashi, K.; Kato, M.; Ma, T.; Miyata, R.; et al. Mechanically stable solvent-free lipid bilayers in nano- and micro-tapered apertures for reconstitution of cell-free synthesized hERG channels. Sci. Rep. 2017, 7, 17736. [Google Scholar] [CrossRef]
- Komiya, M.; Kato, M.; Tadaki, D.; Ma, T.; Yamamoto, H.; Tero, R.; Tozawa, Y.; Niwano, M.; Hirano-Iwata, A. Advances in Artificial Cell Membrane Systems as a Platform for Reconstituting Ion Channels. Chem. Rec. 2020, 20, 730–742. [Google Scholar] [CrossRef]
- Hirano-Iwata, A.; Ishinari, Y.; Yoshida, M.; Araki, S.; Tadaki, D.; Miyata, R.; Ishibashi, K.; Yamamoto, H.; Kimura, Y.; Niwano, M. Reconstitution of Human Ion Channels into Solvent-free Lipid Bilayers Enhanced by Centrifugal Forces. Biophys. J. 2016, 110, 2207–2215. [Google Scholar] [CrossRef]
- Oshima, A.; Hirano-Iwata, A.; Mozumi, H.; Ishinari, Y.; Kimura, Y.; Niwano, M. Reconstitution of human ether-a-go-go-related gene channels in microfabricated silicon chips. Anal. Chem. 2013, 85, 4363–4369. [Google Scholar] [CrossRef]
- Tero, R.; Fukumoto, K.; Motegi, T.; Yoshida, M.; Niwano, M.; Hirano-Iwata, A. Formation of Cell Membrane Component Domains in Artificial Lipid Bilayer. Sci. Rep. 2017, 7, 17905. [Google Scholar] [CrossRef]
- Goh, M.W.S.; Hirano-Iwata, A.; Niwano, M.; Tero, R. Proteoliposome fusion to artificial lipid bilayer promoted by domains of polyunsaturated phosphatidylethanolamine. Jpn. J. Appl. Phys. 2019, 58, SIIB13. [Google Scholar] [CrossRef]
- Goh, M.W.S.; Tero, R. Cholesterol-induced microdomain formation in lipid bilayer membranes consisting of completely miscible lipids. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183626. [Google Scholar] [CrossRef] [PubMed]
