Lipid Transporters Beam Signals from Cell Membranes
Abstract
:1. Introduction
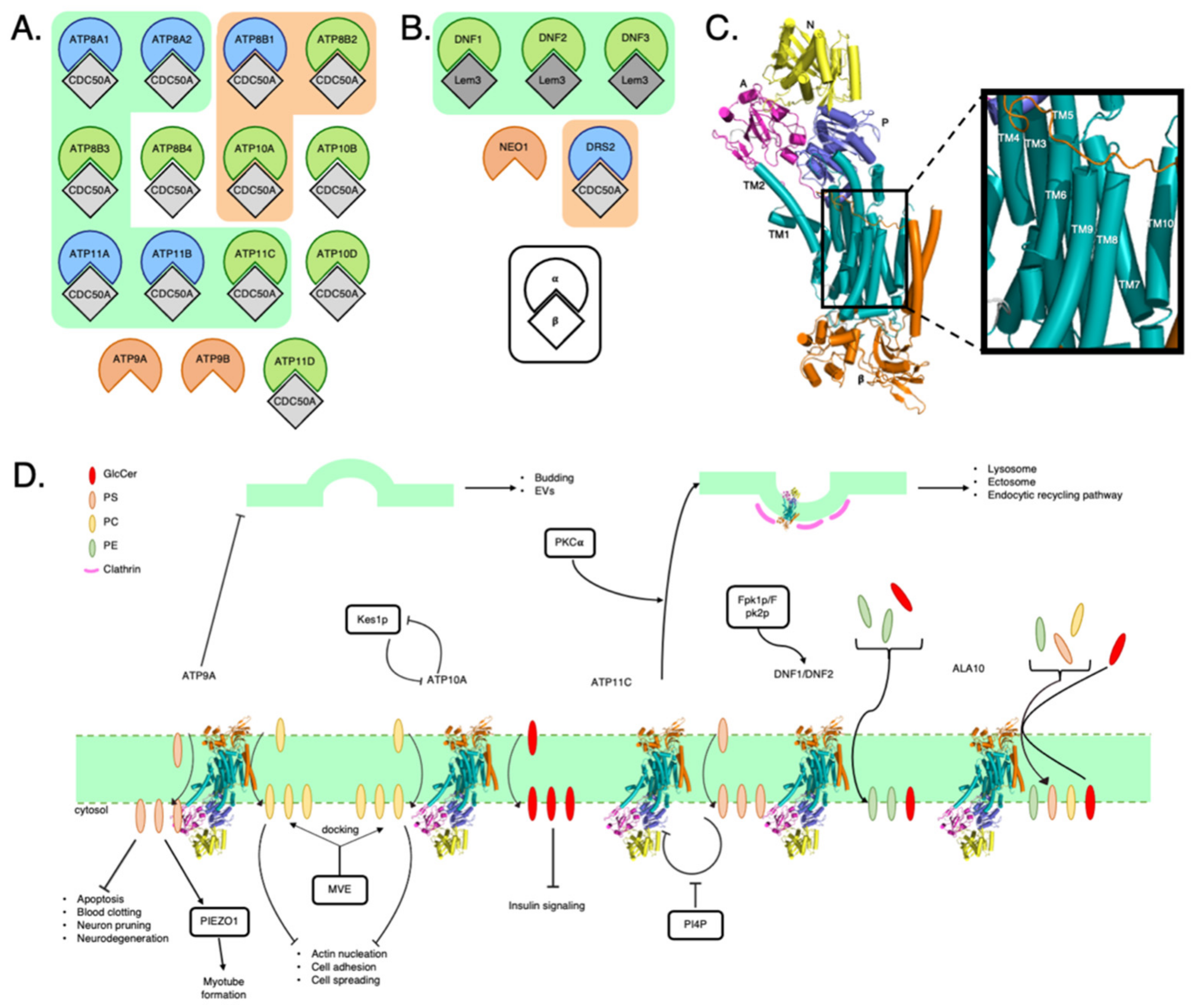
2. P4-ATPase Phospholipid Transporters
2.1. P4-ATPase Structure
2.2. P4-ATPases and How They Affect Membrane Composition
2.3. Lipid Scavenging
2.4. P4-ATPases and Cellular Signaling
2.5. Membrane Curvature
2.6. Coat Protein Recruitment
2.7. Cytoskeleton Modulator
3. ABC Sterol Transporters
3.1. ABC Transporter Structure
3.2. ABC Transporters in the Lipid Raft
3.3. Role of ABCA1 in the Intramembranary Cholesterol Movement
3.4. ABCG Cholesterol Transporters Sinking Rafts
3.5. Twin Brothers in Apoptosis?
3.6. Sterol-Sensing Domain (SSD) Tweaking Protein-Lipid Interactions
3.7. ABC Transporters and Cellular Signaling
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horn, A.; Jaiswal, J.K. Cellular mechanisms and signals that coordinate plasma membrane repair. Cell. Mol. Life Sci. 2018, 75, 3751–3770. [Google Scholar] [CrossRef]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef] [Green Version]
- Sonnino, S.; Prinetti, A. Membrane domains and the lipid raft concept. Curr. Med. Chem. 2013, 20. [Google Scholar] [CrossRef]
- Cheney, P.P.; Weisgerber, A.W.; Feuerbach, A.M.; Knowles, M.K. Single lipid molecule dynamics on supported lipid bilayers with membrane curvature. Membranes 2017, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Kalappurakkal, J.M.; Sil, P.; Mayor, S. Toward a new picture of the living plasma membrane. Protein Sci. 2020, 29, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Spector, A.A.; Yorek, M.A. Membrane lipid composition and cellular function. J. Lipid Res. 1985, 26, 1015–1035. [Google Scholar] [CrossRef]
- Sleight, R.G.; Pagano, R.E. Transport of a fluorescent phosphatidylcholine analog from the plasma membrane to the golgi apparatus. J. Cell Biol. 1984, 99, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.J.; Hossain, K.R.; Cao, K. Physiological roles of transverse lipid asymmetry of animal membranes. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183382. [Google Scholar] [CrossRef] [PubMed]
- Doktorova, M.; Symons, J.L.; Levental, I. Structural and functional consequences of reversible lipid asymmetry in living membranes. Nat. Chem. Biol. 2020, 16, 1321–1330. [Google Scholar] [CrossRef]
- Martin, S.J.; Finucane, D.M.; Amarante-Mendes, G.P.; O’Brien, G.A.; Green, D.R. Phosphatidylserine externalization during CD95-induced apoptosis of cells and cytoplasts requires ICE/CED-3 protease activity. J. Biol. Chem. 1996, 271, 28753–28756. [Google Scholar] [CrossRef] [Green Version]
- Aupeix, K.; Hugel, B.; Martin, T.; Bischoff, P.; Lill, H.; Pasquali, J.L.; Freyssinet, J.M. The significance of shed membrane particles during programmed cell death in vitro, and in vivo, in HIV-1 infection. J. Clin. Investig. 1997, 99, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Bevers, E.M.; Comfurius, P.; Zwaal, R.F.A. Changes in membrane phospholipid distribution during platelet activation. BBA Biomembr. 1983, 736, 57–66. [Google Scholar] [CrossRef]
- Lentz, B.R. Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog. Lipid Res. 2003, 42, 423–438. [Google Scholar] [CrossRef]
- Jones, M.E.; Lentz, B.R.; Dombrose, F.A.; Sandberg, H. Comparison of the abilities of synthetic and platelet-derived membranes to enhance thrombin formation. Thromb. Res. 1985, 39, 711–724. [Google Scholar] [CrossRef]
- Nieuwland, R.; Berckmans, R.J.; Rotteveel-Eijkman, R.C.; Maquelin, K.N.; Roozendaal, K.J.; Jansen, P.G.M.; Ten Have, K.T.; Eijsman, L.; Hack, C.E.; Sturk, A. Cell-derived microparticles generated in patients during cardiopulmonary bypass are highly procoagulant. Circulation 1997, 96, 3534–3541. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.G.; Monteiro, R.Q. Activation of blood coagulation in cancer: Implications for tumour progression. Biosci. Rep. 2013, 33, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Reid, V.L.; Webster, N.R. Role of microparticles in sepsis. Br. J. Anaesth. 2012, 109, 503–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tormoen, G.W.; Cianchetti, F.A.; Bock, P.E.; McCarty, O.J.T. Development of coagulation factor probes for the identification of procoagulant circulating tumor cells. Front. Oncol. 2012, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berny-Lang, M.A.; Aslan, J.E.; Tormoen, G.W.; Patel, I.A.; Bock, P.E.; Gruber, A.; McCarty, O.J.T. Promotion of experimental thrombus formation by the procoagulant activity of breast cancer cells. Phys. Biol. 2011, 8, 15014. [Google Scholar] [CrossRef] [Green Version]
