Detection of Prostate Cancer via IR Spectroscopic Analysis of Urinary Extracellular Vesicles: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Isolation of EVs
2.3. Characterization of EVs
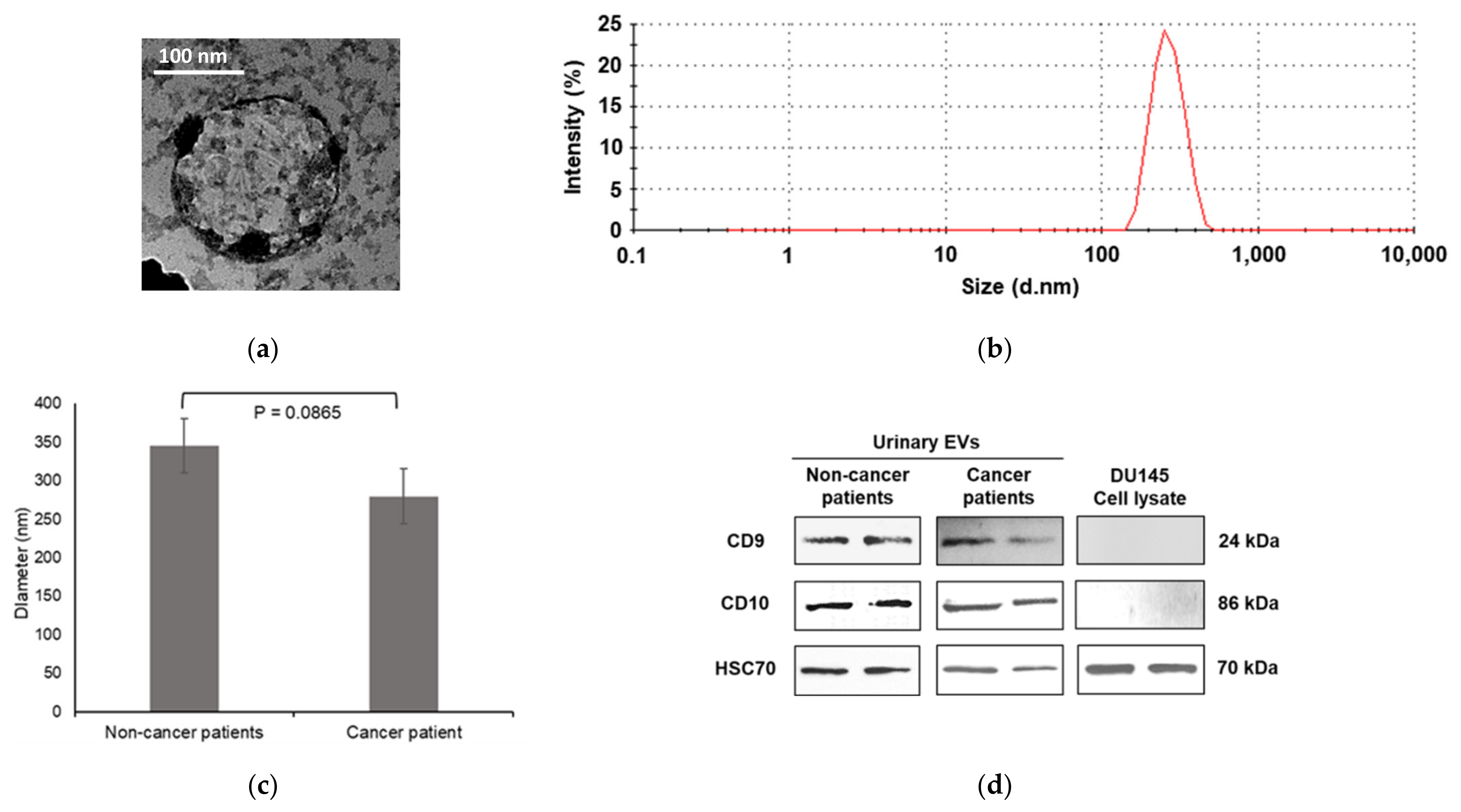
2.3.1. Transmission Electron Microscopy (TEM)
2.3.2. Particle Size Analysis (Zetasizer)
2.3.3. Immunoblotting
2.4. Attenuated Total Reflection-Fourier Transform Infrared Spectroscopy (ATR-FTIR)
2.5. Statistical Analysis
3. Results
3.1. Characterization of Urinary EVs
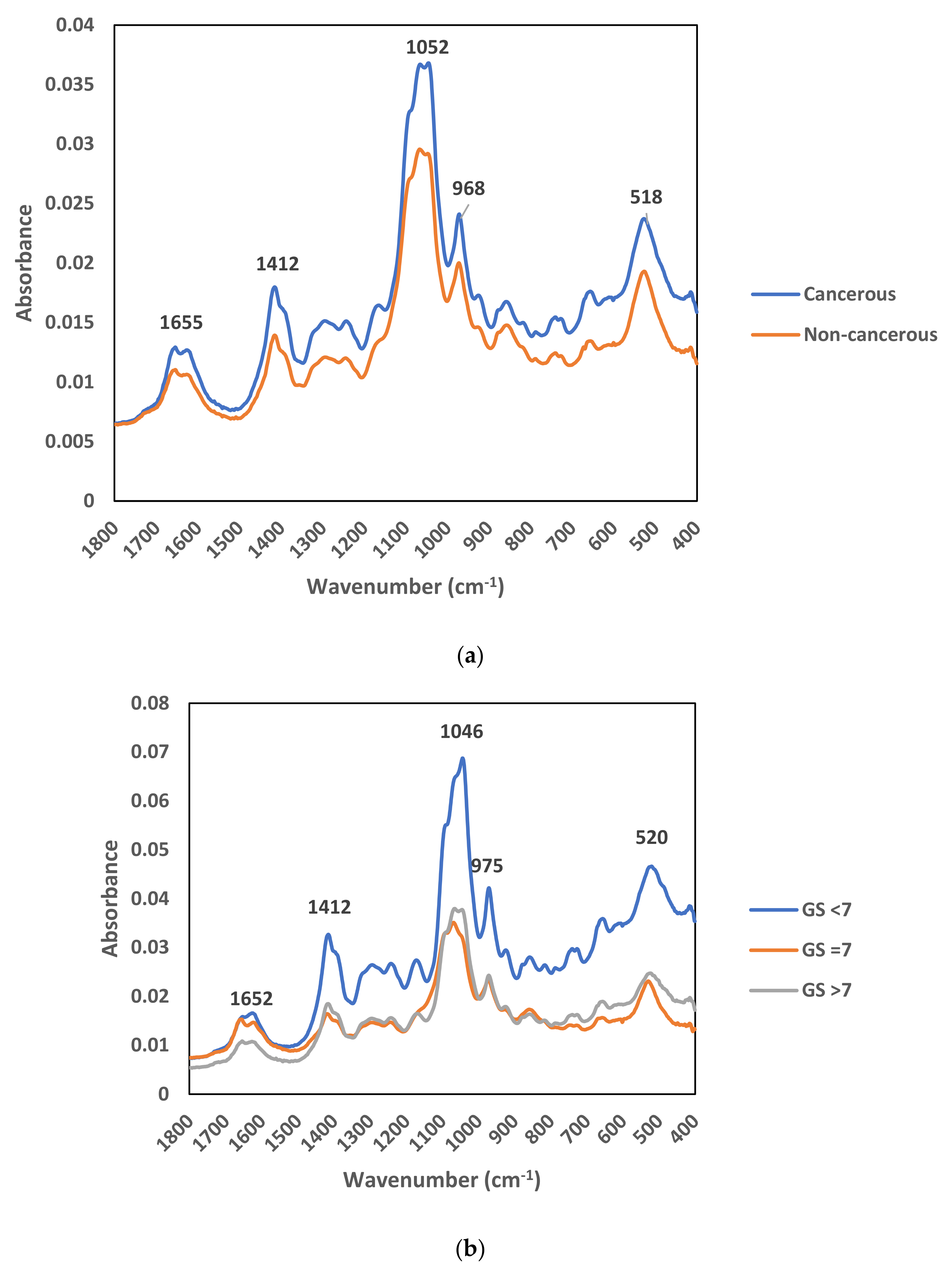
3.2. FTIR Analysis of Urinary EVs
3.3. Principle Component Analysis (PCA)
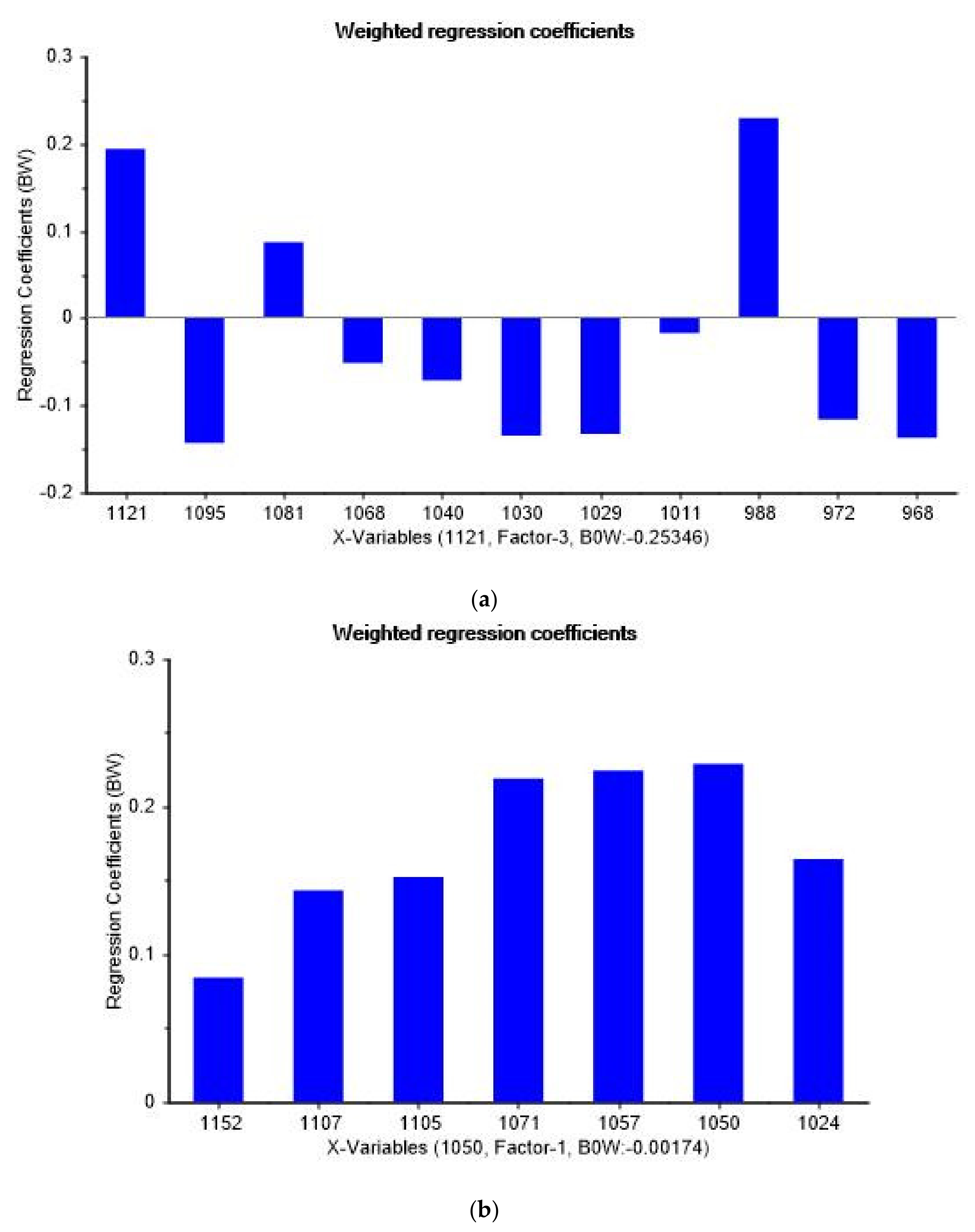
3.4. Spectral Peaks Responsible for Prostate Cancer Discrimination
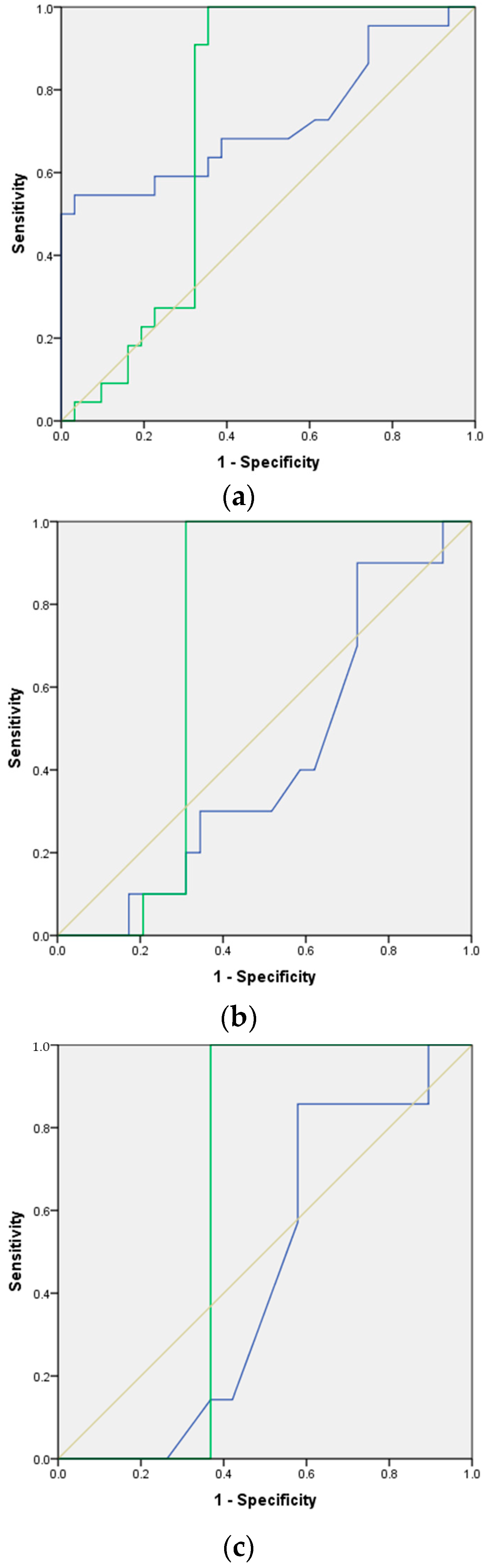
3.5. Accuray of FTIR Analysis of Urinary EVs in Prostate Cancer Detection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Patient | PSA (ng/mL) | Patient | PSA (ng/mL) | Patient | PSA (ng/mL) |
| 1 | 7 | 11 | 19.69 | 21 | 12 |
| 2 | 15 | 12 | 8.27 | 22 | 8 |
| 3 | 6 | 13 | 30 | 23 | 10 |
