Bioinformatic Analysis of ABCA1 Gene Expression in Smoking and Chronic Obstructive Pulmonary Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Extraction
2.3. Differential Expression Analysis
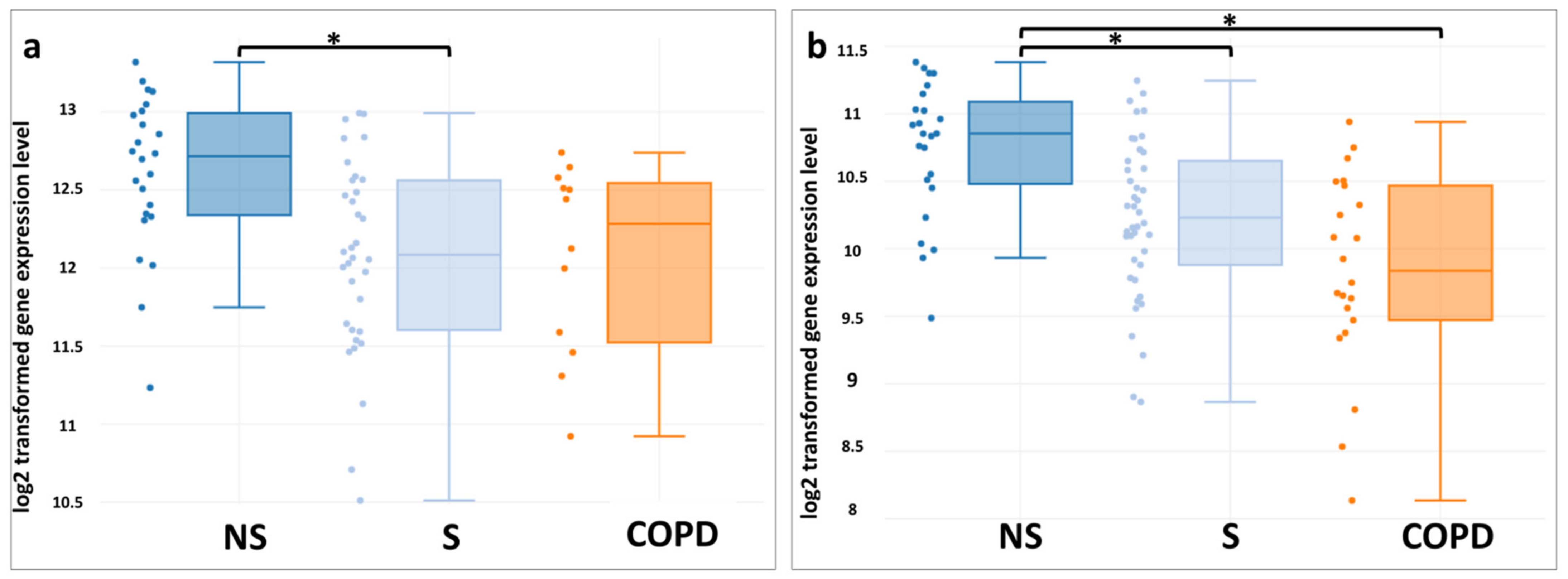
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Proctor, R.N. Tobacco and the global lung cancer epidemic. Nat. Rev. Cancer 2001, 1, 82–86. [Google Scholar] [CrossRef]
- World Health Organization. WHO Report on the Global Tobacco Epidemic, 2019: Offer Help to quit Tobacco Use; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Jafari, A.; Rajabi, A.; Gholian-Aval, M.; Peyman, N.; Mahdizadeh, M.; Tehrani, H. National, regional, and global prevalence of cigarette smoking among women/females in the general population: A systematic review and meta-analysis. Environ. Health Prev. Med. 2021, 26, 1–13. [Google Scholar] [CrossRef]
- Spira, A.; Beane-Ebel, J.; Shah, V.; Liu, G.; Schembri, F.; Yang, X.; Palma, J.; Brody, J.S. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc. Natl. Acad. Sci. USA 2004, 101, 10143–10148. [Google Scholar] [CrossRef] [Green Version]
- Vogelmeier, C.F.; Criner, G.J.; Martínez, F.J.; Anzueto, A.; Barnes, P.J.; Bourbeau, J.; Celli, B.R.; Chen, R.; Decramer, M.; Fabbri, L.M.; et al. Informe 2017 de la iniciativa global para el diagnóstico, tratamiento y prevención de la enfermedad pulmonar obstructiva crónica: Resumen ejecutivo de gold. Am. J. Respir. Crit. Care Med. 2017, 53, 128–149. [Google Scholar] [CrossRef]
- Soriano, J.B.; Kendrick, P.J.; Paulson, K.R.; Gupta, V.; Abrams, E.M.; Adedoyin, R.A.; Adhikari, T.B.; Advani, S.M.; Agrawal, A.; Ahmadian, E.; et al. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet Respir. Med. 2020, 8, 585–596. [Google Scholar] [CrossRef]
- Quaderi, S.; Hurst, J. The unmet global burden of COPD. Glob. Health Epidemiol. Genom. 2018, 3, e4. [Google Scholar] [CrossRef] [Green Version]
- Blanco, I.; Diego, I.; Bueno, P.; Casas-Maldonado, F.; Miravitlles, M. Geographic distribution of COPD prevalence in the world displayed by geographic information system maps. Eur. Respir. J. 2019, 54, 1900610. [Google Scholar] [CrossRef]
- Løkke, A.; Lange, P.; Lykkegaard, J.; Ibsen, R.; Andersson, M.; Licht, S.D.F.; Hilberg, O. Economic Burden of COPD by disease severity—A nationwide cohort study in Denmark. Int. J. Chronic Obstr. Pulm. Dis. 2021, 16, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Van Eeden, S.F. Lung macrophage phenotypes and functional responses: Role in the pathogenesis of COPD. Int. J. Mol. Sci. 2018, 19, 582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, P.J. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2016, 138, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Russell, R.; Culpitt, S.V.; DeMatos, C.; Donnelly, L.; Smith, M.; Wiggins, J.; Barnes, P.J. Release and activity of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 by alveolar macrophages from patients with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2002, 26, 602–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batista-Gonzalez, A.; Vidal, R.; Criollo, A.; Carreño, L.J. New insights on the role of lipid metabolism in the metabolic reprogramming of macrophages. Front. Immunol. 2020, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Bossche, J.V.D.; O’Neill, L.; Menon, D. Macrophage immunometabolism: Where are we (going)? Trends Immunol. 2017, 38, 395–406. [Google Scholar] [CrossRef]
- Remmerie, A.; Scott, C.L. Macrophages and lipid metabolism. Cell. Immunol. 2018, 330, 27–42. [Google Scholar] [CrossRef]
- Izquierdo, E.; Cuevas, V.D.; Fernández-Arroyo, S.; Riera-Borrull, M.; Orta-Zavalza, E.; Joven, J.; Rial, E.; Corbi, A.L.; Escribese, M.M. Reshaping of human macrophage polarization through modulation of glucose catabolic pathways. J. Immunol. 2015, 195, 2442–2451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, A.R.; Kadam, S.; Brahme, A.; Agrawal, M.; Apte, K.; Narke, G.; Kekan, K.; Madas, S.; Salvi, S. Systemic immuno-metabolic alterations in chronic obstructive pulmonary disease (COPD). Respir. Res. 2019, 20, 171. [Google Scholar] [CrossRef]
- Angela, M.; Endo, Y.; Asou, H.K.; Yamamoto, T.; Tumes, D.J.; Tokuyama, H.; Yokote, K.; Nakayama, T. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells. Nat. Commun. 2016, 7, 13683. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Rahman, I.; Van Schadewijk, A.A.M.; Crowther, A.J.L.; Hiemstra, P.S.; Stolk, J.; MacNee, W.; de Boer, W. 4-Hydroxy-2-nonenal, a specific lipid peroxidation product, is elevated in lungs of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2002, 166, 490–495. [Google Scholar] [CrossRef]
- Aoshiba, K.; Koinuma, M.; Yokohori, N.; Nagai, A. Immunohistochemical evaluation of oxidative stress in murine lungs after cigarette smoke exposure. Inhal. Toxicol. 2003, 15, 1029–1038. [Google Scholar] [CrossRef]
- Malhotra, D.; Thimmulappa, R.; Navas-Acien, A.; Sandford, A.; Elliott, M.; Singh, A.; Chen, L.; Zhuang, X.; Hogg, J.; Pare, P.; et al. Decline in NRF2-regulated antioxidants in chronic obstructive pulmonary disease lungs due to loss of its positive regulator, DJ-1. Am. J. Respir. Crit. Care Med. 2008, 178, 592–604. [Google Scholar] [CrossRef] [Green Version]