- Utsugi, T.; Schroit, A.J.; Connor, J.; Bucana, C.D.; Fidler, I.J. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res. 1991, 51, 3062–3066. [Google Scholar]
- Daleke, D.L. Phospholipid flippases. J. Biol. Chem. 2007, 282, 821–825. [Google Scholar] [CrossRef] [Green Version]
- Menon, A.K.; Watkins, W.E.; Hrafnsdóttir, S. Specific proteins are required to translocate phosphatidylcholine bidirectionally across the endoplasmic reticulum. Curr. Biol. 2000, 10, 241–252. [Google Scholar] [CrossRef] [Green Version]
- Seigneuret, M.; Devauxt, P.F. ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: Relation to shape changes (phospholipid asymmetry/transverse diffusion/erythrocyte shape/bilayer couple/electron spin resonance). Proc. Natl. Acad. Sci. USA 1984, 81, 3751–3755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, W.R.; Bell, R.M. Assembly of the endoplasmic reticulum phospholipid bilayer: The phosphatidylcholine transporter. Cell 1985, 42, 51–60. [Google Scholar] [CrossRef]
- Devaux, P.F.; Dan, B.S. Transmembrane lipid trafficking facts and speculations. Traffic 1998, 49, 195–202. [Google Scholar]
- Montigny, C.; Lyons, J.; Champeil, P.; Nissen, P.; Lenoir, G. On the molecular mechanism of flippase- and scramblase-mediated phospholipid transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 767–783. [Google Scholar] [CrossRef] [PubMed]
- Williamson, P.; Bevers, E.M.; Smeets, E.F.; Comfurius, P.; Schlegel, R.A.; Zwaal, R.F.A. Continuous analysis of the mechanism of activated transbilayer lipid movement in platelets. Biochemistry 1995, 34, 10448–10455. [Google Scholar] [CrossRef]
- Coleman, J.A.; Quazi, F.; Molday, R.S. Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 555–574. [Google Scholar] [CrossRef] [Green Version]
- Zwaal, R.F.A.; Comfurius, P.; Bevers, E.M. Surface exposure of phosphatidylserine in pathological cells. Cell. Mol. Life Sci. 2005, 62, 971–988. [Google Scholar] [CrossRef]
- Folmer, D.E.; Elferink, R.P.J.O.; Paulusma, C.C. P4 ATPases-Lipid flippases and their role in disease. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2009, 1791, 628–635. [Google Scholar] [CrossRef]
- Chaudhry, F.; Kawai, H.; Johnson, K.W.; Narula, N.; Shekhar, A.; Nakahara, T.; Tanimoto, T.; Kim, D.; Adapoe, M.K.M.Y.; Blankenberg, F.G.; et al. Molecular imaging of apoptosis in atherosclerosis by targeting cell membrane phospholipid asymmetry. J. Am. Coll. Cardiol. 2020, 76, 1862–1874. [Google Scholar] [CrossRef]
- Bai, L.; You, Q.; Jain, B.K.; Duan, H.D.; Kovach, A.; Graham, T.R.; Li, H. Transport mechanism of P4 ATPase phosphatidylcholine flippases. Elife 2020, 9. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004, 5, 282–295. [Google Scholar] [CrossRef]
- Okamoto, S.; Naito, T.; Shigetomi, R.; Kosugi, Y.; Nakayama, K.; Takatsu, H.; Shin, H.W. The N- or C-terminal cytoplasmic regions of P4-ATPases determine their cellular localization. Mol. Biol. Cell 2020, 31, 2115–2124. [Google Scholar] [CrossRef]
- Andersen, J.P.; Vestergaard, A.L.; Mikkelsen, S.A.; Mogensen, L.S.; Chalat, M.; Molday, R.S. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front. Physiol. 2016, 7, 275. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, A.L.; Coleman, J.A.; Lemmin, T.; Mikkelsen, S.A.; Molday, L.L.; Vilsen, B.; Molday, R.S.; Dal Peraro, M.; Andersen, J.P. Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. USA 2014, 111, e1334. [Google Scholar] [CrossRef] [Green Version]
- Sebastian, T.T.; Baldridge, R.D.; Xu, P.; Graham, T.R. Phospholipid flippases: Building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2012, 1821, 1068–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiraizumi, M.; Yamashita, K.; Nishizawa, T.; Nureki, O. Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 2021, 365, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Timcenko, M.; Lyons, J.A.; Januliene, D.; Ulstrup, J.J.; Dieudonné, T.; Montigny, C.; Ash, M.R.; Karlsen, J.L.; Boesen, T.; Kühlbrandt, W.; et al. Structure and autoregulation of a P4-ATPase lipid flippase. Nature 2019, 571, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Kovach, A.; You, Q.; Hsu, H.C.; Zhao, G.; Li, H. Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Xu, J.; Wu, X.; Li, L. Structures of a P4-ATPase lipid flippase in lipid bilayers. Protein Cell 2020, 11, 458–463. [Google Scholar] [CrossRef] [Green Version]
- Palmgren, M.; Østerberg, J.T.; Nintemann, S.J.; Poulsen, L.R.; López-Marqués, R.L. Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1135–1151. [Google Scholar] [CrossRef]