| 4 | 8 | 14 | 11.95 | 24 | 12 |
| 5 | 14 | 15 | 4.3 | 25 | 6 |
| 6 | 5 | 16 | 5.77 | 26 | 12 |
| 7 | 17 | 17 | 20 | 27 | 9 |
| 8 | 11 | 18 | 6.8 | 28 | 24 |
| 9 | 8 | 19 | 6 | 29 | 4 |
| 10 | 9 | 20 | 6.16 | 30 | 8 |
| 31 | 7 |


Appendix B. Suggested Workflow for FTIR PCA Model of Urinary EV Analysis
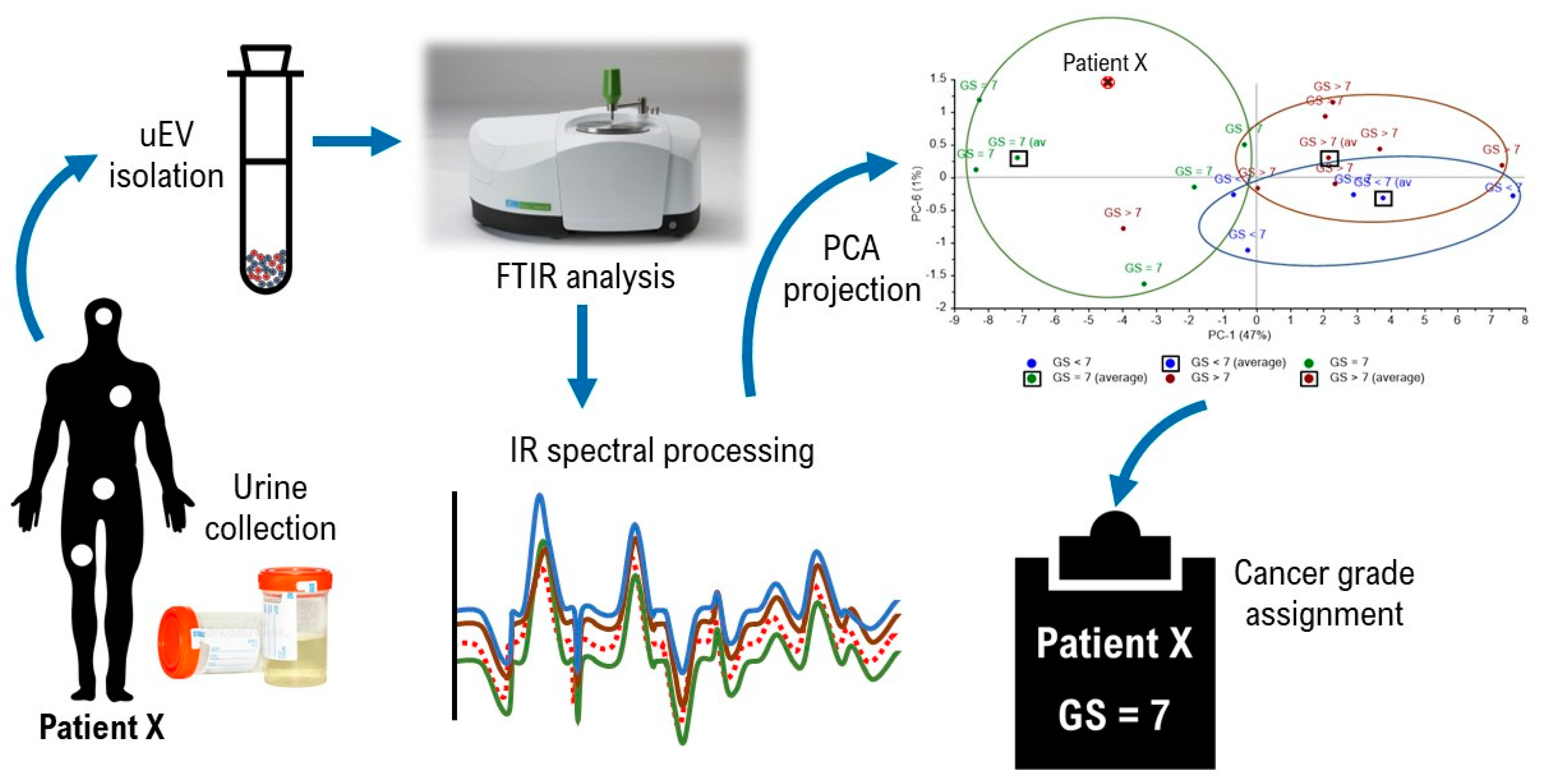
- 1 .
- Isolate extracellular vesicles (EVs) from urine samples
- Urine preserved in sodium azide (20 mM)
- Phosphate-buffered saline (PBS)
- Dithiothreitol (DTT) (1 M) in ultrapure water
- 50-mL polypropylene centrifuge tubes
- Ultracentrifuge and fixed-angle or swinging-bucket rotor
- Polyallomer tubes or polycarbonate bottles
- Vortex the urine samples for 30 s each. Transfer the fluid into 50-mL tubes.
- Centrifuge the urine samples at 400× g and 15,500× g for 20 min respectively.
- Carefully transfer the supernatant to ultracentrifuge tubes and spin at 200,000× g using a fixed-angle rotor for 90 min at 18 °C.
- Discard the supernatant and resuspend the pellet using 0.5 mL of 1M DTT and 1.5 mL of PBS.
