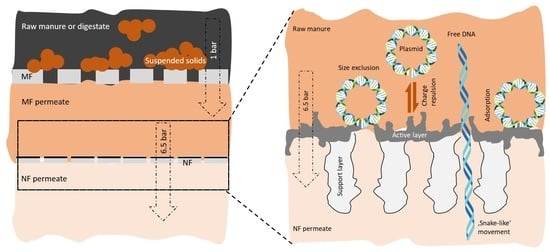
Removal of Diverse and Abundant ARGs by MF-NF Process from Pig Manure and Digestate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pig Manure and Digestate Sample Collection
2.2. Characteristics Analysis
2.3. Filtration Protocols
2.3.1. Pre-Treatment
2.3.2. Nanofiltration
2.4. DNA Extraction
2.5. Smart Chip qPCR Analysis Description
2.6. ARG Retention Calculations
3. Results and Discussion
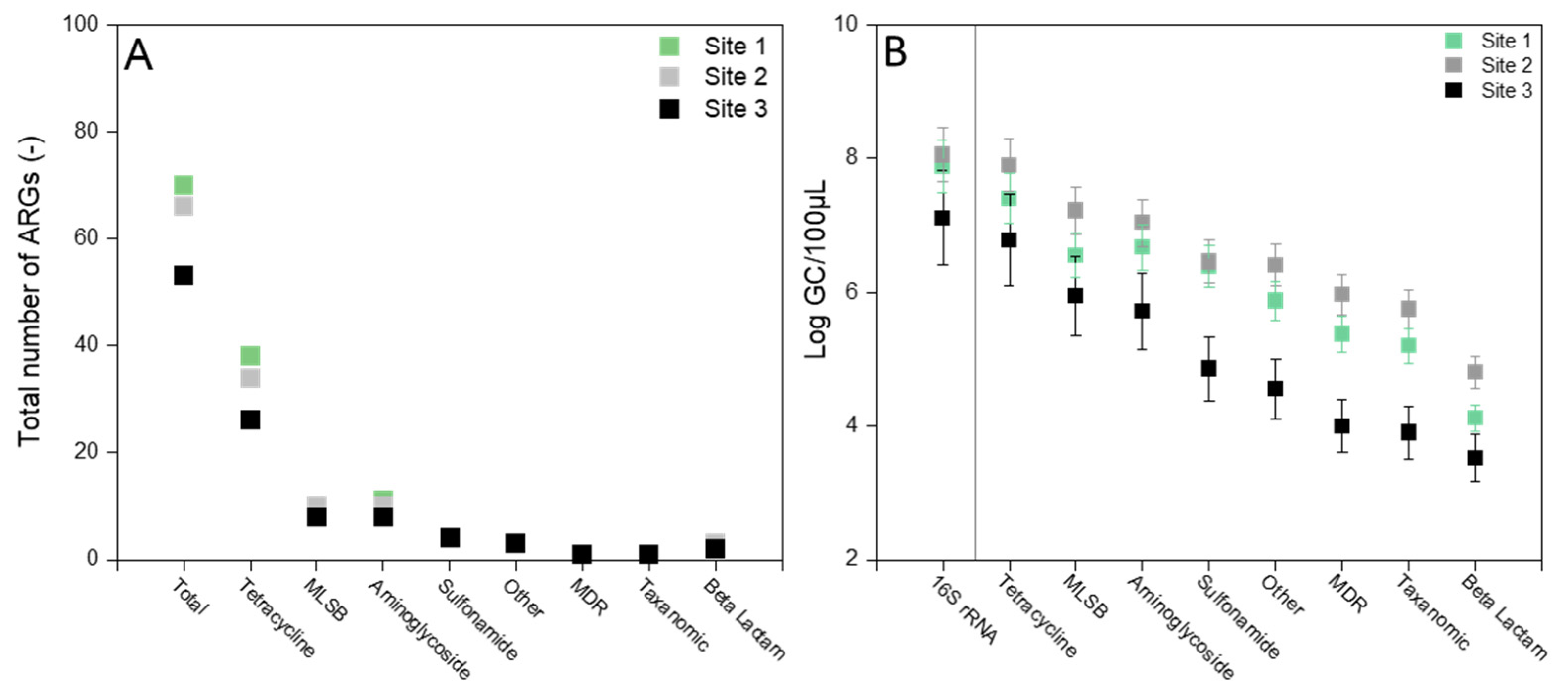
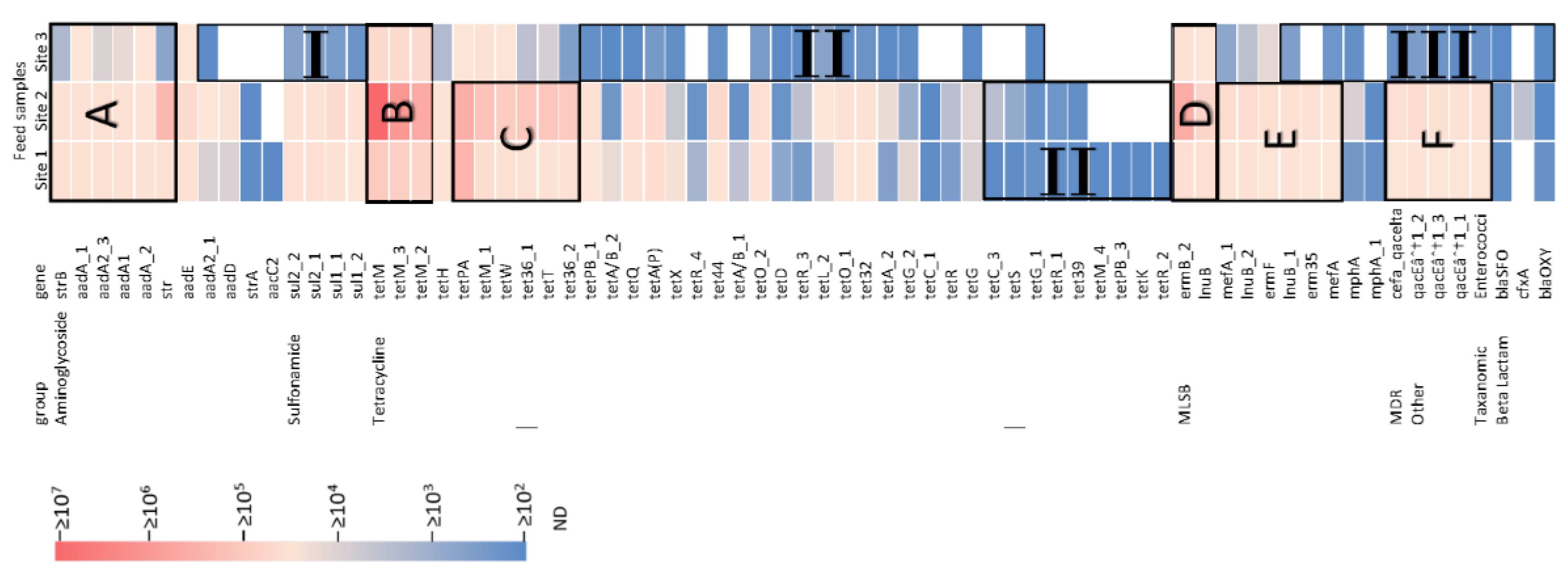
3.1. Presence of Diverse ARGs in Pig Manure and Digestate
3.2. Absolute ARG Abundances in Raw Manure and Digestate
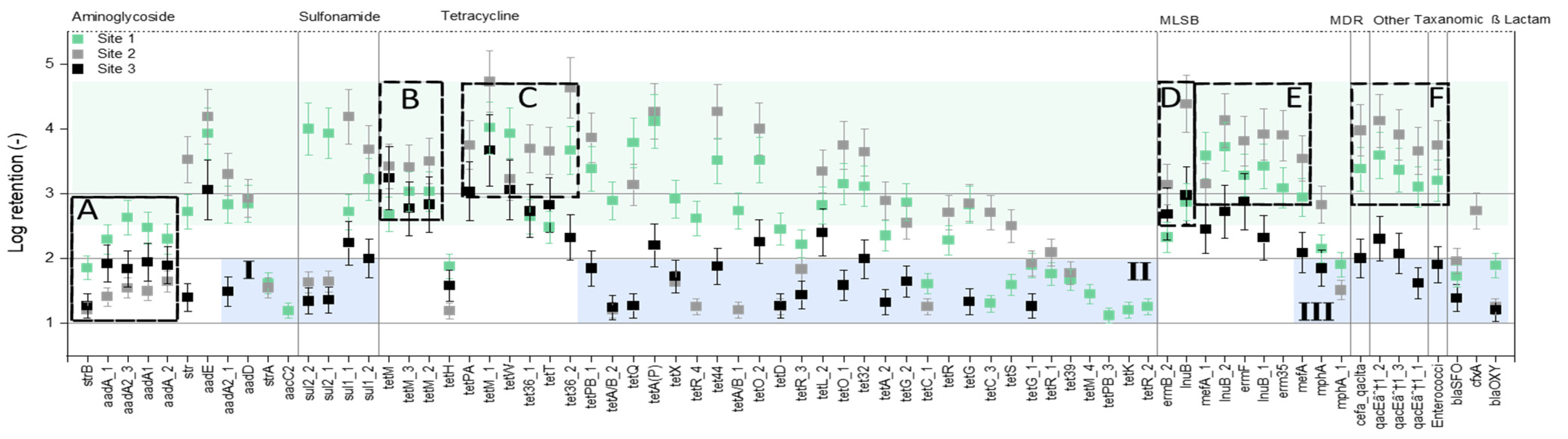
3.3. Removal of ARGs from Raw Manure and Digestate by Nanofiltration
4. Conclusions
- Further removal of some ARGs (e.g., tetH, strB) which were present at a concentration of 103 to 104 copies per 100 µL in NF permeate.
- Better pretreatment of the raw manure and digestate samples for further fouling reduction of NF270.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arias, C.A.; Murray, B.E. Antibiotic-resistant bugs in the 21st century—A clinical super-challenge. N. Engl. J. Med. 2009, 360, 439–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udwadia, Z.F.; Amale, R.A.; Ajbani, K.K.; Rodrigues, C. Totally drug-resistant tuberculosis in India. Clin. Infect. Dis. 2012, 54, 579–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nat. News 2017, 543, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knapp, C.W.; Dolfing, J.; Ehlert, P.A.; Graham, D.W. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2010, 44, 580–587. [Google Scholar] [CrossRef]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235, 178–185. [Google Scholar] [CrossRef]
- Whitehead, T.; Cotta, M. Stored swine manure and swine faeces as reservoirs of antibiotic resistance genes. Lett. Appl. Microbiol. 2013, 56, 264–267. [Google Scholar] [CrossRef]
- Spielmeyer, A.; Höper, H.; Hamscher, G. Long-term monitoring of sulfonamide leaching from manure amended soil into groundwater. Chemosphere 2017, 177, 232–238. [Google Scholar] [CrossRef]
- Kumar, K.; Gupta, S.C.; Chander, Y.; Singh, A.K. Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 2005, 87, 1–54. [Google Scholar]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef]
- Beattie, R.E.; Walsh, M.; Cruz, M.C.; McAliley, L.R.; Dodgen, L.; Zheng, W.; Hristova, K.R. Agricultural contamination impacts antibiotic resistance gene abundances in river bed sediment temporally. FEMS Microbiol. Ecol. 2018, 94, fiy131. [Google Scholar] [CrossRef]
- Huang, L.; Xu, Y.; Xu, J.; Ling, J.; Zheng, L.; Zhou, X.; Xie, G. Dissemination of antibiotic resistance genes (ARGs) by rainfall on a cyclic economic breeding livestock farm. Int. Biodeterior. Biodegrad. 2019, 138, 114–121. [Google Scholar] [CrossRef]
