A Self-Controlled and Self-Healing Model of Bacterial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Computational Methdology and Simulation
- Floating objects are small shapeless atomic objects of positive volume floating freely within the environment and occupying a specific position in space at every moment (akin to a center of gravity or centroid). They can be carried through tiles via protion channels and participate in mutual reactions with other types of objects, as specified by the production rules.
- Tiles are obstructive (like solid) objects of dimension 1 or 2. Each tile has its own pre-defined shape and size. Unlike other models of self-assembly systems, such as the aTAM model [23,24], tiles are not present implicitly in arbitrarily many copies, but can only be created via reactions involving floating objects. Selected points or segments on tiles called connectors are covered with certain types of glues. Another tile can stick to a connector by its own connector with a matching glue, where a match is defined by a glue relation pre-specified in the model. In this way, larger interconnected structures of tiles can gradually self-assemble into more complex objects.
- Protions are regulators or catalysts that (i) allow for certain reactions and (ii) transport floating objects through a tile (which may be part of a wall of a self-assembled closed compartment, akin to an ion channel in a biological membrane). They have a certain unchangeable position on a tile. Protions follow the notion in P systems as abstractions of proteins on membranes [20]. The connected tiles can be also disconnected and/or destroyed by rules under certain conditions requiring the presence of specific floating objects.