- Incubate the suspensions at 37 °C for 10 min.
- Top up the suspensions with PBS and centrifuge the samples at 200,000× g using a swinging-bucket rotor for 70 min at 25 °C.
- Pour away the supernatant and resuspend the pellet using 100 µL of PBS.
- Quantify the protein content of resuspended EVs using Bradford assay.
- Store at −80 °C until further analysis.
- 2 .
- Acquire infrared (IR) spectra of urinary EVs using attenuated total reflection-Fourier transform infrared (ATR-FTIR) spectroscopy
- 0.45 µm Ultrafree-MC Centrifugal Filter (Merck)
- Phosphate-buffered saline (PBS)
- Ultrapure water
- Blow dryer
- FTIR spectrometer
- Dilute thawed urinary EVs to 0.1 μg/500 μL per tube and filter the samples using a 0.45 µm Ultrafree-MC Centrifugal Filter (Merck).
- Set the settings of the FTIR machine to 32 scans, 4 cm−1 resolution and wavelength from 4000 cm−1 to 400 cm−1.
- Acquire a background spectrum.
- Acquire the spectrum of ultrapure water and PBS.
- Clean the diamond with isopropanol.
- Thaw urinary EV sample and load 2 µL of the filtrate on the ATR diamond.
- Blow dry the solution on the diamond using a blow dryer.
- Run triplicates for each sample.
- Save the spectrum files (.sp format)
- 3 .
- Process acquired IR spectra and apply multivariate statistical analysis
- Spectrum files of urinary EVs, water and PBS
- The Unscrambler X 10.5.1 software
- Import all the extracellular vesicles spectra into the software (.sp format).
- Correct baseline by performing baseline correction.
- Perform unit vector normalization on the spectra.
- Apply derivative Savitsky−Golay transformation at the following parameters:
- Derivative order: 1
- Polynomial order: 2
- Smoothing points: 21
- Left point: 10
- Right points: 10
- Apply standard normal variate transformation.
- Apply principal component analysis (PCA) to the wavelength ranged between 1794–813 cm−1 at the following parameters:
- Number of principal components (PCs) included in analysis: 7
- Validation type: cross validation
- Generate PCA score plot using the first and second principal components (PC1 and PC2).