- Szekeres, E.; Chiriac, C.M.; Baricz, A.; Szőke-Nagy, T.; Lung, I.; Soran, M.-L.; Rudi, K.; Dragos, N.; Coman, C. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas. Environ. Pollut. 2018, 236, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binh, C.T.T.; Heuer, H.; Kaupenjohann, M.; Smalla, K. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 2008, 66, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Li, R.; Cui, Y.; Duan, Y. Utilization and develop strategy of organic fertilizer resources in China. Soil Fertil. Sci. China 2010, 4, 77–82. [Google Scholar]
- Ma, Y.; Wilson, C.A.; Novak, J.T.; Riffat, R.; Aynur, S.; Murthy, S.; Pruden, A. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons. Environ. Sci. Technol. 2011, 45, 7855–7861. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [Green Version]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3, e00305-11. [Google Scholar] [CrossRef] [Green Version]
- Barancheshme, F.; Munir, M. Strategies to combat antibiotic resistance in the wastewater treatment plants. Front. Microbiol. 2017, 8, 2603. [Google Scholar] [CrossRef] [Green Version]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef] [PubMed]
- Wernli, D.; Jørgensen, P.S.; Harbarth, S.; Carroll, S.P.; Laxminarayan, R.; Levrat, N.; Røttingen, J.-A.; Pittet, D. Antimicrobial resistance: The complex challenge of measurement to inform policy and the public. PLoS Med. 2017, 14, e1002378. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wu, H.; Zhang, K.; Xu, X.; Wang, C.; Zhu, W.; Wu, W. Long-term low dissolved oxygen accelerates the removal of antibiotics and antibiotic resistance genes in swine wastewater treatment. Chem. Eng. J. 2018, 334, 630–637. [Google Scholar] [CrossRef]
- Yi, Q.; Zhang, Y.; Gao, Y.; Tian, Z.; Yang, M. Anaerobic treatment of antibiotic production wastewater pretreated with enhanced hydrolysis: Simultaneous reduction of COD and ARGs. Water Res. 2017, 110, 211–217. [Google Scholar] [CrossRef]
- Li, N.; Sheng, G.-P.; Lu, Y.-Z.; Zeng, R.J.; Yu, H.-Q. Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res. 2017, 111, 204–212. [Google Scholar] [CrossRef]
- Michael-Kordatou, I.; Karaolia, P.; Fatta-Kassinos, D. The role of operating parameters and oxidative damage mechanisms of advanced chemical oxidation processes in the combat against antibiotic-resistant bacteria and resistance genes present in urban wastewater. Water Res. 2018, 129, 208–230. [Google Scholar] [CrossRef]
- Fiorentino, A.; Esteban, B.; Garrido-Cardenas, J.A.; Kowalska, K.; Rizzo, L.; Aguera, A.; Pérez, J.A.S. Effect of solar photo-Fenton process in raceway pond reactors at neutral pH on antibiotic resistance determinants in secondary treated urban wastewater. J. Hazard. Mater. 2019, 378, 120737. [Google Scholar] [CrossRef]