- Davies, P.; Sornberger, G.C.; Huber, G.L. The stereology of pulmonary alveolar macrophages after prolonged experimental exposure to tobacco smoke. Lab. Investig. 1977, 37, 297–306. [Google Scholar]
- Hannan, S.E.; Harris, J.O.; Sheridan, N.P.; Patel, J.M. Cigarette smoke alters plasma membrane fluidity of rat alveolar macrophages. Am. Rev. Respir. Dis. 1989, 140, 1668–1673. [Google Scholar] [CrossRef]
- Azzam, K.M.; Fessler, M.B. Crosstalk between reverse cholesterol transport and innate immunity. Trends Endocrinol. Metab. 2012, 23, 169–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tall, A.R.; Yvan-Charvet, L. Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 2015, 15, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Ouimet, M.; Barrett, T.; Fisher, E.A. HDL and reverse cholesterol transport. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef]
- Jessup, W.; Gelissen, I.C.; Gaus, K.; Kritharides, L. Roles of ATP binding cassette transporters A1 and G1, scavenger receptor BI and membrane lipid domains in cholesterol export from macrophages. Curr. Opin. Lipidol. 2006, 17, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Kotlyarov, S.; Kotlyarova, A. The role of ABC transporters in lipid metabolism and the comorbid course of chronic obstructive pulmonary disease and atherosclerosis. Int. J. Mol. Sci. 2021, 22, 6711. [Google Scholar] [CrossRef] [PubMed]
- Kotlyarov, S. Participation of ABCA1 transporter in pathogenesis of chronic obstructive pulmonary disease. Int. J. Mol. Sci. 2021, 22, 3334. [Google Scholar] [CrossRef]
- Aguiar, J.A.; Tamminga, A.; Lobb, B.; Huff, R.D.; Nguyen, J.P.; Kim, Y.; Dvorkin-Gheva, A.; Stampfli, M.R.; Doxey, A.C.; Hirota, J.A. The impact of cigarette smoke exposure, COPD, or asthma status on ABC transporter gene expression in human airway epithelial cells. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaykhiev, R.; Krause, A.; Salit, J.; Strulovici-Barel, Y.; Harvey, B.-G.; O’Connor, T.P.; Crystal, R.G. Smoking-dependent reprogramming of alveolar macrophage polarization: Implication for pathogenesis of chronic obstructive pulmonary disease. J. Immunol. 2009, 183, 2867–2883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Beirne, S.L.; Kikkers, S.A.; Oromendia, C.; Salit, J.; Rostmai, M.R.; Ballman, K.V.; Kaner, R.J.; Crystal, R.G.; Cloonan, S.M. Alveolar macrophage immunometabolism and lung function impairment in smoking and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2020, 201, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Harvey, B.-G.; Heguy, A.; Leopold, P.L.; Carolan, B.J.; Ferris, B.; Crystal, R.G. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J. Mol. Med. 2006, 85, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Tilley, A.E.; Harvey, B.-G.; Heguy, A.; Hackett, N.R.; Wang, R.; O’Connor, T.P.; Crystal, R.G. Down-regulation of the notch pathway in human airway epithelium in association with smoking and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2009, 179, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Zhou, H.; Strulovici-Barel, Y.; Al-Hijji, M.; Ou, X.; Salit, J.; Walters, M.S.; Staudt, M.; Kaner, R.J.; Crystal, R.G. Role of OSGIN1 in mediating smoking-induced autophagy in the human airway epithelium. Autophagy 2017, 13, 1205–1220. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wang, R.; Ferris, B.; Salit, J.; Strulovici-Barel, Y.; Hackett, N.R.; Crystal, R.G. Smoking-mediated up-regulation of GAD67 expression in the human airway epithelium. Respir. Res. 2010, 11, 150. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Wang, G.; Ricard, M.J.; Ferris, B.; Strulovici-Barel, Y.; Salit, J.; Hackett, N.R.; Gudas, L.J.; Crystal, R.G. Smoking-induced upregulation of AKR1B10 expression in the airway epithelium of healthy individuals. Chest 2010, 138, 1402–1410. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zuo, W.-L.; Fukui, T.; Chao, I.; Gomi, K.; Lee, B.; Staudt, M.; Kaner, R.J.; Strulovici-Barel, Y.; Salit, J.; et al. Smoking-dependent distal-to-proximal repatterning of the adult human small airway epithelium. Am. J. Respir. Crit. Care Med. 2017, 196, 340–352. [Google Scholar] [CrossRef]
- Raman, T.; O’Connor, T.P.; Hackett, N.R.; Wang, W.; Harvey, B.-G.; Attiyeh, M.; Dang, D.T.; Teater, M.; Crystal, R.G. Quality control in microarray assessment of gene expression in human airway epithelium. BMC Genom. 2009, 10, 493. [Google Scholar] [CrossRef] [Green Version]
- Tilley, A.E.; O’Connor, T.P.; Hackett, N.R.; Strulovici-Barel, Y.; Salit, J.; Amoroso, N.; Zhou, X.K.; Raman, T.; Omberg, L.; Clark, A.; et al. Biologic phenotyping of the human small airway epithelial response to cigarette smoking. PLoS ONE 2011, 6, e22798. [Google Scholar] [CrossRef]
- Gindele, J.A.; Kiechle, T.; Benediktus, K.; Birk, G.; Brendel, M.; Heinemann, F.; Wohnhaas, C.T.; Leblanc, M.; Zhang, H.; Strulovici-Barel, Y.; et al. Intermittent exposure to whole cigarette smoke alters the differentiation of primary small airway epithelial cells in the air-liquid interface culture. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zenkova, D.K.V.; Sablina, R.; Artyomov, M.; Sergushichev, A. Phantasus: Visual and Interactive Gene Expression Analysis. Available online: https://genome.ifmo.ru/phantasus (accessed on 30 July 2021).