- Chalat, M.; Moleschi, K.; Molday, R.S.; Martin, T.F.J. C-terminus of the P4-ATPase ATP8A2 functions in protein folding and regulation of phospholipid flippase activity. Mol. Biol. Cell 2017, 28, 452–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roland, B.P.; Naito, T.; Best, J.T.; Arnaiz-Yépez, C.; Takatsu, H.; Yu, R.J.; Shin, H.W.; Graham, T.R. Yeast and human P4-ATPases transport glycosphingolipids using conserved structural motifs. J. Biol. Chem. 2019, 294, 1794–1806. [Google Scholar] [CrossRef] [Green Version]
- Westerlund, B.; Slotte, J.P. How the molecular features of glycosphingolipids affect domain formation in fluid membranes. Biochim. Biophys. Acta Biomembr. 2009, 1788, 194–201. [Google Scholar] [CrossRef] [Green Version]
- Reza, S.; Ugorski, M.; Suchański, J. Glucosylceramide and galactosylceramide, small glycosphingolipids with significant impact on health and disease. Glycobiology 2021. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, R.D.; Xu, P.; Graham, T.R. Type IV p-type ATPases distinguish mono-versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J. Biol. Chem. 2013, 288, 19516–19527. [Google Scholar] [CrossRef] [Green Version]
- Riekhof, W.R.; Wu, J.; Gijó, M.A.; Zarini, S.; Murphy, R.C.; Voelker, D.R. Lysophosphatidylcholine Metabolism in Saccharomyces cerevisiae. The role of p-type atpases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 2007, 282, 36853–36861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riekhof, W.R.; Voelker, D.R. Uptake and Utilization of Lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 36588–36596. [Google Scholar] [CrossRef] [Green Version]
- Roland, B.P.; Graham, T.R. Decoding P4-ATPase substrate interactions. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Kilian, N.; Choi, J.Y.; Voelker, D.R.; Mamoun, C. Ben Role of phospholipid synthesis in the development and differentiation of malaria parasites in the blood. J. Biol. Chem. 2018, 293, 17308–17316. [Google Scholar] [CrossRef] [Green Version]
- Voelker, D.R. Interorganelle transport of aminoglycerophospholipids. Biochim. Biophys. Acta 2000, 1486, 97–107. [Google Scholar] [CrossRef]
- Nintemann, S.J.; Palmgren, M.; López-Marqués, R.L. Catch you on the flip side: A critical review of flippase mutant phenotypes. Trends Plant Sci. 2019, 24, 468–478. [Google Scholar] [CrossRef]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Wang, W.; Xie, Q.; Liu, N.; Liu, L.; Wang, D.; Zhang, X.; Yang, C.; Chen, X.; Tang, D.; et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, B.K.; Roland, B.P.; Graham, T.R. Exofacial membrane composition and lipid metabolism regulates plasma membrane P4-ATPase substrate specificity. J. Biol. Chem. 2020, 295, 17997–18009. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, B.P.; Raychaudhuri, S.; Natarajan, P.; Abe, F.; Liu, K.; Prinz, W.A.; Graham, T.R. Control of protein and sterol trafficking by antagonistic activities of a type IV P-type ATPase and oxysterol binding protein homologue. Mol. Biol. Cell 2009, 20, 2920–2931. [Google Scholar] [CrossRef] [Green Version]
- Hankins, H.M.; Sere, Y.Y.; Diab, N.S.; Menon, A.K.; Graham, T.R. Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol. Biol. Cell 2015, 26, 4674–4685. [Google Scholar] [CrossRef]
- Lyssenko, N.N.; Miteva, Y.; Gilroy, S.; Hanna-Rose, W.; Schlegel, R.A. An unexpectedly high degree of specialization and a widespread involvement in sterol metabolism among the C. elegans putative aminophospholipid translocases. BMC Dev. Biol. 2008, 8, 96. [Google Scholar] [CrossRef] [Green Version]
- Ruaud, A.F.; Nilsson, L.; Richard, F.; Larsen, M.K.; Bessereau, J.L.; Tuck, S. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic 2009, 10, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Tuck, S. Extracellular vesicles: Budding regulated by a phosphatidylethanolamine translocase. Curr. Biol. 2011, 21, R988–R990. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Xu, L.; Zhang, P.; Ouyang, K.; Xiao, Y.; Xiong, J.; Wang, D.; Liang, Y.; Duan, L. Effects of ATP9A on extracellular vesicle release and exosomal lipid composition. Oxid. Med. Cell. Longev. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Norris, A.; Graham, T.; Stafford, J.; Zhu, L. Exploring the role of ATP10A in diet-induced obesity, insulin resistance, and type 2 diabetes. FASEB J. 2021, 35. [Google Scholar] [CrossRef]