- Reaction rules can only be of four types: metabolic, creation, destruction, and division rules, as defined by the kind of result they may produce when applied to objects in the system.
- Brownian motion moves objects by a certain distance (as specified by a parameter in the system, the interaction radius) at every iteration, before applying any rules. It ensures that further interaction is eventually possible, even if no rule was applied at a given time.
2.2. Comparison to Other Approaches to Cell Growth Modeling
2.3. Implementation of Bacterial Growth Simulation
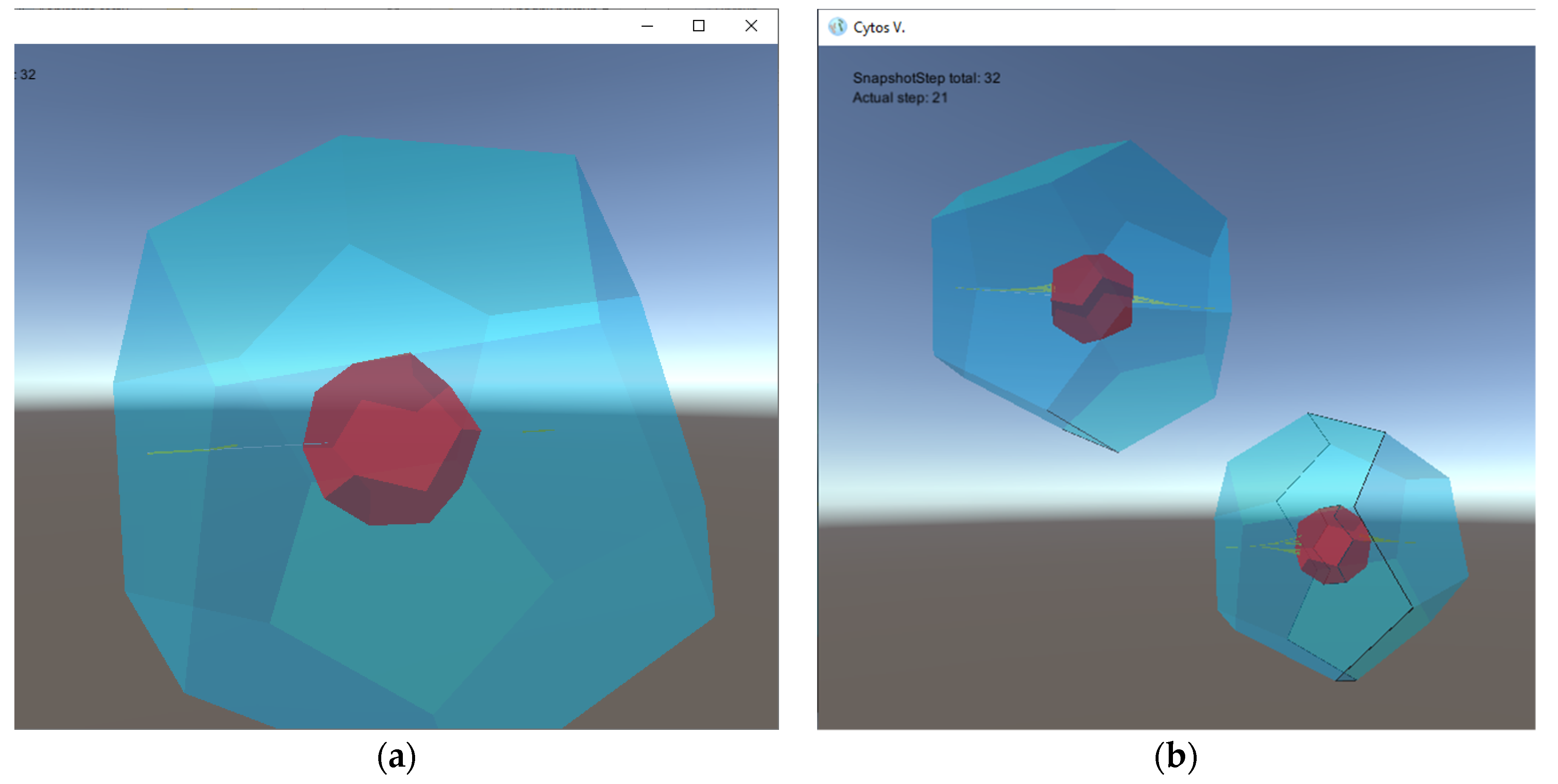
- four types of 2D tiles that build cell walls and septum components; for the sake of simplicity, these tiles are given polygonal shapes, and hence the resulting cells membranes are shaped as dodecahedra with octagonal sides, as illustrated in Figure 2 (other tiles can be used to produce assemblies with more complex shapes, if desirable);
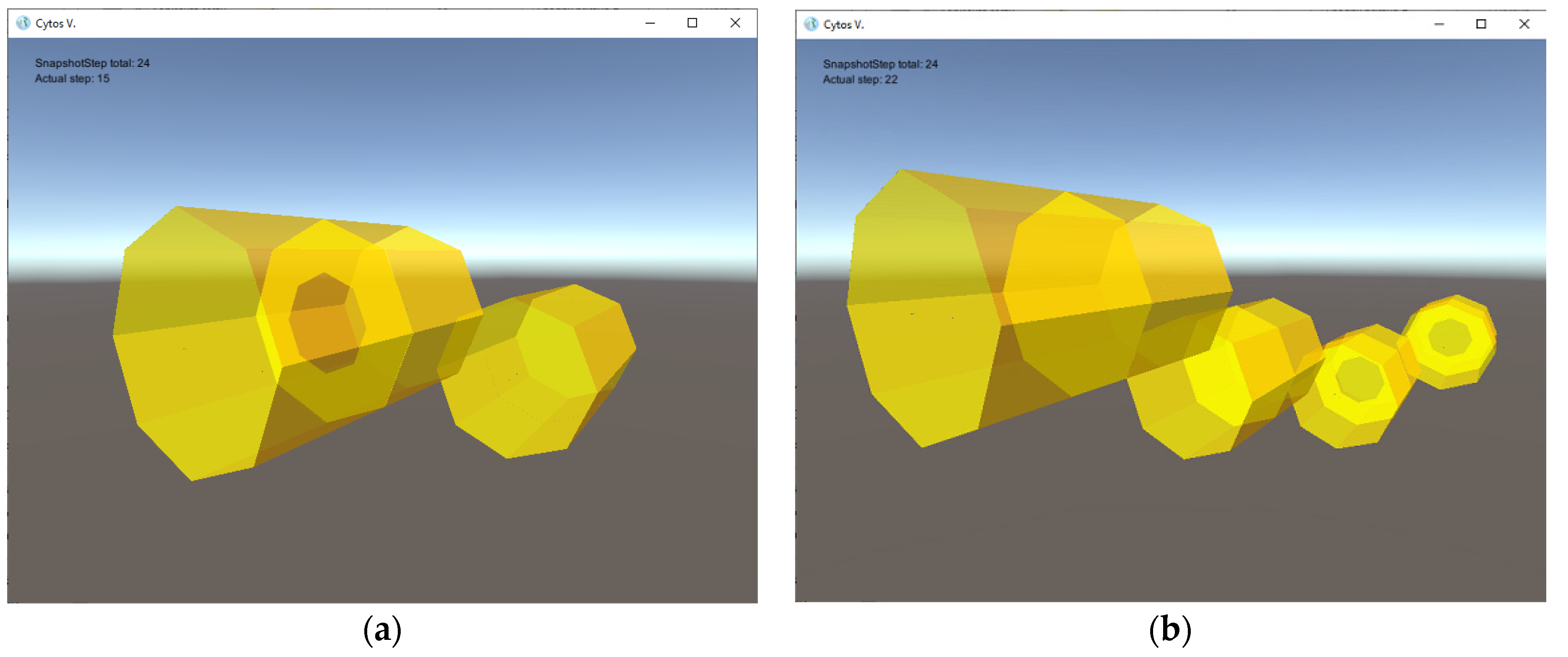
- three types of small auxiliary rod-shaped tiles controlling cell division;
- two types of floating objects: the first one (denoted by a) contained in the environment with a pre-defined concentration and serving as a nutrient, and the second one (denoted by s) serving as a signal molecule controlling the cell division process;
- one type of protion located in the tiles of the simulated cells controls the flow of nutrients from the environment into the interior of a simulated cell;
- Nine rules: one metabolic rule enables the transport of nutrients into cell interiors; six creation rules build tiles and consume nutrients; one destruction rule annihilates small auxiliary rods; and one division rule concludes the process of cell division when the septum formation is complete.