- Le, T.-H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y.-H. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J.; Wang, J.; Cai, Y. Fate of antibiotic resistance genes in reclaimed water reuse system with integrated membrane process. J. Hazard. Mater. 2020, 382, 121025. [Google Scholar] [CrossRef]
- Lan, L.; Kong, X.; Sun, H.; Li, C.; Liu, D. High removal efficiency of antibiotic resistance genes in swine wastewater via nanofiltration and reverse osmosis processes. J. Environ. Manag. 2019, 231, 439–445. [Google Scholar] [CrossRef]
- Tuczinski, M.; Saravia, F.; Horn, H. Treatment of thermophilic hydrolysis reactor effluent with ceramic microfiltration membranes. Bioprocess Biosyst. Eng. 2018, 41, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Ravi, P.P.; Merkle, W.; Tuczinski, M.; Saravia, F.; Horn, H.; Lemmer, A. Integration of membrane filtration in two-stage anaerobic digestion system: Specific methane yield potentials of hydrolysate and permeate. Bioresour. Technol. 2019, 275, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Al Seadi, T.; Drosg, B.; Fuchs, W.; Rutz, D.; Janssen, R. Biogas digestate quality and utilization. In The Biogas Handbook; Elsevier: Amsterdam, The Netherlands, 2013; pp. 267–301. [Google Scholar]
- Slipko, K.; Reif, D.; Woegerbauer, M.; Hufnagl, P.; Krampe, J.; Kreuzinger, N. Removal of extracellular free DNA and antibiotic resistance genes from water and wastewater by membranes ranging from microfiltration to reverse osmosis. Water Res. 2019, 164, 114916. [Google Scholar] [CrossRef]
- Gros, M.; Marti, E.; Balcázar, J.L.; Boy-Roura, M.; Busquets, A.; Colon, J.; Sanchez-Melsio, A.; Lekunberri, I.; Borrego, C.M.; Ponsá, S. Fate of pharmaceuticals and antibiotic resistance genes in a full-scale on-farm livestock waste treatment plant. J. Hazard. Mater. 2019, 378, 120716. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.L.; Hjorth, M.; Keiding, K. Characterization of pig slurry with reference to flocculation and separation. Water Res. 2009, 43, 773–783. [Google Scholar] [CrossRef]
- Ott, A.; O’Donnell, G.; Tran, N.H.; Mohd Haniffah, M.R.; Su, J.-Q.; Zealand, A.M.; Gin, K.Y.-H.; Goodson, M.L.; Zhu, Y.-G.; Graham, D.W. Developing Surrogate Markers for Predicting Antibiotic Resistance “Hot Spots” in Rivers Where Limited Data Are Available. Environ. Sci. Technol. 2021, 55, 7466–7478. [Google Scholar] [CrossRef] [PubMed]
- Ros, M.; Hendriks, C.; Sigurnjak, I.; Aguilar, A.R.; Meers, E.; Hajdu, Z.; Prado, J.; Guerra, H.P.; Fangueiro, J.D. 1.4 Effects of Current Techniques and Management Systems on CNP Flows in Europe. 2020. Available online: https://www.nutri2cycle.eu/wp-content/uploads/2020/03/D1.4-Nutri2Cycle-techniques-and-management-systems.pdf (accessed on 1 January 2021).