- Van Niel, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Publ. Gr. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Naik, J.; Hau, C.M.; ten Bloemendaal, L.; Mok, K.S.; Hajji, N.; Wehman, A.M.; Meisner, S.; Muncan, V.; Paauw, N.J.; de Vries, H.E.; et al. The P4-ATPase ATP9A is a novel determinant of exosome release. PLoS ONE 2019, 14, e0213069. [Google Scholar] [CrossRef]
- Mattioli, F.; Darvish, H.; Paracha, S.A.; Tafakhori, A.; Firouzabadi, S.G.; Chapi, M.; Muhammad, H.; Baig, A.; Reymond, A.; Antonarakis, S.E.; et al. Biallelic truncation variants in ATP9A are associated with a novel autosomal recessive neurodevelopmental disorder authors. medRxiv 2021. [Google Scholar] [CrossRef]
- Takatsu, H.; Takayama, M.; Naito, T.; Takada, N.; Tsumagari, K.; Ishihama, Y.; Nakayama, K.; Shin, H.W. Phospholipid flippase ATP11C is endocytosed and downregulated following Ca2+ mediated protein kinase C activation. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirnikjoo, B.; Balasubramanian, K.; Schroit, A.J. Suicidal membrane repair regulates phosphatidylserine externalization during apoptosis. J. Biol. Chem. 2009, 284, 22512–22516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapar, M.L.; Ji, H.; Wang, B.; Poe, A.R.; Dubey, K.; Ren, X.; Ni, J.Q.; Han, C. Phosphatidylserine externalization results from and causes neurite degeneration in drosophila. Cell Rep. 2018, 24, 2273–2286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchiya, M.; Hara, Y.; Okuda, M.; Itoh, K.; Nishioka, R.; Shiomi, A.; Nagao, K.; Mori, M.; Mori, Y.; Ikenouchi, J.; et al. Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Yamamoto, T.; Kishimoto, T.; Noji, T.; Tanaka, K. Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol. Biol. Cell 2008, 19, 1783–1797. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Mioka, T.; Tanaka, K.; Nagatani, A. An optogenetic system to control membrane phospholipid asymmetry through flippase activation in budding yeast. Sci. Rep. 2020, 10, 12474. [Google Scholar] [CrossRef] [PubMed]
- Siggs, O.M.; Schnabl, B.; Webb, B.; Beutler, B. X-linked cholestasis in mouse due to mutations of the P4-ATPase ATP11C. Proc. Natl. Acad. Sci. USA 2011, 108, 7890–7895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platre, M.P.; Noack, L.C.; Doumane, M.; Bayle, V.; Simon, M.L.A.; Maneta-Peyret, L.; Fouillen, L.; Stanislas, T.; Armengot, L.; Pejchar, P.; et al. A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev. Cell 2018, 45, 465–480.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takada, N.; Naito, T.; Inoue, T.; Nakayama, K.; Takatsu, H.; Shin, H. Phospholipid-flipping activity of P4- ATP ase drives membrane curvature. EMBO J. 2018, 37, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Furuta, N.; Fujimura-Kamada, K.; Saito, K.; Yamamoto, T.; Tanaka, K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell 2007, 18, 295–312. [Google Scholar] [CrossRef]
- Kook, S.; Wang, P.; Meng, S.; Jetter, C.S.; Sucre, J.M.S.; Benjamin, J.T.; Gokey, J.J.; Hanby, H.A.; Jaume, A.; Goetzl, L.; et al. AP-3-dependent targeting of flippase ATP8A1 to lamellar bodies suppresses activation of YAP in alveolar epithelial type 2 cells. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Naito, T.; Takatsu, H.; Miyano, R.; Takada, N.; Nakayama, K.; Shin, H.W. Phospholipid flippase ATP10A translocates phosphatidylcholine and is involved in plasma membrane dynamics. J. Biol. Chem. 2015, 290, 15004–15017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, H.T.; Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011, 12, 517–533. [Google Scholar] [CrossRef]
- Hinners, I.; Tooze, S.A. Changing directions: Clathrin-mediated transport between the Golgi and endosomes. J. Cell Sci. 2003, 116, 763–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Surendhran, K.; Nothwehr, S.F.; Graham, T.R. P4-ATPase requirement for AP-1/Clathrin function in protein transport from the trans-golgi network and early endosomes. Mol. Biol. Cell 2008, 19, 3526–3535. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Sun, K.; Liu, W.; Li, X.; Tian, W.; Shuai, P.; Zhu, X. The phosphatidylserine flippase β-subunit Tmem30a is essential for normal insulin maturation and secretion. Mol. Ther. 2021. [Google Scholar] [CrossRef]
- Chen, C.Y.; Ingram, M.F.; Rosal, P.H.; Graham, T.R. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 1999, 147, 1223–1236. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, T.; Yamamoto, T.; Tanaka, K. Defects in structural integrity of ergosterol and the Cdc50p-Drs2p putative phospholipid translocase cause accumulation of endocytic membranes, onto which actin patches are assembled in yeast. Mol. Biol. Cell 2005, 16, 5592–5609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Ding, X. Characterization and expression of mouse Cdc50c during spermatogenesis. Acta Biochim. Biophys. Sin. 2007, 39, 739–744. [Google Scholar] [CrossRef] [Green Version]