3. Results
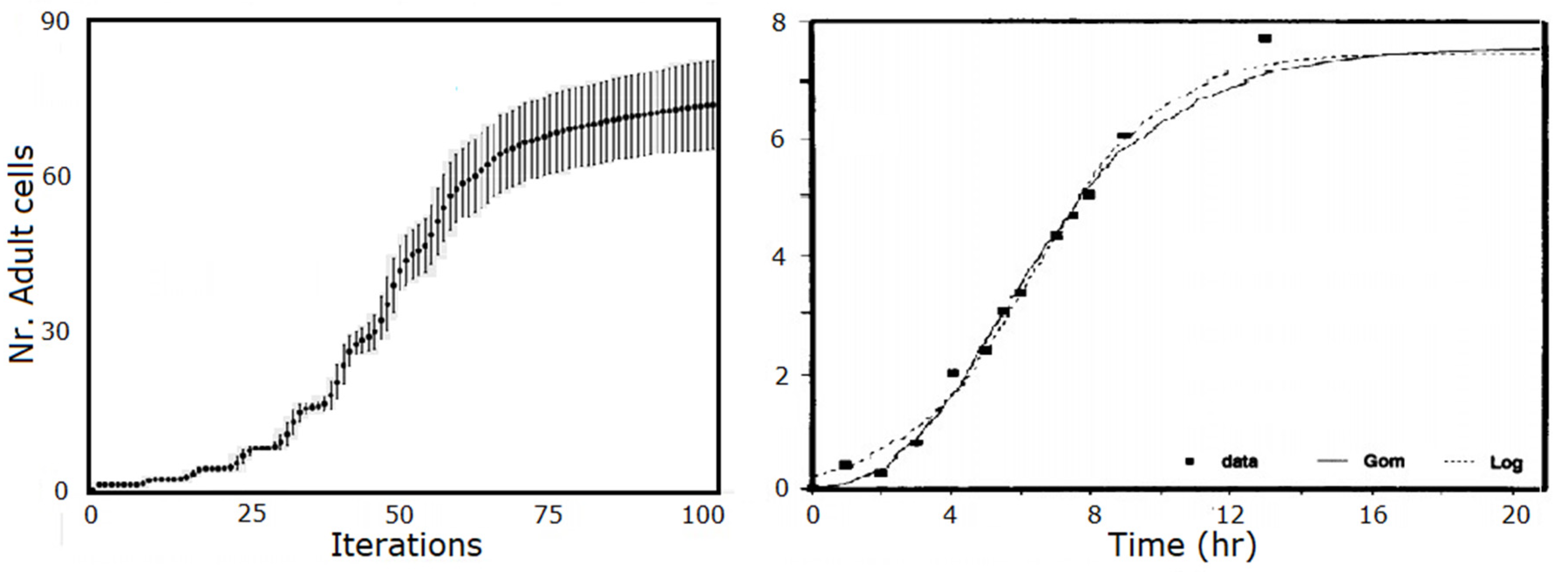
3.1. Bacterial Growth Profiles
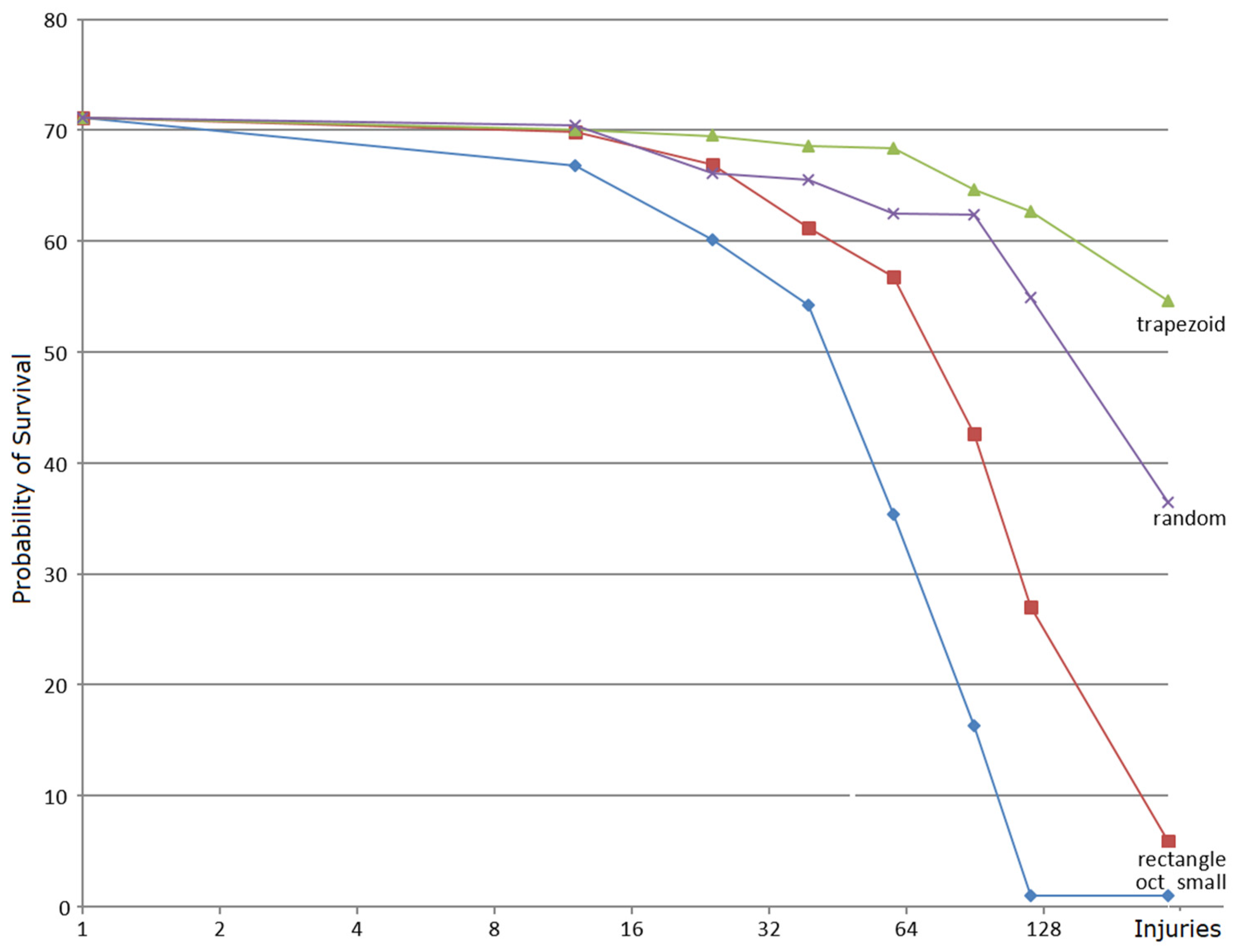
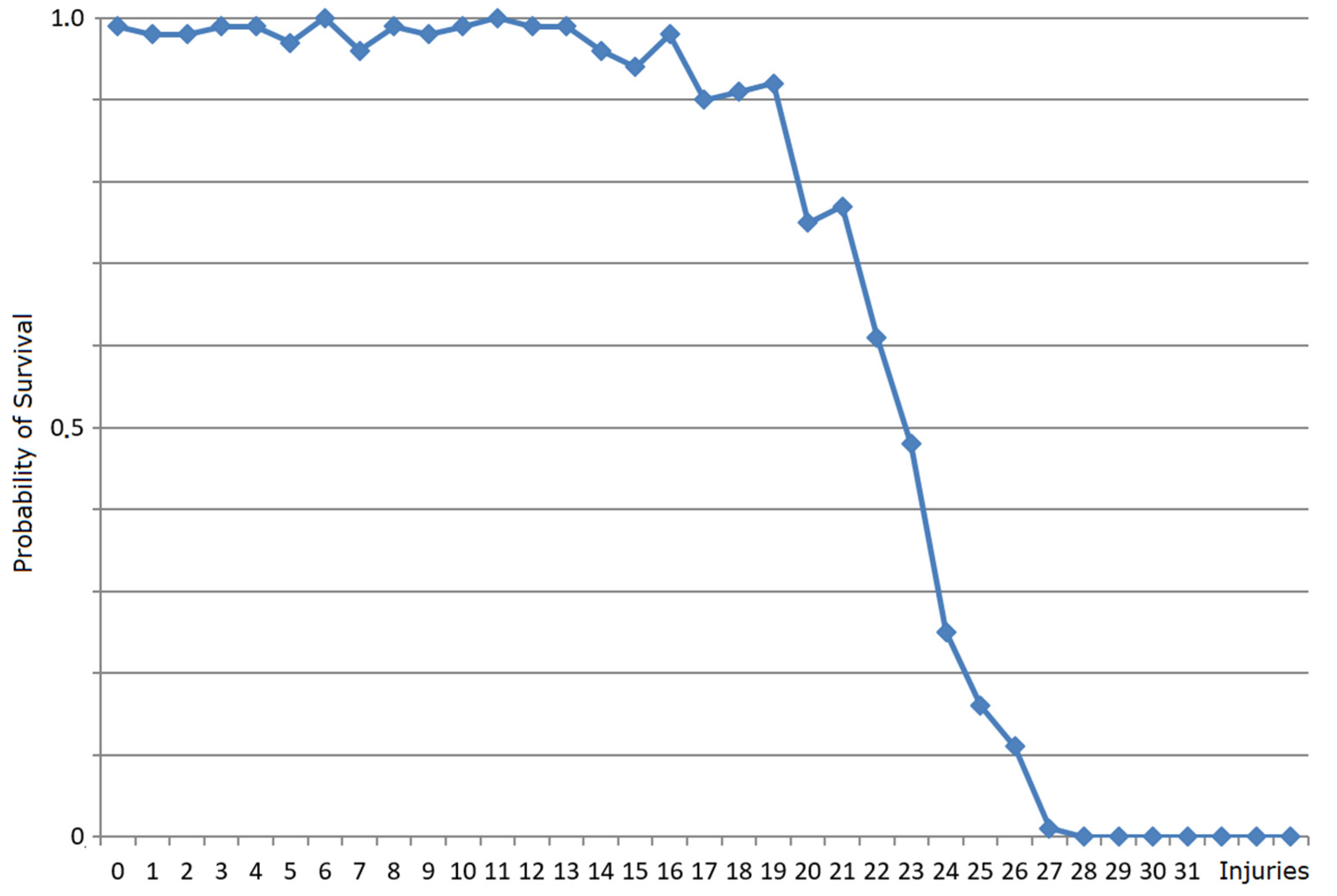
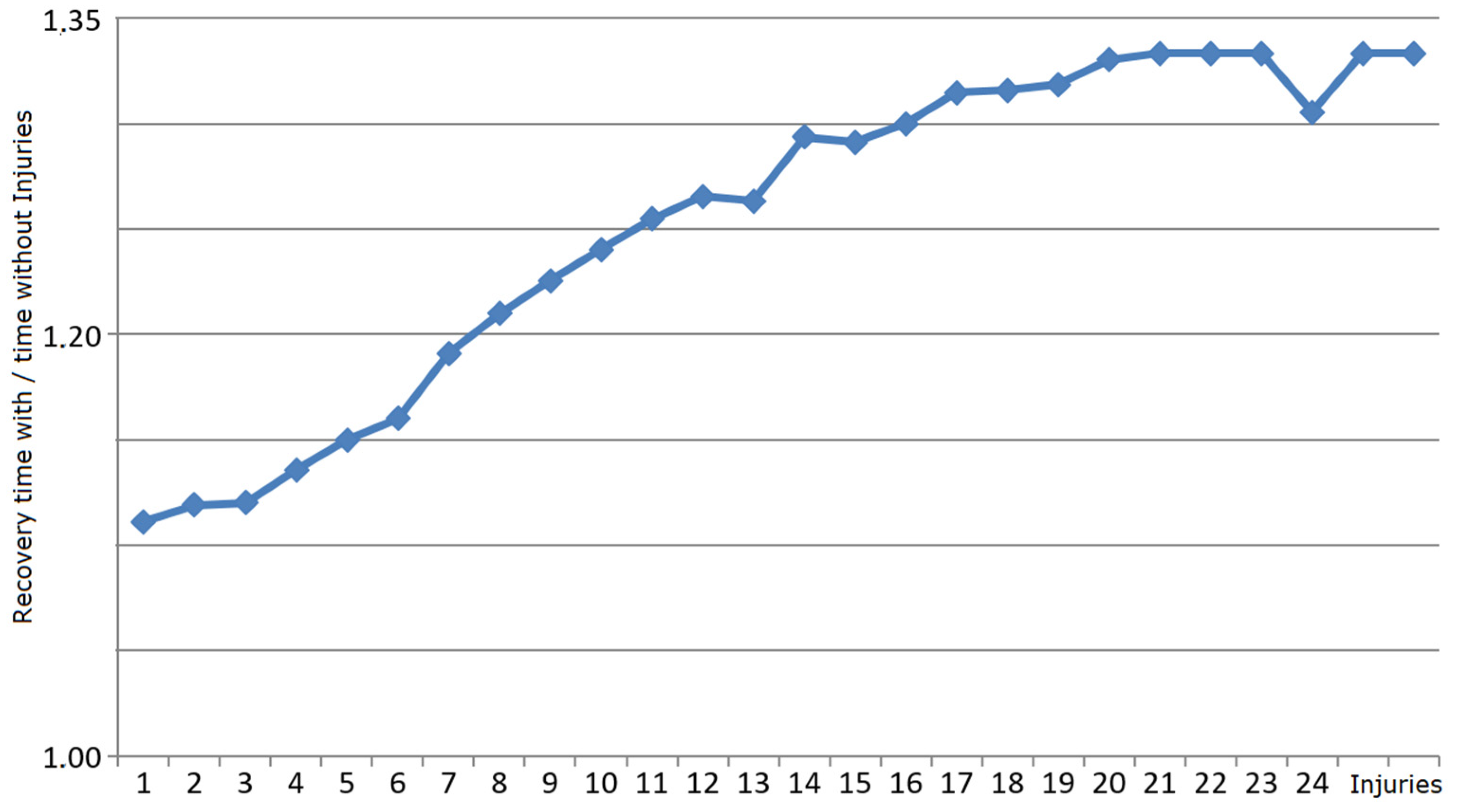