- Bonmatí-Blasi, A.; Cerrillo-Moreno, M.; Riau-Arenas, V. Systems Based on Physical-Chemical Processes: Nutrient Recovery for Cycle Closure. In Waste Management: Concepts, Methodologies, Tools, and Applications; IGI Global: Hershey, PA, USA, 2020; pp. 526–558. [Google Scholar]
- Cerrillo, M.; Palatsi, J.; Comas, J.; Vicens, J.; Bonmatí, A. Struvite precipitation as a technology to be integrated in a manure anaerobic digestion treatment plant–removal efficiency, crystal characterization and agricultural assessment. J. Chem. Technol. Biotechnol. 2015, 90, 1135–1143. [Google Scholar] [CrossRef]
- Tampio, E.; Marttinen, S.; Rintala, J. Liquid fertilizer products from anaerobic digestion of food waste: Mass, nutrient and energy balance of four digestate liquid treatment systems. J. Clean. Prod. 2016, 125, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Ledda, C.; Schievano, A.; Salati, S.; Adani, F. Nitrogen and water recovery from animal slurries by a new integrated ultrafiltration, reverse osmosis and cold stripping process: A case study. Water Res. 2013, 47, 6157–6166. [Google Scholar] [CrossRef]
- American Public Health Association; American Water Work Association. Standard Methods for the Examination of Water and Wastewater, 20th ed.; American Public Health Association: Washington, DC, USA; American Water Work Association: Washington, DC, USA; Water Environment Federation: Washington, DC, USA, 1998. [Google Scholar]
- Wei, C.-H.; Laborie, S.; Aim, R.B.; Amy, G. Full utilization of silt density index (SDI) measurements for seawater pre-treatment. J. Membr. Sci. 2012, 405, 212–218. [Google Scholar] [CrossRef]
- Mänttäri, M.; Pekuri, T.; Nyström, M. NF270, a new membrane having promising characteristics and being suitable for treatment of dilute effluents from the paper industry. J. Membr. Sci. 2004, 242, 107–116. [Google Scholar] [CrossRef]
- Dang, H.Q.; Price, W.E.; Nghiem, L.D. The effects of feed solution temperature on pore size and trace organic contaminant rejection by the nanofiltration membrane NF270. Sep. Purif. Technol. 2014, 125, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Stedtfeld, R.D.; Guo, X.; Stedtfeld, T.M.; Sheng, H.; Williams, M.R.; Hauschild, K.; Gunturu, S.; Tift, L.; Wang, F.; Howe, A. Primer set 2.0 for highly parallel qPCR array targeting antibiotic resistance genes and mobile genetic elements. FEMS Microbiol. Ecol. 2018, 94, fiy130. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Pitkänen, L.K.; Sørum, H.; Stedtfeld, R.D.; Tiedje, J.M.; Virta, M. The resistome of farmed fish feces contributes to the enrichment of antibiotic resistance genes in sediments below Baltic Sea fish farms. Front. Microbiol. 2017, 7, 2137. [Google Scholar] [CrossRef]