- 4 .
- Establish reference matrix and prostate cancer classification
- Urinary EVs of participants in transrectal ultrasound (TRUS)-guided prostate biopsy
- Excel/data science program e.g., Phyton for derivation of Euclidean distances
- Isolate EVs from urine samples collected from healthy individuals and patients diagnosed with different stages of prostate cancers (Gleason scores <7, =7 & >7).
- Acquire and process IR spectra from all the collected EV samples.
- Analyze all the processed spectra and generate a PCA score plot. This is the reference plot for sample analysis.
- Export the datasets of PC1 and PC2 from Unscrambler software.
- Import the datasets into Excel/Phyton for derivation of average points (centroids) in PCA score plots based on the mean of all samples in the respective clusters. These centroids are reference points for sample classification.
- Collect urinary EV samples from patients suspected of prostate cancer.
- Acquire and process IR spectra from the collected samples.
- Add the processed sample’s spectral data to the reference PCA dataset to obtain the coordination of the sample in score plot.
- Calculate the Euclidean distances between the sample’s point and the centroids of respective clusters.
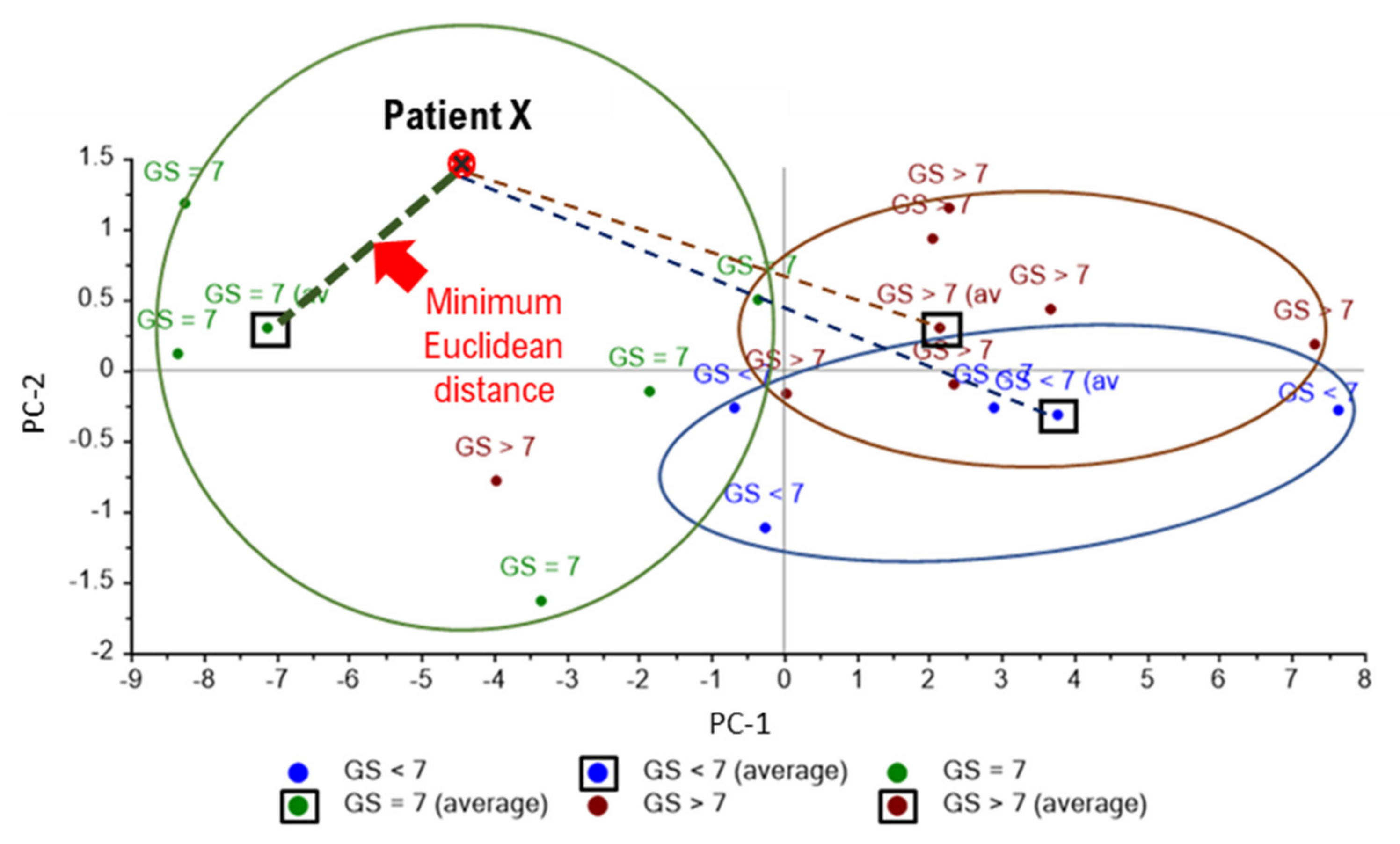
- Classify the sample base on minimum Euclidean distance, e.g., FTIR PCA analysis of urinary EVs shown in Figure A7 classified patient X in the cluster of Gleason score = 7
References
- GLOBOCAN 2020: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2020. Available online: http://globocan.iarc.fr/Default.aspx (accessed on 31 July 2021).