3.2. Robustness Properties of M Systems
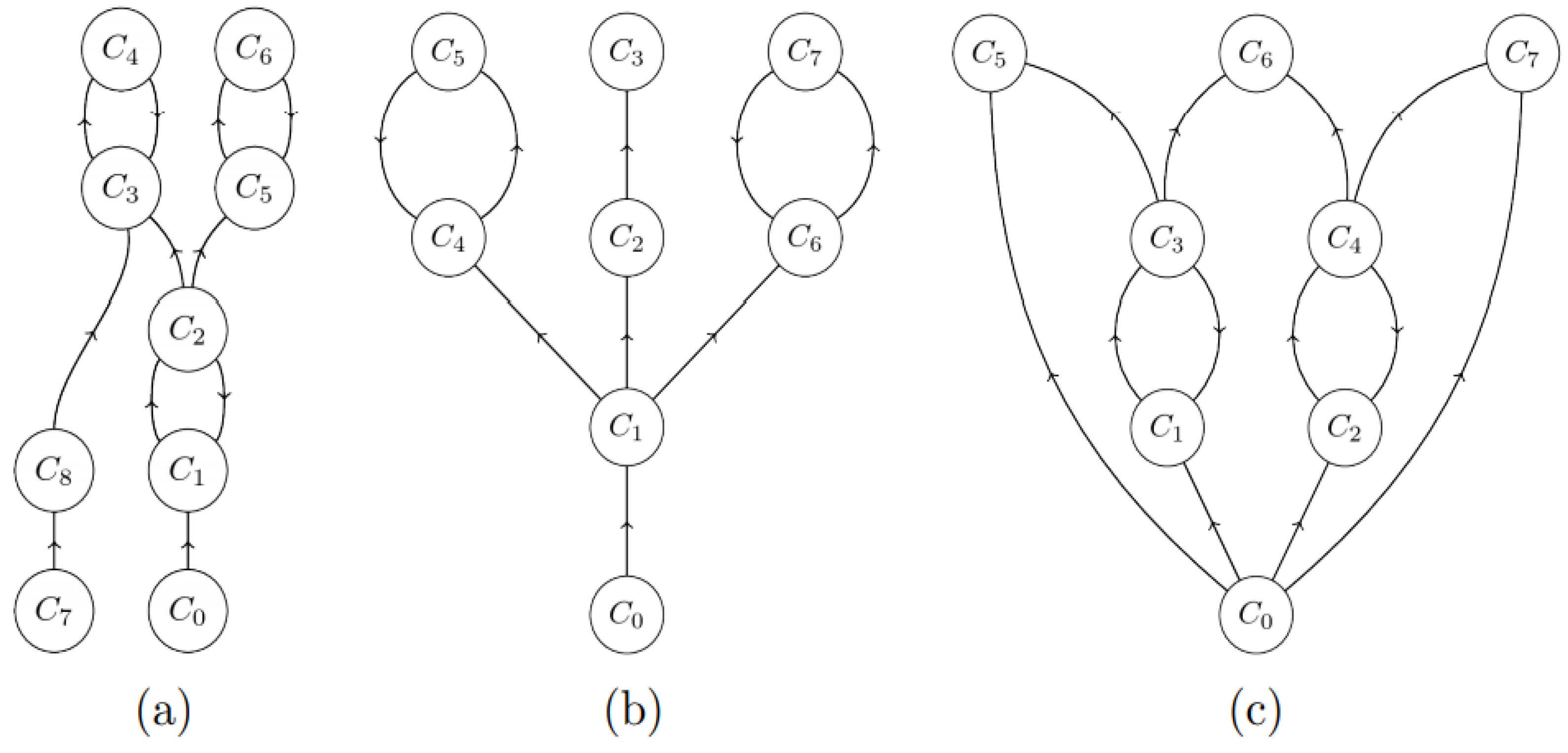
3.2.1. What Is Self-Healing?
- the transitive closure C^ of each directed cycle C belongs to an h-component, i.e., it consists of all nodes that are reachable from a node in the cycle following arcs in C using the successor relationship. For example, the closure of C3-C4 is itself in Figure 4a, while it is the entire graph in Figure 4c.
- each leaf node (one not containing any successors, such as C5, C6, C7 in Figure 4c) in M* belongs to an h-component;
- each node x whose transitive closure x^ intersects with a single h-component belongs to this h-component.