- Muziasari, W.I.; Pärnänen, K.; Johnson, T.A.; Lyra, C.; Karkman, A.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol. Ecol. 2016, 92, fiw052. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.-H.; Qiao, M.; Su, J.-Q.; Chen, Z.; Zhou, X.; Zhu, Y.-G. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ. Sci. Technol. 2014, 48, 9079–9085. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Pu, C.; Liu, H.; Ding, G.; Sun, Y.; Yu, X.; Chen, J.; Ren, J.; Gong, X. Impact of direct application of biogas slurry and residue in fields: In situ analysis of antibiotic resistance genes from pig manure to fields. J. Hazard. Mater. 2018, 344, 441–449. [Google Scholar] [CrossRef]
- Zhang, R.-M.; Liu, X.; Wang, S.-L.; Fang, L.-X.; Sun, J.; Liu, Y.-H.; Liao, X.-P. Distribution patterns of antibiotic resistance genes and their bacterial hosts in pig farm wastewater treatment systems and soil fertilized with pig manure. Sci. Total Environ. 2021, 758, 143654. [Google Scholar] [CrossRef]
- Krishnasamy, V.; Otte, J.; Silbergeld, E. Antimicrobial use in Chinese swine and broiler poultry production. Antimicrob. Resist. Infect. Control 2015, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agersø, Y.; Jensen, L.B.; Givskov, M.; Roberts, M.C. The identification of a tetracycline resistance gene tet (M), on a Tn 916-like transposon, in the Bacillus cereus group. FEMS Microbiol. Lett. 2002, 214, 251–256. [Google Scholar] [CrossRef]
- Agersø, Y.; Sengeløv, G.; Jensen, L.B. Development of a rapid method for direct detection of tet (M) genes in soil from Danish farmland. Environ. Int. 2004, 30, 117–122. [Google Scholar] [CrossRef]
- Cheng, W.; Li, J.; Wu, Y.; Xu, L.; Su, C.; Qian, Y.; Zhu, Y.-G.; Chen, H. Behavior of antibiotics and antibiotic resistance genes in eco-agricultural system: A case study. J. Hazard. Mater. 2016, 304, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Li, B.; Li, L.-G.; Zhang, T.; Angelidaki, I. Antibiotic resistance genes and correlations with microbial community and metal resistance genes in full-scale biogas reactors as revealed by metagenomic analysis. Environ. Sci. Technol. 2017, 51, 4069–4080. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Stokes, H. Are humans increasing bacterial evolvability? Trends Ecol. Evol. 2012, 27, 346–352. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use; AGISAR: Copenhagen, Denmark, 2017; Volume 3, pp. 1–26.
- Tian, Z.; Zhang, Y.; Yu, B.; Yang, M. Changes of resistome, mobilome and potential hosts of antibiotic resistance genes during the transformation of anaerobic digestion from mesophilic to thermophilic. Water Res. 2016, 98, 261–269. [Google Scholar] [CrossRef]
- Sun, W.; Qian, X.; Gu, J.; Wang, X.-J.; Duan, M.-L. Mechanism and effect of temperature on variations in antibiotic resistance genes during anaerobic digestion of dairy manure. Sci. Rep. 2016, 6, 30237. [Google Scholar] [CrossRef] [Green Version]