- López-Marqués, R.L.; Poulsen, L.R.; Hanisch, S.; Meffert, K.; Buch-Pedersen, M.J.; Jakobsen, M.K.; Pomorski, T.G.; Palmgren, M.G. Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA α-subunit. Mol. Biol. Cell 2010, 21, 791. [Google Scholar] [CrossRef] [Green Version]
- Hua, Z.; Fatheddin, P.; Graham, T.R. An essential subfamily of Drs2p-related P-Type ATPases Is required for protein trafficking between golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 2002, 13, 3162. [Google Scholar] [CrossRef] [Green Version]
- Pomorski, T.; Lombardi, R.; Riezman, H.; Devaux, P.F.; Van Meer, G.; Holthuis, J.C.M. Drs2p-related P-type ATPases Dnflp and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 2003, 14, 1240–1254. [Google Scholar] [CrossRef] [Green Version]
- Paulusma, C.C.; Folmer, D.E.; Ho-Mok, K.S.; de Waart, D.R.; Hilarius, P.M.; Verhoeven, A.J.; Oude Elferink, R.P. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 2008, 47, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Takatsu, H.; Baba, K.; Shima, T.; Umino, H.; Kato, U.; Umeda, M.; Nakayama, K.; Shin, H.-W. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 2011, 286, 38159. [Google Scholar] [CrossRef] [Green Version]
- Fairn, G.D.; Hermansson, M.; Somerharju, P.; Grinstein, S. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat. Cell Biol. 2011, 13, 1424–1430. [Google Scholar] [CrossRef]
- Dean, M.; Allikmets, R. Evolution of ATP-binding cassette transporter genes. Curr. Opin. Genet. Dev. 1995, 5, 779–785. [Google Scholar] [CrossRef]
- Wu, A.; Wojtowicz, K.; Savary, S.; Hamon, Y.; Trombik, T. Do ABC transporters regulate plasma membrane organization? Cell. Mol. Biol. Lett. 2020, 25. [Google Scholar] [CrossRef]
- Zolnerciks, J.K.; Andress, E.J.; Nicolaou, M.; Linton, K.J. Structure of ABC transporters. Essays Biochem. 2011, 50. [Google Scholar] [CrossRef] [Green Version]
- Xavier, B.M.; Jennings, W.J.; Zein, A.A.; Wang, J.; Lee, J.Y. Structural snapshot of the cholesterol-transport ATP-binding cassette proteins. Biochem. Cell Biol. 2019, 97, 224–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, G.; Liebisch, G.; Langmann, T. Lipidomic strategies to study structural and functional defects of ABC-transporters in cellular lipid trafficking. FEBS Lett. 2006, 580, 5597–5610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Small, D.M. Role of ABC transporters in secretion of cholesterol from liver into bile. Proc. Natl. Acad. Sci. USA 2003, 100, 4–6. [Google Scholar] [CrossRef] [Green Version]
- Ismair, M.G.; Häusler, S.; Stuermer, C.A.; Guyot, C.; Meier, P.J.; Roth, J.; Stieger, B. ABC-transporters are localized in caveolin-1-positive and reggie-1-negative and reggie-2-negative microdomains of the canalicular membrane in rat hepatocytes. Hepatology 2009, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, S.M.; Weber, C.C.; Franke, C.; Müller, W.E.; Eckert, G.P. Cholesterol: Coupling between membrane microenvironment and ABC transporter activity. Biochem. Biophys. Res. Commun. 2007, 354. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef]
- Marquardt, D.; Kučerka, N.; Wassall, S.R.; Harroun, T.A.; Katsaras, J. Cholesterol’s location in lipid bilayers. Chem. Phys. Lipids 2016, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundner, M.; Panevska, A.; Sepčić, K.; Skočaj, M. What can mushroom proteins teach us about lipid rafts? Membranes 2021, 11, 264. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and physiological relevance of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Brown, M.S.; Anderson, D.D.; Goldstein, J.L.; Radhakrishnan, A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife 2014, 2014, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.A.; Vasudevan, S.; Chiu, S.W.; Mashl, R.J.; Jakobsson, E.; Scott, H.L. Sphingomyelin-cholesterol domains in phospholipid membranes: Atomistic simulation. Biophys. J. 2004, 87, 1092–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landry, Y.D.; Denis, M.; Nandi, S.; Bell, S.; Vaughan, A.M.; Zha, X. ATP-binding cassette transporter A1 expression disrupts raft membrane microdomains through its ATPase-related functions. J. Biol. Chem. 2006, 281, 36091–36101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parton, R.G.; Del Pozo, M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013, 14, 98–112. [Google Scholar] [CrossRef]