- Chou, R.; Croswell, J.M.; Dana, T.; Bougatsos, C.; Blazina, I.; Fu, R.; Gleitsmann, K.; Koenig, H.C.; Lam, C.; Maltz, A.; et al. Screening for prostate cancer: A review of the evidence for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2011, 155, 762–771. [Google Scholar] [CrossRef]
- Malati, T.; Kumari, G.R.; Murthy, P.V.L.N.; Reddy, C.R.; Prakash, B.S. Prostate specific antigen in patients of benign prostate hypertrophy and carcinoma prostate. Indian J. Clin. Biochem. 2006, 21, 34–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Dong, B.; Qu, W.; Wang, J.; Xu, Y.; Yu, S.; Zhang, X. Using clinical parameters to predict prostate cancer and reduce the unnecessary biopsy among patients with PSA in the gray zone. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.P.; Balaj, L.; Zaborowski, M.P.; Breakefield, X.O. Extracellular vesicles: Composition, biological relevance, and methods of study. BioScience 2015, 65, 783–797. [Google Scholar]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Øverbye, A.; Skotland, T.; Koehler, C.J.; Thiede, B.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015, 6, 30357–30376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, K.; Kume, H.; Matsuzaki, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Uemura, M.; Miyagawa, Y.; Tomonaga, T.; Nonomura, N. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci. Rep. 2017, 7, 42961. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef]
- Bryzgunova, O.E.; Zaripov, M.M.; Skvortsova, T.E.; Lekchnov, E.A.; Grigor’eva, A.E.; Zaporozhchenko, I.A.; Morozkin, E.S.; Ryabchikova, E.I.; Yurchenko, Y.B.; Voitsitskiy, V.E.; et al. Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS ONE 2016, 11, e0157566. [Google Scholar] [CrossRef]
- Foj, L.; Ferrer, F.; Serra, M.; Arévalo, A.; Gavagnach, M.; Giménez, N.; Filella, X. Exosomal and non-exosomal urinary miRNAs in prostate cancer detection and prognosis. Prostate 2017, 77, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; McCague, S.B.; Munro, B.R.; Predebon, M.J.; Scarlett, C.J.; Skelding, K.A.; Weidenhofer, J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018, 17, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavoosidana, G.; Ronquist, G.; Darmanis, S.; Yan, J.; Carlsson, L.; Wu, D.; Conze, T.; Ek, P.; Semjonow, A.; Eltze, E.; et al. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 8809–8814. [Google Scholar] [CrossRef] [Green Version]
- Motamedinia, P.; Scott, A.N.; Bate, K.L.; Sadeghi, N.; Salazar, G.; Shapiro, E.; Ahn, J.; Lipsky, M.; Lin, J.; Hruby, G.W.; et al. Urine exosomes for non-invasive assessment of gene expression and mutations of prostate cancer. PLoS ONE 2016, 11, e0154507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malins, D.C.; Polissar, N.L.; Gunselman, S.J. Models of DNA structure achieve almost perfect discrimination between normal prostate, benign prostatic hyperplasia (BPH), and adenocarcinoma and have a high potential for predicting BPH and prostate cancer. Proc. Natl. Acad. Sci. USA 1997, 94, 259–264. [Google Scholar] [CrossRef] [Green Version]
- Mihály, J.; Deák, R.; Szigyártó, I.C.; Bóta, A.; Beke-Somfai, T.; Varga, Z. Characterization of extracellular vesicles by IR spectroscopy: Fast and simple classification based on amide and CH stretching vibrations. Biochim. Biophys. Acta Biomembr. 2017, 1859, 459–466. [Google Scholar] [CrossRef]