- Varga, C.; Rajić, A.; McFall, M.E.; Reid-Smith, R.J.; Deckert, A.E.; Checkley, S.L.; McEwen, S.A. Associations between reported on-farm antimicrobial use practices and observed antimicrobial resistance in generic fecal Escherichia coli isolated from Alberta finishing swine farms. Prev. Vet. Med. 2009, 88, 185–192. [Google Scholar] [CrossRef]
- Barlow, M. What antimicrobial resistance has taught us about horizontal gene transfer. Horiz. Gene Transf. 2009, 532, 397–411. [Google Scholar]
- Heuer, H.; Kopmann, C.; Binh, C.T.; Top, E.M.; Smalla, K. Spreading antibiotic resistance through spread manure: Characteristics of a novel plasmid type with low% G + C content. Environ. Microbiol. 2009, 11, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Binh, C.T.T.; Jechalke, S.; Kopmann, C.; Zimmerling, U.; Krögerrecklenfort, E.; Ledger, T.; González, B.; Top, E.M.; Smalla, K. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: Diversification driven by class 1 integron gene cassettes. Front. Microbiol. 2012, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Binh, C.T.; Heuer, H.; Kaupenjohann, M.; Smalla, K. Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res. Microbiol. 2009, 160, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Wang, X.; Gu, J.; Zhang, S.; Yin, Y.; Li, Y.; Qian, X.; Sun, W. Effects of different swine manure to wheat straw ratios on antibiotic resistance genes and the microbial community structure during anaerobic digestion. Bioresour. Technol. 2017, 231, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sui, Q.; Zhang, J.; Chen, M.; Tong, J.; Wang, R.; Wei, Y. Distribution of antibiotic resistance genes (ARGs) in anaerobic digestion and land application of swine wastewater. Environ. Pollut. 2016, 213, 751–759. [Google Scholar] [CrossRef]
- Tao, C.-W.; Hsu, B.-M.; Ji, W.-T.; Hsu, T.-K.; Kao, P.-M.; Hsu, C.-P.; Shen, S.-M.; Shen, T.-Y.; Wan, T.-J.; Huang, Y.-L. Evaluation of five antibiotic resistance genes in wastewater treatment systems of swine farms by real-time PCR. Sci. Total Environ. 2014, 496, 116–121. [Google Scholar] [CrossRef]
- Jackson, C.R.; Fedorka-Cray, P.J.; Barrett, J.B.; Ladely, S.R. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 2004, 70, 4205. [Google Scholar] [CrossRef] [Green Version]
- Enne, V.I.; Cassar, C.; Sprigings, K.; Woodward, M.J.; Bennett, P.M. A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol. Lett. 2008, 278, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef]
- Pärnänen, K.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, W.; Xu, L.; Strong, P.; Chen, H. Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. Environ. Sci. Pollut. Res. 2015, 22, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Hammarén, R.; Pal, C.; Östman, M.; Björlenius, B.; Flach, C.-F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718. [Google Scholar] [CrossRef] [Green Version]