- Goh, M.W.S.; Tero, R. Non-raft submicron domain formation in cholesterol-containing lipid bilayers induced by polyunsaturated phosphatidylethanolamine. Colloids Surf. B Biointerfaces 2022, 210, 112235. [Google Scholar] [CrossRef] [PubMed]
- Castellana, E.T.; Cremer, P.S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006, 61, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.P.; Bérat, R.; Brisson, A.R. Formation of Solid-Supported Lipid Bilayers: An Integrated View. Langmuir 2006, 22, 3497–3505. [Google Scholar] [CrossRef]
- Katsaras, J.; Kučerka, N.; Nieh, M.-P. Structure from substrate supported lipid bilayers (Review). Biointerphases 2008, 3, FB55–FB63. [Google Scholar] [CrossRef]
- Morigaki, K.; Tanimoto, Y. Evolution and development of model membranes for physicochemical and functional studies of the membrane lateral heterogeneity. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2012–2017. [Google Scholar] [CrossRef]
- Miyata, R.; Tadaki, D.; Yamaura, D.; Araki, S.; Sato, M.; Komiya, M.; Ma, T.; Yamamoto, H.; Niwano, M.; Hirano-Iwata, A. Parallel Recordings of Transmembrane hERG Channel Currents Based on Solvent-Free Lipid Bilayer Microarray. Micromachines 2021, 12, 98. [Google Scholar] [CrossRef]
- Uchihashi, T.; Ganser, C. Recent advances in bioimaging with high-speed atomic force microscopy. Biophys. Rev. 2020, 12, 363–369. [Google Scholar] [CrossRef]
- Fotiadis, D. Atomic force microscopy for the study of membrane proteins. Curr. Opin. Biotechnol. 2012, 23, 510–515. [Google Scholar] [CrossRef]
- Sumino, A.; Yamamoto, D.; Iwamoto, M.; Dewa, T.; Oiki, S. Gating-Associated Clustering–Dispersion Dynamics of the KcsA Potassium Channel in a Lipid Membrane. J. Phys. Chem. Lett. 2014, 5, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Ido, S.; Kobayashi, K.; Oyabu, N.; Hirata, Y.; Matsushige, K.; Yamada, H. Structured Water Molecules on Membrane Proteins Resolved by Atomic Force Microscopy. Nano Lett. 2022, 22, 2391–2397. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, Y.; Sumitomo, K.; Furukawa, K.; Miyashita, H.; Tamba, Y.; Kasai, N.; Nakashima, H.; Torimitsu, K. Visualization of Single Membrane Protein Structure in Stretched Lipid Bilayer Suspended over Nanowells. Appl. Phys. Express 2010, 3, 027002. [Google Scholar] [CrossRef]
- Suzuki, K.; Inoue, H.; Matsuoka, S.; Tero, R.; Hirano-Iwata, A.; Tozawa, Y. Establishment of a cell-free translation system from rice callus extracts. Biosci. Biotechnol. Biochem. 2020, 84, 2028–2036. [Google Scholar] [CrossRef]
- Leonenko, Z.; Carnini, A.; Cramb, D. Supported planar bilayer formation by vesicle fusion: The interaction of phospholipid vesicles with surfaces and the effect of gramicidin on bilayer properties using atomic force microscopy. Biochim. Biophys. Acta Biomembr. 2000, 1509, 131–147. [Google Scholar] [CrossRef]
- Attwood, S.J.; Choi, Y.; Leonenko, Z. Preparation of DOPC and DPPC supported planar lipid bilayers for atomic force microscopy and atomic force spectroscopy. Int. J. Mol. Sci. 2013, 14, 3514–3539. [Google Scholar] [CrossRef]
- Thoma, J.; Burmann, B.M. Fake It ‘Till You Make It—The Pursuit of Suitable Membrane Mimetics for Membrane Protein Biophysics. Int. J. Mol. Sci. 2021, 22, 50. [Google Scholar] [CrossRef]
- Fukui, K.; Sugiyama, S.; Iwasawa, Y. Atomic force microscopic study on thermal and UV-irradiative formation and control of Au nano-particles on TiO2(110) from Au(PPh3)(NO3). Phys. Chem. Chem. Phys. 2001, 3, 3871–3877. [Google Scholar] [CrossRef]