- Kneipp, J.; Beekes, M.; Lasch, P.; Naumann, D. Molecular changes of preclinical scrapie can be detected by infrared spectroscopy. J. Neurosci. 2002, 22, 2989–2997. [Google Scholar] [CrossRef] [Green Version]
- Heraud, P.; Caine, S.; Campanale, N.; Karnezis, T.; McNaughton, D.; Wood, B.R.; Tobin, M.J.; Bernard, C.C. Early detection of the chemical changes occurring during the induction and prevention of autoimmune-mediated demyelination detected by FT-IR imaging. Neuroimage 2010, 49, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Yap, X.L.; Ong, T.A.; Lim, J.; Wood, B.; Lee, W.L. Study of prostate cancer-derived extracellular vesicles in urine using IR spectroscopy. Prog. Drug Dis. Biomed. Sci. 2019, 2, 1. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Medipally, D.K.; Nguyen, T.N.Q.; Bryant, J.; Untereiner, V.; Sockalingum, G.D.; Cullen, D.; Noone, E.; Bradshaw, S.; Finn, M.; Dunne, M.; et al. Monitoring radiotherapeutic response in prostate cancer patients using high throughput FTIR spectroscopy of liquid biopsies. Cancers 2019, 11, 925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonova, D.; Karamancheva, I. Application of Fourier transform infrared spectroscopy for tumor diagnosis. Biotechnol. Biotechnol. Equip. 2013, 27, 4200–4207. [Google Scholar] [CrossRef]
- Gazi, E.; Dwyer, J.; Gardner, P.; Ghanbari-Siahkali, A.; Wade, A.P.; Miyan, J.; Lockyer, N.P.; Vickerman, J.C.; Clarke, N.W.; Shanks, J.H.; et al. Applications of Fourier transform infrared microspectroscopy in studies of benign prostate and prostate cancer. A pilot study. J. Pathol. J. Pathol. Soc. Great Br. Irel. 2003, 201, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Martins, Á.M.; Ramos, C.C.; Freitas, D.; Reis, C.A. Glycosylation of cancer extracellular vesicles: Capture strategies, functional roles and potential clinical applications. Cells 2021, 10, 109. [Google Scholar] [CrossRef]
- Carnino, J.M.; Ni, K.; Jin, Y. Post-translational modification regulates formation and cargo-loading of extracellular vesicles. Front. Immunol. 2020, 11, 948. [Google Scholar] [CrossRef] [PubMed]
- Su, K.Y.; Lee, W.L. Fourier transform infrared spectroscopy as a cancer screening and diagnostic tool: A review and prospects. Cancers 2020, 12, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Patient | Histologic Gleason Grade | Histologic Gleason Score | Actual Gleason Score Grouping | Predicted Gleason Score Grouping | Tumor Stage | PSA (ng/mL) |
|---|---|---|---|---|---|---|
| 32 | 5 + 4 | 9 | GS > 7 | GS < 7 | T4N1M1b | 806 |
| 33 | 3 + 3 | 6 | GS < 7 | GS < 7 | T2aN0M0 | 7 |
| 34 | 3 + 4 | 7 | GS = 7 | GS = 7 | T2cN0M1b | 73 |
| 35 | 4 + 4 | 8 | GS > 7 | GS > 7 | T3aN0M0 | 7 |
| 36 | 3 + 4 | 7 | GS = 7 | GS = 7 | T2cNxMx | 25 |
| 37 | 3 + 4 | 7 | GS = 7 | GS = 7 | T2aN0M0 | 4 |
| 38 | 4 + 4 | 8 | GS > 7 | GS < 7 | T2bN0M0 | 121 |
| 39 | 3 + 4 | 7 | GS = 7 | GS = 7 | T1aNxM0 | 13 |
| 40 | 3 + 4 | 7 | GS = 7 | GS = 7 | T2aN0Mx | 6 |
| 41 | 3 + 3 | 6 | GS < 7 | GS > 7 | T2aN0M0 | 10 |
| 42 | 3 + 3 | 6 | GS < 7 | GS < 7 | T2cN0M0 | 33 |
| 43 | 5 + 4 | 9 | GS > 7 | GS < 7 | T3bN0M0 | 7 |
| 44 | 4 + 5 | 9 | GS > 7 | GS = 7 | T4N1M1b | 7 |
| 45 | 4 + 4 | 8 | GS > 7 | GS > 7 | T2cN0M1b | 188 |
| 46 | 5 + 4 | 9 | GS > 7 | GS = 7 | TxNxM1b | 11 |
| 47 | 3 + 3 | 6 | GS < 7 | GS = 7 | T2cN0M0 | 8 |
| 48 | 5 + 5 | 10 | GS > 7 | GS > 7 | T4N1M1c | 231 |
| 49 | 4 + 4 | 8 | GS > 7 | GS > 7 | TxNxM1b | 1178 |
| 50 | 4 + 5 | 9 | GS > 7 | GS = 7 | TxNxM1b | 260 |
| 51 | 5 + 5 | 10 | GS > 7 | GS = 7 | T4N1M1b | 256 |