- Barnes, P.J. Alveolar macrophages in Chronic Obstructive Pulmonary Disease (COPD). Cell. Mol. Boil. 2004, 50. [Google Scholar]
- Vlahos, R. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front. Immunol. 2014, 5, 435. [Google Scholar] [CrossRef] [Green Version]
- Soumian, S.; Albrecht, C.; Davies, A.H.; Gibbs, R.G.J. ABCA1 and atherosclerosis. Vasc. Med. 2005, 10, 109–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, A.B.; Ammit, A.J.; Gelissen, I.C. Examining the role of ABC lipid transporters in pulmonary lipid homeostasis and inflammation. Respir. Res. 2017, 18, 1–9. [Google Scholar] [CrossRef] [Green Version]
- He, P.; Smith, A.; Gelissen, I.C.; Ammit, A.J. The effect of statins and the synthetic LXR agonist T0901317 on expression of ABCA1 transporter protein in human lung epithelial cell lines in vitro. Pharmacol. Rep. 2019, 71, 1219–1226. [Google Scholar] [CrossRef]
- Jacobo-Albavera, L.; Domínguez-Pérez, M.; Medina-Leyte, D.; González-Garrido, A.; Villarreal-Molina, T. The role of the ATP-binding cassette A1 (ABCA1) in human disease. Int. J. Mol. Sci. 2021, 22, 1593. [Google Scholar] [CrossRef]
- McNeish, J.; Aiello, R.J.; Guyot, D.; Turi, T.; Gabel, C.; Aldinger, C.; Hoppe, K.; Roach, M.L.; Royer, L.J.; de Wet, J.; et al. High density lipoprotein deficiency and foam cell accumulation in mice with targeted disruption of ATP-binding cassette transporter-1. Proc. Natl. Acad. Sci. USA 2000, 97, 4245–4250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, S.R.; Tao, J.-Q.; Collins, H.L.; Francone, O.L.; Rothblat, G.H. Pulmonary abnormalities due to ABCA1 deficiency in mice. Am. J. Physiol. Cell. Mol. Physiol. 2005, 289, L980–L989. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Liao, D.-F.; Tang, C.-K. ATP-binding membrane cassette transporter A1 (ABCA1): A possible link between inflammation and reverse cholesterol transport. Mol. Med. 2010, 16, 438–449. [Google Scholar] [CrossRef] [Green Version]
- Francone, O.L.; Aiello, R.J. ABCA1: Regulation, function and relationship to atherosclerosis. Curr. Opin. Investig. Drugs 2002, 3. [Google Scholar]
- Zanotti, I.; Potì, F.; Pedrelli, M.; Favari, E.; Moleri, E.; Franceschini, G.; Calabresi, L.; Bernini, F. The LXR agonist T0901317 promotes the reverse cholesterol transport from macrophages by increasing plasma efflux potential. J. Lipid Res. 2008, 49, 954–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussein, M.A.; Shrestha, E.; Ouimet, M.; Barrett, T.; Leone, S.; Moore, K.J.; Herault, Y.; Fisher, E.A.; Garabedian, M.J. LXR-mediated ABCA1 expression and function are modulated by high glucose and PRMT2. PLoS ONE 2015, 10, e0135218. [Google Scholar] [CrossRef] [Green Version]
- Chawla, A.; Boisvert, W.A.; Lee, C.-H.; Laffitte, B.A.; Barak, Y.; Joseph, S.; Liao, D.; Nagy, L.; Edwards, P.A.; Curtiss, L.K.; et al. A PPARγ-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 2001, 7, 161–171. [Google Scholar] [CrossRef]
- Park, Y.; Pham, T.X.; Lee, J. Lipopolysaccharide represses the expression of ATP-binding cassette transporter G1 and scavenger receptor class B, type I in murine macrophages. Inflamm. Res. 2012, 61, 465–472. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Gelissen, I.C.; Ammit, A.J. Regulation of ATP binding cassette transporter A1 (ABCA1) expression: Cholesterol-dependent and independent signaling pathways with relevance to inflammatory lung disease. Respir. Res. 2020, 21, 1–11. [Google Scholar] [CrossRef]