- WHO. Antimicrobial Resistance Global Report on Surveillance: 2014 Summary; World Health Organization: Geneva, Switzerland, 2014.
- Chen, J.; Michel, F.C.; Sreevatsan, S.; Morrison, M.; Yu, Z. Occurrence and persistence of erythromycin resistance genes (erm) and tetracycline resistance genes (tet) in waste treatment systems on swine farms. Microb. Ecol. 2010, 60, 479–486. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X. Removal of antibiotic resistance genes (ARGs) in various wastewater treatment processes: An overview. Crit. Rev. Environ. Sci. Technol. 2020, 52, 571–630. [Google Scholar] [CrossRef]
- Cheng, W.; Chen, H.; Su, C.; Yan, S. Abundance and persistence of antibiotic resistance genes in livestock farms: A comprehensive investigation in eastern China. Environ. Int. 2013, 61, 1–7. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, M. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ. Int. 2013, 55, 9–14. [Google Scholar] [CrossRef]
- Subirats, J.; Timoner, X.; Sànchez-Melsió, A.; Balcázar, J.L.; Acuña, V.; Sabater, S.; Borrego, C.M. Emerging contaminants and nutrients synergistically affect the spread of class 1 integron-integrase (intI1) and sul1 genes within stable streambed bacterial communities. Water Res. 2018, 138, 77–85. [Google Scholar] [CrossRef]
- Cristóvão, M.B.; Tela, S.; Silva, A.F.; Oliveira, M.; Bento-Silva, A.; Bronze, M.R.; Crespo, M.T.B.; Crespo, J.G.; Nunes, M.; Pereira, V.J. Occurrence of Antibiotics, Antibiotic Resistance Genes and Viral Genomes in Wastewater Effluents and Their Treatment by a Pilot Scale Nanofiltration Unit. Membranes 2021, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Arkhangelsky, E.; Sefi, Y.; Hajaj, B.; Rothenberg, G.; Gitis, V. Kinetics and mechanism of plasmid DNA penetration through nanopores. J. Membr. Sci. 2011, 371, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Breazeal, M.V.R.; Novak, J.T.; Vikesland, P.J.; Pruden, A. Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res. 2013, 47, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Latulippe, D.R.; Ager, K.; Zydney, A.L. Flux-dependent transmission of supercoiled plasmid DNA through ultrafiltration membranes. J. Membr. Sci. 2007, 294, 169–177. [Google Scholar] [CrossRef]
- Latulippe, D.; Zydney, A. Elongational flow model for transmission of supercoiled plasmid DNA during membrane ultrafiltration. J. Membr. Sci. 2009, 329, 201–208. [Google Scholar] [CrossRef]
- Cheng, H.; Hong, P.-Y. Removal of antibiotic-resistant bacteria and antibiotic resistance genes affected by varying degrees of fouling on anaerobic microfiltration membranes. Environ. Sci. Technol. 2017, 51, 12200–12209. [Google Scholar] [CrossRef] [Green Version]