3.2.2. Self-Healing Properties of Mbac
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Turing, A.M. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B 1950, 237, 71–72. [Google Scholar]
- Watson, J.; Crick, F. A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Das, P.N.; Kumar, A.; Bairaji, N.; Chatterje, S. Restoring calcium homeostasis in diabetic cardiomyocites: An investigation through mathematical modeling. Mol. Biosyst. 2017, 13, 2672–2686. [Google Scholar] [CrossRef]
- Cui, J.; Kaandorp, J.A. Mathematical modeling of calcium homeostasis in yeast cells. Calcium 2006, 39, 337–348. [Google Scholar] [CrossRef]
- Alicea, B. The emergent connectome in in Caenorhabditis elegans embryogenesis. Biosystems 2018, 173, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Huang, B.; Kuang, Y.; Liu, G. Molecular dynamics simulations elucidate conformational selection and induced fit mechanisms in the binding of PD-1 and PD-L1. Mol. Biosyst. 2017, 13, 892–900. [Google Scholar] [CrossRef]
- Lecca, P.; Bagagiolo, F.; Scarpa, M. Hybrid deterministic/stochastic simulation of complex biochemical systems. Mol. Biosyst. 2017, 13, 2672–2686. [Google Scholar] [CrossRef]
- Tomita, M. Whole-cell simulation: A grand challenge of the 21st century. Trends Biotechnol. 2001, 19, 205–210. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Gordon, R.; Alicea, B.; Cherdantsev, V.C. Computational, theoretical, and experimental approaches to morphogenesis. Biosystems 2018, 173, 1–3. [Google Scholar] [CrossRef]
- Siregar, P.; Julen, N.; Hufnag, P.; Muttern, G. A General Framework dedicated to computational Morphogenesis Part I—Knowledge Representation and Architecture. Biosystems 2018, 173, 314–334. [Google Scholar] [CrossRef]
- Mazzarello, P. A unifying concept: The history of the cell theory. Nat. Cell Biol. 1999, 1, E13–E15. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; W.W Norton & Co.: New York, NY, USA, 2016. [Google Scholar]
- Eswara, P.J.; Ramamurthi, K.S. Bacterial Cell Division: Nonmodels Poised to Take the Spotlight. Annu. Rev. Microbiol. 2017, 71, 393–411. [Google Scholar] [CrossRef]
- Errington, J.; Wu, L.J. Cell Cycle Machinery in Bacillus subtilis. Biochemistry 2017, 84, 67–101. [Google Scholar]
- Li, S.; Brazhnik, P.; Sobral, B.; Tyson, J.J. A Quantitative Study of the Division Cycle of Caulobacter crescentus Stalked Cells. PLOS Comput. Biol. 2008, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; de la Torre, D.; Robertson, W.E.; Chin, J.W. Programmed chromosome fission and fusion enable precise large-scale genome rearrangements and assembly. Science 2019, 365, 922–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sosik, P.; Smolka, V.; Drastik, J.; Moore, T.; Garzon, M. Morphogenetic and homeostatic self-assembled systems. Lect. Notes Comput. Sci. 2017, 10240, 144–159. [Google Scholar]
- Sosik, P.; Smolka, V.; Drastik, J.; Bradik, J.; Garzon, M. On the robust power of morphogenetic systems for time bounded computation. Lect. Notes Comput. Sci. 2018, 10725, 270–292. [Google Scholar]
- Păun, G.; Rozenberg, G.; Salomaa, A. (Eds.) The Oxford Handbook of Membrane Computing; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Păun, A.; Popa, B. P systems with proteins on membranes. Fundam. Inform. 2006, 72, 467–483. [Google Scholar]
- Yuan, J.; Guo, D.; Zhang, G.; Paul, P.; Zhu, M.; Yang, Q. A resolution-free parallel algorithm for image edge detection within the framework of enzymatic numerical P systems. Molecules 2019, 24, 1235. [Google Scholar] [CrossRef] [Green Version]
- Smolka, V.; Drastík, J.; Garzon, M.; Sosík, P. Cytos: Morphogenetic (M) systems for modeling and experimentation. In Proceedings of the 20th International Conference of Membrane Computing (CMC20), Bibliostar, Râmnicu Vâlcea, Romania, 5–8 August 2019; pp. 475–496. [Google Scholar]
- Winfree, E. Algorithmic Self-Assembly of DNA. Ph.D. Dissertation, The California Institute of Technology, Pasadena, CA, USA, 1998. [Google Scholar]
- Krasnogor, N.; Gustafson, S.D.A.; Pelta, J.L.V. Systems Self-Assembly: Multidisciplinary Snapshots. In Studies in Multidisciplinarity 5; Elsevier Science: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Drastik, J. M System Models of Self-Reproduction of Eukaryotic Cells. 2020. Available online: https://www.youtube.com/watch?v=mvBLeUHCfW8 (accessed on 31 April 2022).