- Sonett, J.; Goldklang, M.; Sklepkiewicz, P.; Gerber, A.; Trischler, J.; Zelonina, T.; Westerterp, M.; Lemaître, V.; Okada, Y.; Armiento, J.D. A critical role for ABC transporters in persistent lung inflammation in the development of emphysema after smoke exposure. FASEB J. 2018, 32, 6724–6736. [Google Scholar] [CrossRef]
- Landry, Y.D.; Denis, M.; Nandi, S.; Bell, S.; Vaughan, A.M.; Zha, X. ATP-binding cassette transporter A1 expression disrupts raft membrane microdomains through its ATPase-related functions. J. Biol. Chem. 2006, 281, 36091–36101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Owen, J.S.; Wilson, M.D.; Li, H.; Griffiths, G.L.; Thomas, M.J.; Hiltbold, E.M.; Fessler, M.; Parks, J.S. Macrophage ABCA1 reduces MyD88-dependent toll-like receptor trafficking to lipid rafts by reduction of lipid raft cholesterol. J. Lipid Res. 2010, 51, 3196–3206. [Google Scholar] [CrossRef] [Green Version]
- Bi, X.; Vitali, C.; Cuchel, M. ABCA1 and inflammation. Arter. Thromb. Vasc. Biol. 2015, 35, 1551–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, K.; Ehehalt, R. Cholesterol, lipid rafts, and disease. J. Clin. Investig. 2002, 110, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Epand, R.M.; Barrantes, F.J. Cholesterol-recognition motifs in membrane proteins. Adv. Exp. Med. Biol. 2019, 1135, 3–25. [Google Scholar] [CrossRef]
- Ruysschaert, J.-M.; Lonez, C. Role of lipid microdomains in TLR-mediated signalling. Biochim. Biophys. Acta Biomembr. 2015, 1848, 1860–1867. [Google Scholar] [CrossRef]
- Francone, O.L.; Royer, L.; Boucher, G.; Haghpassand, M.; Freeman, A.; Brees, D.; Aiello, R.J. Increased cholesterol deposition, expression of scavenger receptors, and response to chemotactic factors in abca1 deficient macrophages. Arter. Thromb. Vasc. Biol. 2005, 25, 1198–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.; Houston, B.A.; Storey, C.; LeBoeuf, R.C. Both STAT3 activation and cholesterol efflux contribute to the anti-inflammatory effect of apoA-I/ABCA1 interaction in macrophages. J. Lipid Res. 2016, 57, 848–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Ishibashi, M.; Seimon, T.; Lee, M.; Sharma, S.; Fitzgerald, K.; Samokhin, A.O.; Wang, Y.; Sayers, S.; Aikawa, M.; et al. Free cholesterol accumulation in macrophage membranes activates toll-like receptors and p38 mitogen-activated protein kinase and induces Cathepsin, K. Circ. Res. 2009, 104, 455–465. [Google Scholar] [CrossRef]
- Zhou, J.; You, Y.; Ryan, A.J.; Mallampalli, R.K. Upregulation of surfactant synthesis triggers ABCA1-mediated basolateral phospholipid efflux. J. Lipid Res. 2004, 45, 1758–1767. [Google Scholar] [CrossRef] [Green Version]
- Bates, S.R.; Tao, J.-Q.; Yu, K.J.; Borok, Z.; Crandall, E.D.; Collins, H.L.; Rothblat, G.H. Expression and biological activity of ABCA1 in alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 2008, 38, 283–292. [Google Scholar] [CrossRef] [Green Version]
- Jubinville, E.; Talbot, M.; Bérubé, J.-C.; Hamel-Auger, M.; Maranda-Robitaille, M.; Beaulieu, M.-J.; Aubin, S.; Paré, M.; Kallend, D.G.; Arsenault, B.; et al. Interplay between cigarette smoking and pulmonary reverse lipid transport. Eur. Respir. J. 2017, 50, 1700681. [Google Scholar] [CrossRef] [Green Version]
- Kotlyarov, S.; Kotlyarova, A. Molecular mechanisms of lipid metabolism disorders in infectious exacerbations of chronic obstructive pulmonary disease. Int. J. Mol. Sci. 2021, 22, 7634. [Google Scholar] [CrossRef] [PubMed]



| DataSet | Characteristic | Method of Obtaining the Material | Microarray Platform | References |
|---|---|---|---|---|