- Nghiem, L.D.; Hawkes, S. Effects of membrane fouling on the nanofiltration of pharmaceutically active compounds (PhACs): Mechanisms and role of membrane pore size. Sep. Purif. Technol. 2007, 57, 176–184. [Google Scholar] [CrossRef]
- Westhof, E. Water: An integral part of nucleic acid structure. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 125–144. [Google Scholar] [CrossRef]
- Ager, K.; Latulippe, D.R.; Zydney, A.L. Plasmid DNA transmission through charged ultrafiltration membranes. J. Membr. Sci. 2009, 344, 123–128. [Google Scholar] [CrossRef]
- Schwermer, C.U.; Krzeminski, P.; Wennberg, A.C.; Vogelsang, C.; Uhl, W. Removal of antibiotic resistant E. coli in two Norwegian wastewater treatment plants and by nano-and ultra-filtration processes. Water Sci. Technol. 2018, 77, 1115–1126. [Google Scholar] [CrossRef]
- Lamba, M.; Ahammad, S.Z. Performance comparison of secondary and tertiary treatment systems for treating antibiotic resistance. Water Res. 2017, 127, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.Y.; Kwon, Y.-N.; Leckie, J.O. Characterization of humic acid fouled reverse osmosis and nanofiltration membranes by transmission electron microscopy and streaming potential measurements. Environ. Sci. Technol. 2007, 41, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xiong, B.; Yin, C.; Zhang, X.; Zhou, Y.; Wang, Z.; Fang, P.; He, C. Free volume characteristics on water permeation and salt rejection of polyamide reverse osmosis membranes investigated by a pulsed slow positron beam. J. Mater. Sci. 2018, 53, 16132–16145. [Google Scholar] [CrossRef]
- Song, X.; Smith, J.W.; Kim, J.; Zaluzec, N.J.; Chen, W.; An, H.; Dennison, J.M.; Cahill, D.G.; Kulzick, M.A.; Chen, Q. Unraveling the Morphology–Function Relationships of Polyamide Membranes Using Quantitative Electron Tomography. ACS Appl. Mater. Interfaces 2019, 11, 8517–8526. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samanta, P.; Horn, H.; Saravia, F. Removal of Diverse and Abundant ARGs by MF-NF Process from Pig Manure and Digestate. Membranes 2022, 12, 661. https://doi.org/10.3390/membranes12070661
Samanta P, Horn H, Saravia F. Removal of Diverse and Abundant ARGs by MF-NF Process from Pig Manure and Digestate. Membranes. 2022; 12(7):661. https://doi.org/10.3390/membranes12070661
Chicago/Turabian StyleSamanta, Prantik, Harald Horn, and Florencia Saravia. 2022. "Removal of Diverse and Abundant ARGs by MF-NF Process from Pig Manure and Digestate" Membranes 12, no. 7: 661. https://doi.org/10.3390/membranes12070661
APA StyleSamanta, P., Horn, H., & Saravia, F. (2022). Removal of Diverse and Abundant ARGs by MF-NF Process from Pig Manure and Digestate. Membranes, 12(7), 661. https://doi.org/10.3390/membranes12070661