- Pirt, S.J. A kinetic study of the mode of growth of surface colonies of bacteria and fungi. Microbiology 1967, 47, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, J.A. Bacteria as multicellular organisms. Sci. Am. 1988, 258, 82–89. [Google Scholar] [CrossRef]
- Ben-Jacob, E.; Schochet, O.; Tenenbaum, A.; Cohen, I.; Czirok, A.; Vicsek, T. Generic modelling of cooperative growth patterns in bacterial colonies. Nature 1994, 368, 46–49. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, D.L.; Gross, L.J. Individual-Based Models and Approaches in Ecology: Populations, Communities, and Ecosystems; Chapman & Hall: New York, NY, USA, 1992. [Google Scholar]
- Hellweger, F.J.; Clegg, R.J.; Clark, J.R.; Plugge, C.M.; Kreft, J.-U. Advancing microbial sciences by individual-based modelling. Nat. Rev. Microbiol. 2016, 14, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Palsson, B.O. Escherichia coli growth dynamics: A three-pool biochemically based description. Biotechnol. Bioeng. 1988, 31, 102–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domach, M.M.; Leung, S.K.; Cahn, R.E.; Cocks, G.G.; Shuler, M.L. Computer model for glucose-limited growth of a single cell of Escherichia coli B/r-A. Biotechnol. Bioeng. 2000, 67, 827–840. [Google Scholar] [CrossRef]
- Kreft, J.-U.; Piciorenau, C.; Wimpenny, J.W.T.; van Loosdrecht, M.C.M. Individual-based modelling of biofilms. Microbiology 2001, 147, 2897–2912. [Google Scholar] [CrossRef] [Green Version]
- Gorochowski, T.E.; Matyjaszkiewicz, A.; Todd, T.; Oak, N.; Kowalska, K.; Reid, S.; Savery, N.J. BSim An Agent-Based Tool for Modeling Bacterial Populations in Systems and Synthetic Biology. PLoS ONE 2012, 7, e42790. [Google Scholar] [CrossRef] [Green Version]
- Naylor, J.; Fellermann, H.; Ding, Y.; Mohammed, W.K.; Jakubovics, N.S.; Mukherjee, J.; Biggs, C.A. Simbiotics: A multiscale integrative platform for 3D modeling of bacterial populations. ACS Synth. Biol. 2017, 6, 1194–1210. [Google Scholar] [CrossRef]
- Waclaw, B.; Bozic, I.; Pittman, M.E.; Hruban, R.H.; Vogelstein, B.; Nowak, M.A. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 2015, 525, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Gumbart, J.C.; Beeby, M.; Jensen, G.J. Coarse-grained simulations of bacterial cell wall growth reveal that local coordination alone can be sufficient to maintain rod shape. Proc. Natl. Acad. Sci. USA 2015, 112, E3689–E3698. [Google Scholar] [CrossRef] [Green Version]
- Doursat, R.; Sayama, H.; Michel, O. (Eds.) Morphogenetic Engineering: Toward Programmable Complex Systems; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Tanaka, S. Simulation frameworks for morphogenetic problems. Computation 2015, 3, 197–221. [Google Scholar] [CrossRef] [Green Version]
- Drastik, J. Dynamics of M Systems for Bacterial Growth. 2020. Available online: https://www.youtube.com/watch?v=Mu4nY5yzzhQ (accessed on 31 April 2022).
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; Riet, K.V. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todar, K. Time for Generation of Bacterial Growth. 2021. Available online: http://textbookofbacteriology.net/growth_3.html (accessed on 20 March 2021).
- Winfree, E. Self-healing tile sets. In Nanotechnology: Science and Computation; Chen, J., Jonoska, N., Rozenberg, G., Eds.; Springer: Amsterdam, The Netherlands, 2006; pp. 55–66. [Google Scholar]
- Cowin, A. Wound Repair and Regeneration. Int. J. Mol. Sci. 2018, 453, 314–321. [Google Scholar]
- Coates, J.; Park, B.R.; Le, D.; Simsek, E.; Chaudry, W.; Kim, M. Antibiotic-induced population fluctuations and stochastic clearance of bacteria. eLife 2018, 7, e32976. [Google Scholar] [CrossRef] [PubMed]
- Smolka, V.; Drastik, J.; Bradik, J.; Garzon, M.; Sosik, P. Morphogenetic systems: Models and experiments. Biosystems 2020, 198, 104270. [Google Scholar] [CrossRef]
| Bacterium | Medium | Doubling Time (Mins) |
|---|---|---|
| Escherichia coli | Glucose-salts | 17 |
| Streptococcus lactis | Milk | 26 |
| Lactobacillus acidophilus | Milk | 66–87 |
| Doubling Times Bacteria | Actually Observed (Mins) | Simulation (Iterations/Time Mins) | Simulation Time (Normalized to E. coli) |
|---|---|---|---|
| Escherichia Coli | 1.00 | 100/16.10 | 1.00 |
| Steptococcus lactis | 1.53 | 150/24.01 | 1.49 |
| Lactobaccilus acidophilus | 3.88–5.12 | 400/62.58 | 3.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garzon, M.; Sosik, P.; Drastík, J.; Skalli, O. A Self-Controlled and Self-Healing Model of Bacterial Cells. Membranes 2022, 12, 678. https://doi.org/10.3390/membranes12070678
Garzon M, Sosik P, Drastík J, Skalli O. A Self-Controlled and Self-Healing Model of Bacterial Cells. Membranes. 2022; 12(7):678. https://doi.org/10.3390/membranes12070678
Chicago/Turabian StyleGarzon, Max, Petr Sosik, Jan Drastík, and Omar Skalli. 2022. "A Self-Controlled and Self-Healing Model of Bacterial Cells" Membranes 12, no. 7: 678. https://doi.org/10.3390/membranes12070678
APA StyleGarzon, M., Sosik, P., Drastík, J., & Skalli, O. (2022). A Self-Controlled and Self-Healing Model of Bacterial Cells. Membranes, 12(7), 678. https://doi.org/10.3390/membranes12070678