| GSE13896 | Alveolar macrophages obtained from healthy non-smokers, smokers, and smokers with COPD | Bronchoalveolar lavage | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [32] |
| GSE130928 | Alveolar macrophages obtained from healthy non-smokers, smokers, and smokers with COPD | Bronchoalveolar lavage | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [33] |
| GSE4498 | The bronchial epithelium of the 10th–12th order obtained in healthy non-smokers and phenotypically normal smokers | Bronchoscopy | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [34,35] |
| GSE76324 | Bronchial epithelium of the 3rd–4th and 10th–12th order, obtained in healthy non-smokers and smokers | Bronchoscopy | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [36,37] |
| GSE18385 | Bronchial epithelium of the 3rd–4th and 10th–12th order, obtained in healthy non-smokers and smokers | Bronchoscopy | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [38] |
| GSE64614 | Bronchial epithelium, including trachea and bronchi of the 4th–6th order obtained from healthy non-smokers and epithelium of the distal respiratory tract (bronchi of the 10th–12th order) of healthy non-smokers and healthy smokers | Bronchoscopy | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [39] |
| GSE11906 | Bronchial epithelium of trachea, bronchi of the 2nd–3rd order, and bronchi of the 10th–12th order obtained from healthy non-smokers, smokers, and smokers with COPD | Bronchoscopy | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [40] |
| GSE11784 | Bronchial epithelium of the 10th–12th order obtained in healthy non-smokers, smokers, and patients with COPD | Bronchoscopy | GPL570 Affymetrix Human Genome U133 Plus 2.0 Array | [41,42] |
| DataSet | Characteristic | Comparison Groups | ||
|---|---|---|---|---|
| Non-Smokers | Smokers | COPD | ||
| GSE13896 | Age (year) Pack-years index Sex (M/F) | 40.2 ± 8.3 - 18/6 | 42.1 ± 7.8 27.5 ± 18.1 25/9 | 54.2 ± 9.3 51.5 ± 29.2 10/2 |
| GSE130928 | Age (year) Pack-years index Sex (M/F) | 40.3 ± 8.2 - 18/6 | 42.6 ± 7.8 27.1 ± 17.0 29/13 | 53.8 ± 7.9 43.4 ± 28.0 18/4 |
| GSE4498 | Age (year) Pack-years index Sex (M/F) | 42.3 ± 7.7 - 10/2 | 44.0 ± 3.9 25.8 ± 9.1 7/3 | - |
| GSE18385 | Age (year) Pack-years index Sex (M/F) | 41.1 ± 10.5 - 51/21 | 43.1 ± 7.0 28.1 ± 17.2 59 / 30 | - |
| GSE11906 | Age (year) Pack-years index Sex (M/F) | 42.6 ± 9.7 - 32/10 | 42.9 ± 6.3 26.9 ± 15.9 35/14 | 55.6 ± 7.7 36.4 ± 21.9 26/7 |
| GSE11784 | Age (year) Pack-years index Sex (M/F) | 40.3 ± 12 - 40/23 | 42.5 ± 7.6 27.2 ± 15.9 52/20 | 51.6 ± 8.6 40.9 ± 28.2 18/4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotlyarov, S.; Kotlyarova, A. Bioinformatic Analysis of ABCA1 Gene Expression in Smoking and Chronic Obstructive Pulmonary Disease. Membranes 2021, 11, 674. https://doi.org/10.3390/membranes11090674
Kotlyarov S, Kotlyarova A. Bioinformatic Analysis of ABCA1 Gene Expression in Smoking and Chronic Obstructive Pulmonary Disease. Membranes. 2021; 11(9):674. https://doi.org/10.3390/membranes11090674
Chicago/Turabian StyleKotlyarov, Stanislav, and Anna Kotlyarova. 2021. "Bioinformatic Analysis of ABCA1 Gene Expression in Smoking and Chronic Obstructive Pulmonary Disease" Membranes 11, no. 9: 674. https://doi.org/10.3390/membranes11090674
APA StyleKotlyarov, S., & Kotlyarova, A. (2021). Bioinformatic Analysis of ABCA1 Gene Expression in Smoking and Chronic Obstructive Pulmonary Disease. Membranes, 11(9), 674. https://doi.org/10.3390/membranes11090674