| 52 | 4 + 4 | 8 | GS > 7 | GS = 7 | TxNxM1b | 1178 |
| 53 1 | n.a. 2 | n.a. 2 | GS > 7 | GS = 7 | TxNxM1b | 101 |
| Status | Predicted Group Membership | Total | ||
|---|---|---|---|---|
| Non-Cancerous | Cancerous | |||
| Count | Non-cancerous | 25 | 6 | 31 |
| Cancerous | 9 | 13 | 22 | |
| % | Non-cancerous | 81 | 19 | 100 |
| Cancerous | 41 | 59 | 100 | |
| Gleason Score | Predicted Group Membership | Total | |||
|---|---|---|---|---|---|
| GS < 7 | GS = 7 | GS > 7 | |||
| Count | GS < 7 | 2 | 1 | 1 | 4 |
| GS = 7 | 0 | 5 | 0 | 5 | |
| GS > 7 | 3 | 6 | 4 | 13 | |
| % | GS < 7 | 50 | 25 | 25 | 100 |
| GS = 7 | 0 | 100 | 0 | 100 | |
| GS > 7 | 23 | 46 | 31 | 100 | |
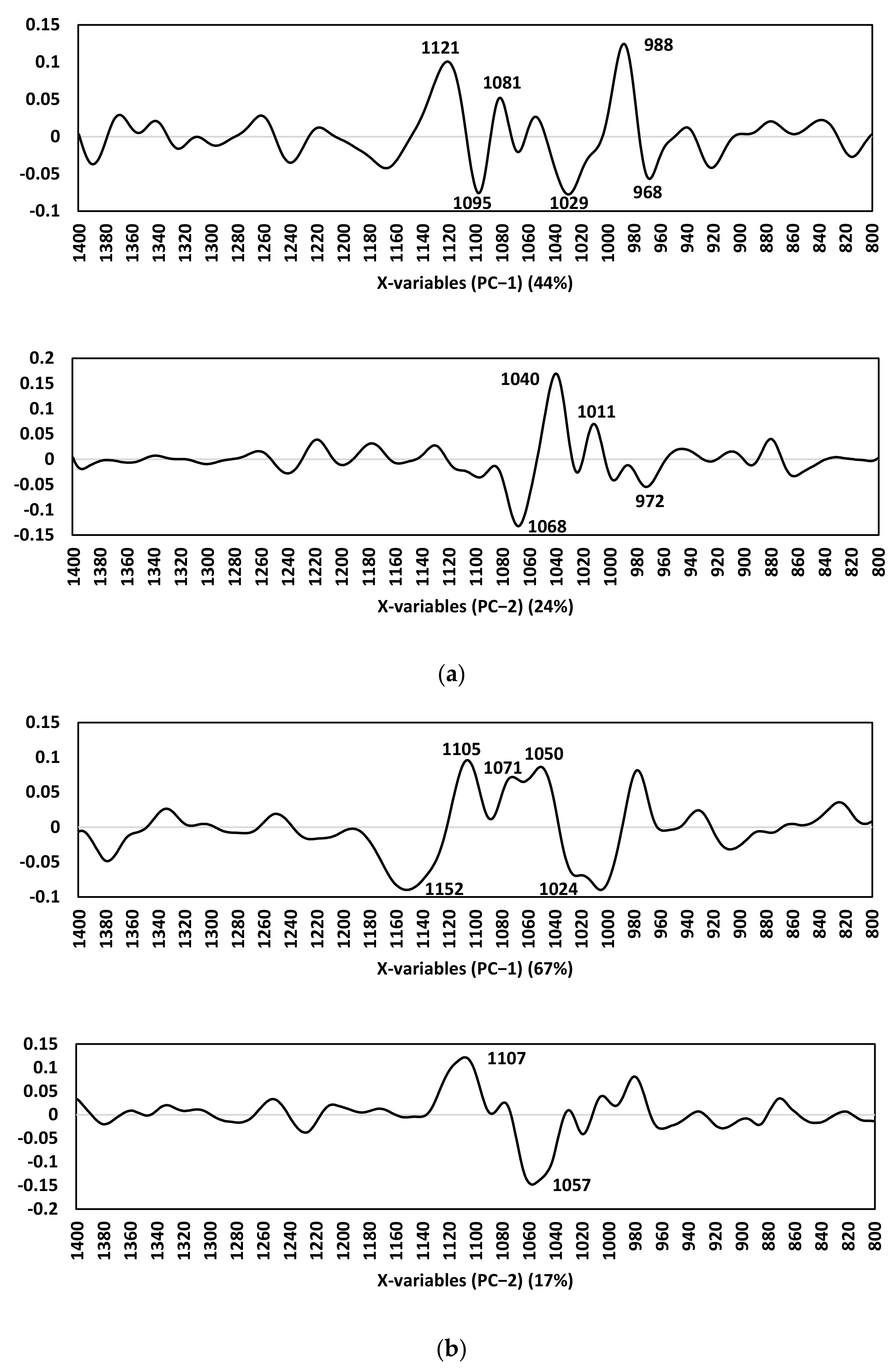
| (a) | Direction | Wavenumber (cm−1) | Proposed Biomolecular Assignment * |
| +ve | 988 | RNA stretching, ring bending of uracil | |
| +ve | 1081 | Symmetric phosphate stretching modes ʋ(PO2−) symmetric stretching of phosphodiesters Phosphate I in RNA One of the triad peaks of nucleic acids | |
| +ve | 1121 | Symmetric phosphodiester stretching band, RNA Shoulder of 1121 cm−1 band due to RNA | |
| −ve | 968 | C-N-C stretch: nucleic acids | |
| −ve | 1029/30 | O-CH3 stretching of methoxy groups | |
| −ve | 1095 | Stretching PO2− symmetric | |
| (b) | Direction | Wavenumber (cm−1) | Proposed Biomolecular Assignment * |
| +ve | 1011 | CHα,α’ out-of-plane bending and Cα = Cα’ torsion | |
| +ve | 1040 | Stretching CO ribose | |
| −ve | 972 | OCH3 (polysaccharides, pectin) | |
| −ve | 1068 | Stretching CO ribose | |
| (c) | Direction | Wavenumber (cm−1) | Proposed Biomolecular Assignment * |
| +ve | 1050 | ʋs CO-O-C C-O stretching coupled with C-O bending of the C-OH of carbohydrates Glycogen | |
| +ve | 1071 | Phosphate I band for 2 different CO vibrations of deoxyribose in DNA in disordering structure | |
| +ve | 1105 | Carbohydrates | |
| −ve | 1024 | Glycogen (CO stretch associated with glycogen) | |
| −ve | 1152 | Glycogen absorption due to CO and CC stretching, and COH deformation motions | |
| (d) | Direction | Wavenumber (cm−1) | Proposed Biomolecular Assignment * |
| +ve | 1107 | ʋ(CO), ʋ(CC), ring (polysaccharides, pectin) | |
| −ve | 1057 | Stretching CO deoxyribose |
| Range of PSA Levels [ng/mL] | Area Under Curve (AUC) | |
|---|---|---|
| PSA | FTIR Analysis of Urinary EVs | |
| 4–1200 | 0.724 | 0.723 |
| 4–20 | 0.437 | 0.700 |
| 4–10 | 0.447 | 0.632 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yap, X.-L.; Wood, B.; Ong, T.-A.; Lim, J.; Goh, B.-H.; Lee, W.-L. Detection of Prostate Cancer via IR Spectroscopic Analysis of Urinary Extracellular Vesicles: A Pilot Study. Membranes 2021, 11, 591. https://doi.org/10.3390/membranes11080591
Yap X-L, Wood B, Ong T-A, Lim J, Goh B-H, Lee W-L. Detection of Prostate Cancer via IR Spectroscopic Analysis of Urinary Extracellular Vesicles: A Pilot Study. Membranes. 2021; 11(8):591. https://doi.org/10.3390/membranes11080591
Chicago/Turabian StyleYap, Xin-Le, Bayden Wood, Teng-Aik Ong, Jasmine Lim, Bey-Hing Goh, and Wai-Leng Lee. 2021. "Detection of Prostate Cancer via IR Spectroscopic Analysis of Urinary Extracellular Vesicles: A Pilot Study" Membranes 11, no. 8: 591. https://doi.org/10.3390/membranes11080591
APA StyleYap, X.-L., Wood, B., Ong, T.-A., Lim, J., Goh, B.-H., & Lee, W.-L. (2021). Detection of Prostate Cancer via IR Spectroscopic Analysis of Urinary Extracellular Vesicles: A Pilot Study. Membranes, 11(8), 591. https://doi.org/10.3390/membranes11080591