- Zhu, D.; Xiong, W.C.; Mei, L. Lipid rafts serve as a signaling platform for nicotinic acetylcholine receptor clustering. J. Neurosci. 2006, 26, 4841–4851. [Google Scholar] [CrossRef] [Green Version]
- Lamerton, R.E.; Lightfoot, A.; Nieves, D.J.; Owen, D.M. The role of protein and lipid clustering in lymphocyte activation. Front. Immunol. 2021, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sano, O.; Ito, S.; Kato, R.; Shimizu, Y.; Kobayashi, A.; Kimura, Y.; Kioka, N.; Hanada, K.; Ueda, K.; Matsuo, M. ABCA1, ABCG1, and ABCG4 are distributed to distinct membrane meso-domains and disturb detergent-resistant domains on the plasma membrane. PLoS ONE 2014, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hisamoto, N.; Tsuge, A.; Pastuhov, S.I.; Shimizu, T.; Hanafusa, H.; Matsumoto, K. Phosphatidylserine exposure mediated by ABC transporter activates the integrin signaling pathway promoting axon regeneration. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quazi, F.; Molday, R.S. Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J. Biol. Chem. 2013, 288, 34414–34426. [Google Scholar] [CrossRef] [Green Version]
- Kiss, R.S.; Elliott, M.R.; Ma, Z.; Marcel, Y.L.; Ravichandran, K.S. Apoptotic cells induce a phosphatidylserine-dependent homeostatic response from phagocytes. Curr. Biol. 2006, 16, 2252–2258. [Google Scholar] [CrossRef] [Green Version]
- Maranghi, M.; Truglio, G.; Gallo, A.; Grieco, E.; Verrienti, A.; Montali, A.; Gallo, P.; Alesini, F.; Arca, M.; Lucarelli, M. A novel splicing mutation in the ABCA1 gene, causing Tangier disease and familial HDL deficiency in a large family. Biochem. Biophys. Res. Commun. 2019, 508, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Mercan, M.; Yayla, V.; Altinay, S.; Seyhan, S. Peripheral neuropathy in Tangier disease: A literature review and assessment. J. Peripher. Nerv. Syst. 2018, 23, 88–98. [Google Scholar] [CrossRef]
- Zarubica, A.; Plazzo, A.P.; Stöckl, M.; Trombik, T.; Hamon, Y.; Müller, P.; Pomorski, T.; Herrmann, A.; Chimini, G. Functional implications of the influence of ABCA1 on lipid microenvironment at the plasma membrane: A biophysical study. FASEB J. 2009, 23, 1775–1785. [Google Scholar] [CrossRef] [Green Version]
- Mayor, S.; Maxfield, F.R. Insolubility and redistribution of GPI-anchored proteins at the cell surface after detergent treatment. Mol. Biol. Cell 1995, 6, 929–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichtenberg, D.; Goñi, F.M.; Heerklotz, H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 2005, 30, 430–436. [Google Scholar] [CrossRef]
- Liu, S.; Sheng, R.; Jung, J.H.; Wang, L.; Stec, E.; Connor, M.J.O.; Song, S.; Bikkavilli, R.K.; Winn, R.A.; Lee, D.; et al. Plasma membrane cholesterol. Elife 2018, 13, 268–274. [Google Scholar] [CrossRef]
- Kobayashi, A.; Takanezawa, Y.; Hirata, T.; Shimizu, Y.; Misasa, K.; Kioka, N.; Arai, H.; Ueda, K.; Matsuo, M. Efflux of sphingomyelin, cholesterol, and phosphatidylcholine by ABCG1. J. Lipid Res. 2006, 47, 1791–1802. [Google Scholar] [CrossRef] [Green Version]
- Sano, O.; Kobayashi, A.; Nagao, K.; Kumagai, K.; Kioka, N.; Hanada, K.; Ueda, K.; Matsuo, M. Sphingomyelin-dependence of cholesterol efflux mediated by ABCG1. J. Lipid Res. 2007, 48, 2377–2384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaughan, A.M.; Oram, J.F. ABCA1 and ABCG1 or ABCG4 act sequentially to remove cellular cholesterol and generate cholesterol-rich HDL. J. Lipid Res. 2006, 47, 2433–2443. [Google Scholar] [CrossRef] [Green Version]
- Storch, C.H.; Ehehalt, R.; Haefeli, W.E.; Weiss, J. Localization of the human breast cancer resistance protein (BCRP/ABCG2) in lipid rafts/caveolae and modulation of its activity by cholesterol in vitro. J. Pharmacol. Exp. Ther. 2007, 323, 257LP–264LP. [Google Scholar] [CrossRef] [Green Version]
- Gu, H.; Wang, F.; Zhang, D. Caveolin-1 interacts with ATP binding cassette transporter G1 (ABCG1) and regulates ABCG1-mediated cholesterol efflux. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 847–858. [Google Scholar] [CrossRef]
- Lu, R.; Tsuboi, T.; Okumura-Noji, K.; Iwamoto, N.; Yokoyama, S. Caveolin-1 facilitates internalization and degradation of ABCA1 and probucol oxidative products interfere with this reaction to increase HDL biogenesis. Atherosclerosis 2016, 253, 54–60. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Gelissen, I.C.; Ammit, A.J. Regulation of ATP binding cassette transporter A1 (ABCA1) expression: Cholesterol-dependent and–independent signaling pathways with relevance to inflammatory lung disease. Respir. Res. 2020, 21, 250. [Google Scholar] [CrossRef]