- Li, K.; Jiang, Q.; Bai, X.; Yang, Y.-F.; Ruan, M.-Y.; Cai, S.-Q. Tetrameric Assembly of K+ Channels Requires ER-Located Chaperone Proteins. Mol. Cell 2017, 65, 52–65. [Google Scholar] [CrossRef]
- Chanturiya, A.; Chernomordik, L.V.; Zimmerberg, J. Flickering fusion pores comparable with initial exocytotic pores occur in protein-free phospholipid bilayers. Proc. Natl. Acad. Sci. USA 1997, 94, 14423–14428. [Google Scholar] [CrossRef]
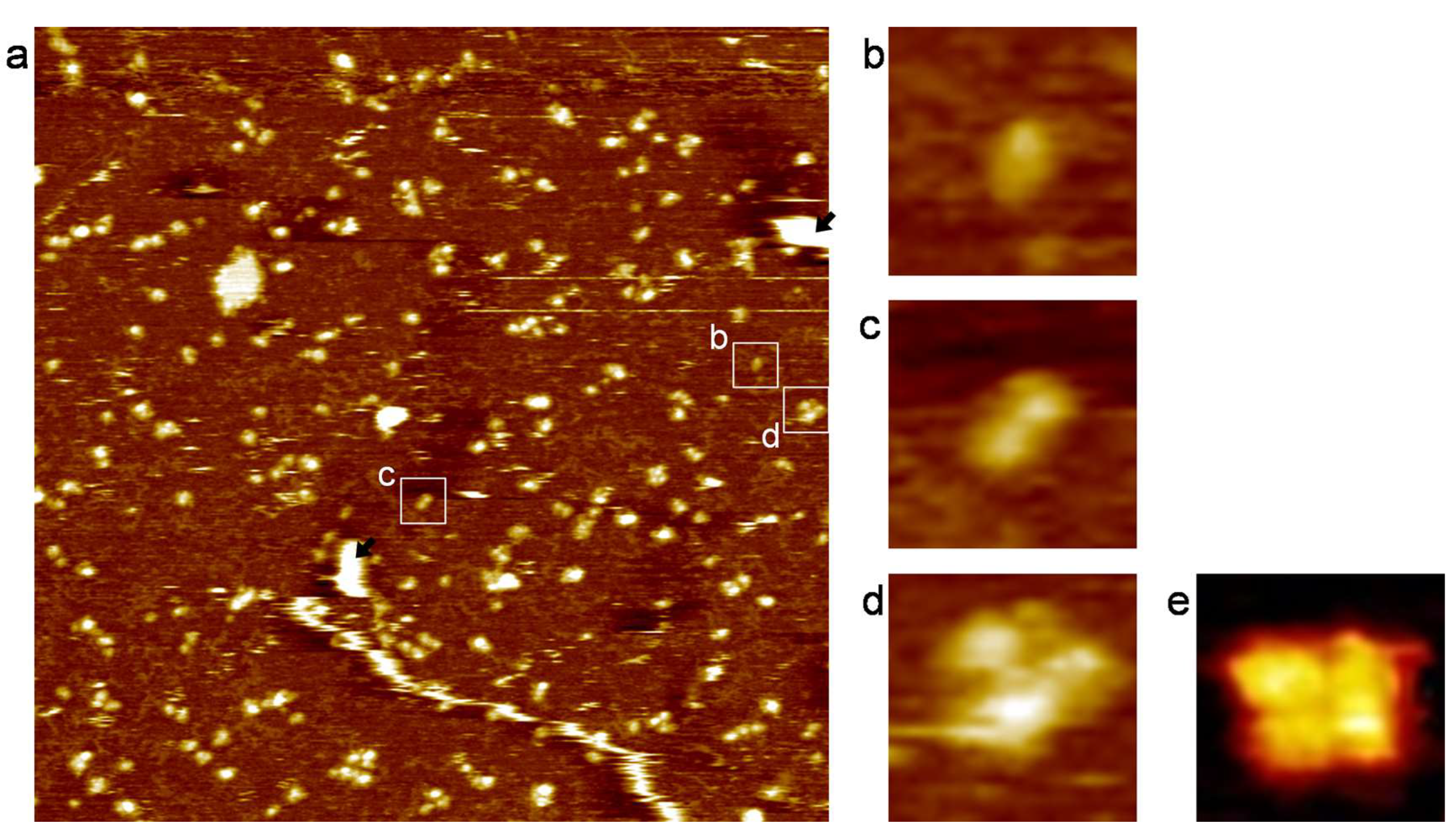

| Monomer | Dimer | Tetramer | |
|---|---|---|---|
| Average area (nm2) | 190 ± 5 | 285 ± 11 | 470 ± 39 |
| Calculated value (nm2) | 180 | 270 | 400 |
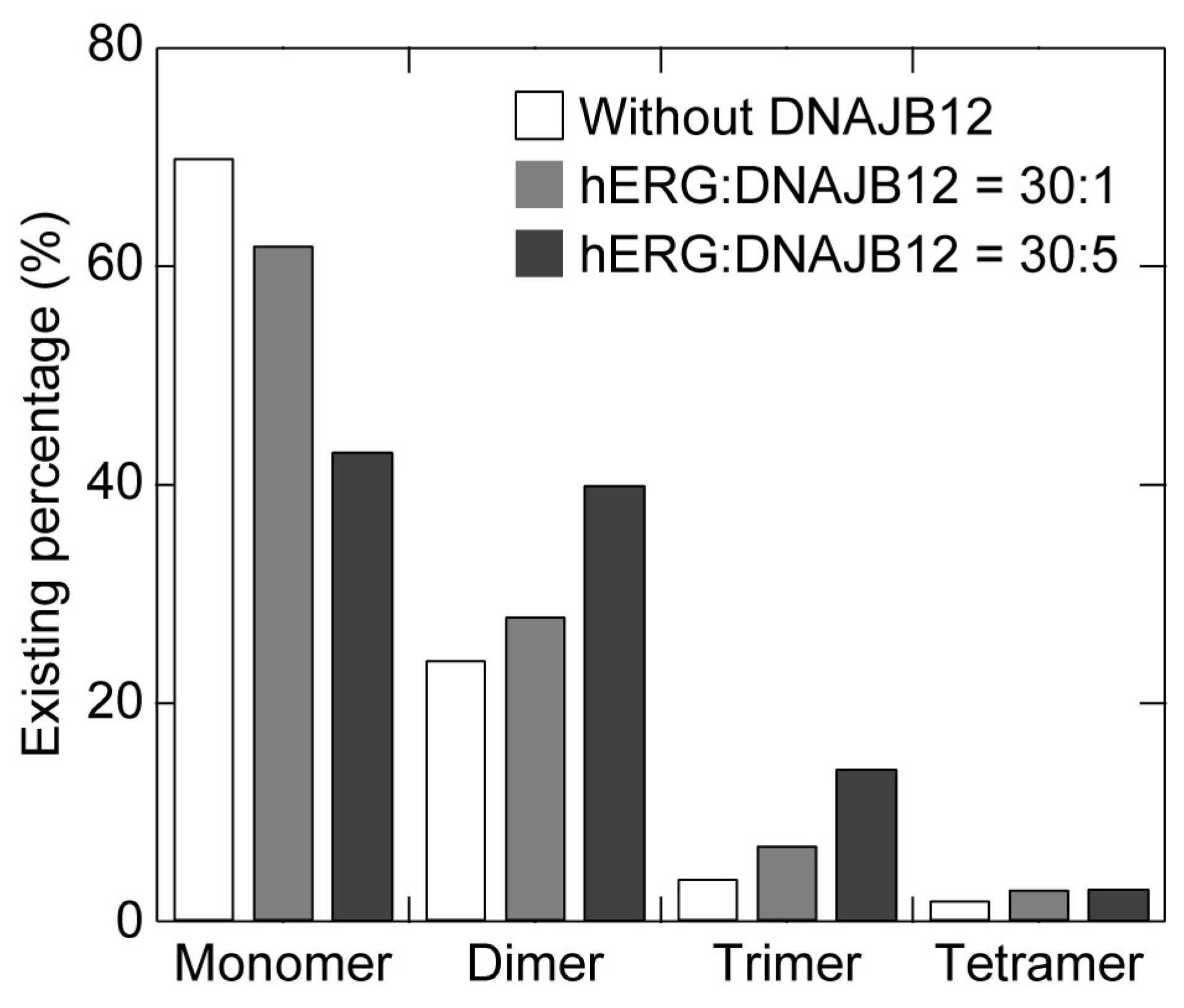
| Expression Ratio (hERG:DNAJB12) | Monomer | Dimer | Trimer | Tetramer |
|---|---|---|---|---|
| 30:0 * | 70 | 24 | 4 | 2 |
| 30:1 | 62 | 28 | 7 | 3 |
| 30:5 | 43 | 40 | 14 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goh, M.W.S.; Tozawa, Y.; Tero, R. Assembly of Cell-Free Synthesized Ion Channel Molecules in Artificial Lipid Bilayer Observed by Atomic Force Microscopy. Membranes 2023, 13, 854. https://doi.org/10.3390/membranes13110854
Goh MWS, Tozawa Y, Tero R. Assembly of Cell-Free Synthesized Ion Channel Molecules in Artificial Lipid Bilayer Observed by Atomic Force Microscopy. Membranes. 2023; 13(11):854. https://doi.org/10.3390/membranes13110854
Chicago/Turabian StyleGoh, Melvin Wei Shern, Yuzuru Tozawa, and Ryugo Tero. 2023. "Assembly of Cell-Free Synthesized Ion Channel Molecules in Artificial Lipid Bilayer Observed by Atomic Force Microscopy" Membranes 13, no. 11: 854. https://doi.org/10.3390/membranes13110854
APA StyleGoh, M. W. S., Tozawa, Y., & Tero, R. (2023). Assembly of Cell-Free Synthesized Ion Channel Molecules in Artificial Lipid Bilayer Observed by Atomic Force Microscopy. Membranes, 13(11), 854. https://doi.org/10.3390/membranes13110854