- Hegyi, Z.; Homolya, L. Functional cooperativity between ABCG4 and ABCG1 isoforms. PLoS ONE 2016, 11, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Seres, L.; Cserepes, J.; Elkind, N.B.; Törőcsik, D.; Nagy, L.; Sarkadi, B.; Homolya, L. Functional ABCG1 expression induces apoptosis in macrophages and other cell types. Biochim. Biophys. Acta Biomembr. 2008, 1778, 2378–2387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Fadok, V.A.; Bratton, D.L.; Frasch, S.C.; Warner, M.L.; Henson, P.M. The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ. 1998, 5, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, T.; Bode, G.; Lueken, A.; Knop, M.; Kannenberg, F.; Nofer, J.-R.; Assmann, G.; Seedorf, U. Expression and functional characterization of ABCG1 splice variant ABCG1(666). FEBS Lett. 2006, 580, 4551–4559. [Google Scholar] [CrossRef] [Green Version]
- Dubey, V.; Bozorg, B.; Wüstner, D.; Khandelia, H. Cholesterol binding to the sterol-sensing region of niemann pick C1 protein confines dynamics of its N-terminal domain. PLoS Comput. Biol. 2020, 16, 1–28. [Google Scholar] [CrossRef]
- Sharpe, L.J.; Rao, G.; Jones, P.M.; Glancey, E.; Aleidi, S.M.; George, A.M.; Brown, A.J.; Gelissen, I.C. Cholesterol sensing by the ABCG1 lipid transporter: Requirement of a CRAC motif in the final transmembrane domain. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 1851, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Ercan, B.; Krshnan, L.; Triebl, A.; Koh, D.H.Z.; Wei, F.Y.; Tomizawa, K.; Torta, F.T.; Wenk, M.R.; Saheki, Y. Movement of accessible plasma membrane cholesterol by GRAMD1 lipid transfer protein complex. Elife 2019, 8, 1–42. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Rohatgi, R.; Siebold, C. Cholesterol access in cellular membranes controls Hedgehog signaling. Nat. Chem. Biol. 2020, 16, 1303–1313. [Google Scholar] [CrossRef]
- Eyster, K.M. The membrane and lipids as integral participants in signal transduction: Lipid signal transduction for the non-lipid biochemist. Am. J. Physiol. Adv. Physiol. Educ. 2007, 31, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Luo, F.; Qin, G.; Xia, T.; Fang, X. Single-molecule imaging of protein interactions and dynamics. Annu. Rev. Anal. Chem. 2020, 13, 337–361. [Google Scholar] [CrossRef]
- Koyama-Honda, I.; Fujiwara, T.K.; Kasai, R.S.; Suzuki, K.G.N.; Kajikawa, E.; Tsuboi, H.; Tsunoyama, T.A.; Kusumi, A. High-speed single-molecule imaging reveals signal transduction by induced transbilayer raft phases. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Sánchez-Aguilera, P.; Diaz-Vega, A.; Campos, C.; Quinteros-Waltemath, O.; Cerda-Kohler, H.; Barrientos, G.; Contreras-Ferrat, A.; Llanos, P. Role of ABCA1 on membrane cholesterol content, insulin-dependent Akt phosphorylation and glucose uptake in adult skeletal muscle fibers from mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863. [Google Scholar] [CrossRef]
- Babashamsi, M.M.; Koukhaloo, S.Z.; Halalkhor, S.; Salimi, A.; Babashamsi, M. ABCA1 and metabolic syndrome; a review of the ABCA1 role in HDL-VLDL production, insulin-glucose homeostasis, inflammation and obesity. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zou, Y.; Hao, S.; Niu, Z.; Wu, L. Polydatin inhibits LPS-induced inflammatory response in BV2 microglia by disrupting the formation of lipid rafts. Immunopharmacol. Immunotoxicol. 2021, 43. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.C.; Kunnumakkara, A.B.; Aggarwal, S.; Aggarwal, B.B. Inflammation, a double-edge sword for cancer and other age-related diseases. Front. Immunol. 2018, 9, 2160. [Google Scholar] [CrossRef]
- Balasuriya, N.; Davey, N.E.; Johnson, J.L.; Liu, H.; Biggar, K.K.; Cantley, L.C.; Li, S.S.C.; O’Donoghue, P. Phosphorylation-dependent substrate selectivity of protein kinase B (AKT1). J. Biol. Chem. 2020, 295. [Google Scholar] [CrossRef]
- Fan, J.; Qi Zhao, R.; Parro, C.; Zhao, W.; Chou, H.Y.; Robert, J.; Deeb, T.Z.; Raynoschek, C.; Barichievy, S.; Engkvist, O.; et al. Small molecule inducers of ABCA1 and apoE that act through indirect activation of the LXR pathway. J. Lipid Res. 2018, 59. [Google Scholar] [CrossRef] [Green Version]
- Hirsch-Reinshagen, V.; Zhou, S.; Burgess, B.L.; Bernier, L.; McIsaac, S.A.; Chan, J.Y.; Tansley, G.H.; Cohn, J.S.; Hayden, M.R.; Wellington, C.L. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J. Biol. Chem. 2004, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackman, N.; Ishii, A.; Bansal, R. Oligodendrocyte development and myelin biogenesis: Parsing out the roles of glycosphingolipids. Physiology 2009, 24, 290–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saher, G.; Brügger, B.; Lappe-Siefke, C.; Möbius, W.; Tozawa, R.I.; Wehr, M.C.; Wieland, F.; Ishibashi, S.; Nave, K.A. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Bazick, H.; Miles, J.R.; Fethiere, A.I.; Salihi, M.O.; Al Fazio, S.; Tavori, H.; Notterpek, L. A neutral lipid-enriched diet improves myelination and alleviates peripheral nerve pathology in neuropathic mice. Exp. Neurol. 2019, 321. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, R.; Zacharek, A.; Wang, F.; Landschoot-Ward, J.; Chopp, M.; Chen, J.; Cui, X. ABCA1/APOE/HDL signaling pathway facilitates myelination and oligodendrogenesis after stroke. Int. J. Mol. Sci. 2020, 21, 4369. [Google Scholar] [CrossRef] [PubMed]
- Dupree, J.L.; Pomicter, A.D. Myelin, DIGs, and membrane rafts in the central nervous system. Prostaglandins Other Lipid Mediat. 2010, 91, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Sviridov, D.; Bukrinsky, M. Interaction of pathogens with host cholesterol metabolism. Curr. Opin. Lipidol. 2014, 25, 333–338. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.L.; Grant, A.; Mukhamedova, N.; Pushkarsky, T.; Jennelle, L.; Dubrovsky, L.; Gaus, K.; Fitzgerald, M.L.; Sviridov, D.; Bukrinsky, M. HIV-1 Nef mobilizes lipid rafts in macrophages through a pathway that competes with ABCA1-dependent cholesterol efflux. J. Lipid Res. 2012, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chichili, G.R.; Rodgers, W. Cytoskeleton-membrane interactions in membrane raft structure. Cell. Mol. Life Sci. 2009, 66, 2319–2328. [Google Scholar] [CrossRef] [Green Version]
- Mukhamedova, N.; Hoang, A.; Dragoljevic, D.; Dubrovsky, L.; Pushkarsky, T.; Low, H.; Ditiatkovski, M.; Fu, Y.; Ohkawa, R.; Meikle, P.J.; et al. Exosomes containing HIV protein Nef reorganize lipid rafts potentiating inflammatory response in bystander cells. PLoS Pathog. 2019, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nofer, J.R.; Remaley, A.T.; Feuerborn, R.; Wolińska, I.; Engel, T.; Von Eckardstein, A.; Assmann, G. Apolipoprotein A-I activates Cdc42 signaling through the ABCA1 transporter. J. Lipid Res. 2006, 47. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhao, Y.F.; Yang, J.; Jing, H.Y.; Liang, W.; Chen, M.Y.; Yang, M.; Wang, Y.; Guo, M.Y. Selenium alleviates lipopolysaccharide-induced endometritis: Via regulating the recruitment of TLR4 into lipid rafts in mice. In Food and Function; Royal Society of Chemistry: Cambridge, UK, 2020; Volume 11. [Google Scholar]
- Baldán, Á.; Bojanic, D.D.; Edwards, P.A. The ABCs of sterol transport. J. Lipid Res. 2009, 50, S80–S85. [Google Scholar] [CrossRef] [Green Version]
- Berridge, M.J.; Irvine, R.F. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature 1984, 312, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Wakelam, M.J. Diacylglycerol--when is it an intracellular messenger? Biochim. Biophys. Acta 1998, 1436, 117–126. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356. [Google Scholar] [CrossRef] [PubMed]
- Postis, V.; Rawson, S.; Mitchell, J.K.; Lee, S.C.; Parslow, R.A.; Dafforn, T.R.; Baldwin, S.A.; Muench, S.P. The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim. Biophys. Acta 2015, 1848, 496–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Q.; Mim, C. Coming of age: Cryo-electron tomography as a versatile tool to generate high-resolution structures at cellular/biological interfaces. Int. J. Mol. Sci. 2021, 22, 6177. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ristovski, M.; Farhat, D.; Bancud, S.E.M.; Lee, J.-Y. Lipid Transporters Beam Signals from Cell Membranes. Membranes 2021, 11, 562. https://doi.org/10.3390/membranes11080562
Ristovski M, Farhat D, Bancud SEM, Lee J-Y. Lipid Transporters Beam Signals from Cell Membranes. Membranes. 2021; 11(8):562. https://doi.org/10.3390/membranes11080562
Chicago/Turabian StyleRistovski, Miliça, Danny Farhat, Shelly Ellaine M. Bancud, and Jyh-Yeuan Lee. 2021. "Lipid Transporters Beam Signals from Cell Membranes" Membranes 11, no. 8: 562. https://doi.org/10.3390/membranes11080562
APA StyleRistovski, M., Farhat, D., Bancud, S. E. M., & Lee, J.-Y. (2021). Lipid Transporters Beam Signals from Cell Membranes. Membranes, 11(8), 562. https://doi.org/10.3390/membranes11080562





