The C-Terminus of Perilipin 3 Shows Distinct Lipid Binding at Phospholipid-Oil-Aqueous Interfaces
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Purification
2.2. Buffer Preparation
2.3. Vesicle Formation
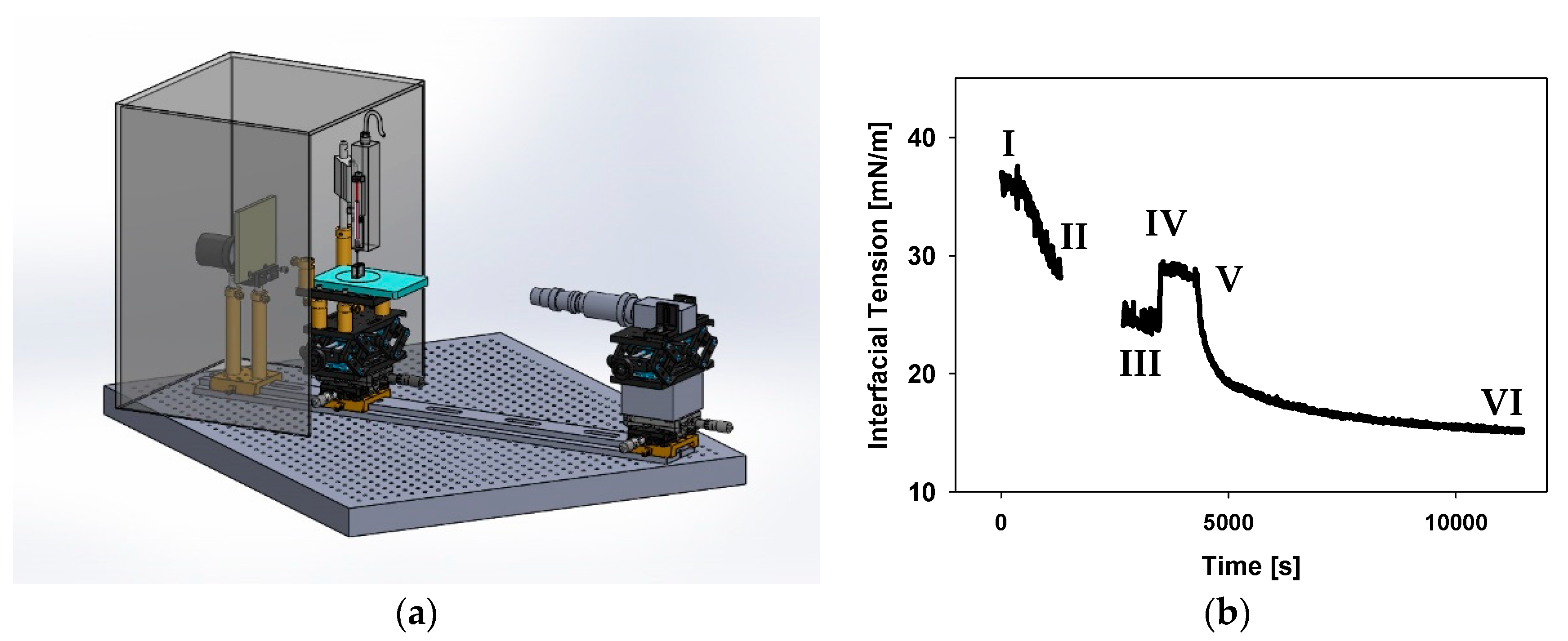
2.4. Pendant Drop Tensiometer Setup
2.5. Lipid Adsorption Protocol
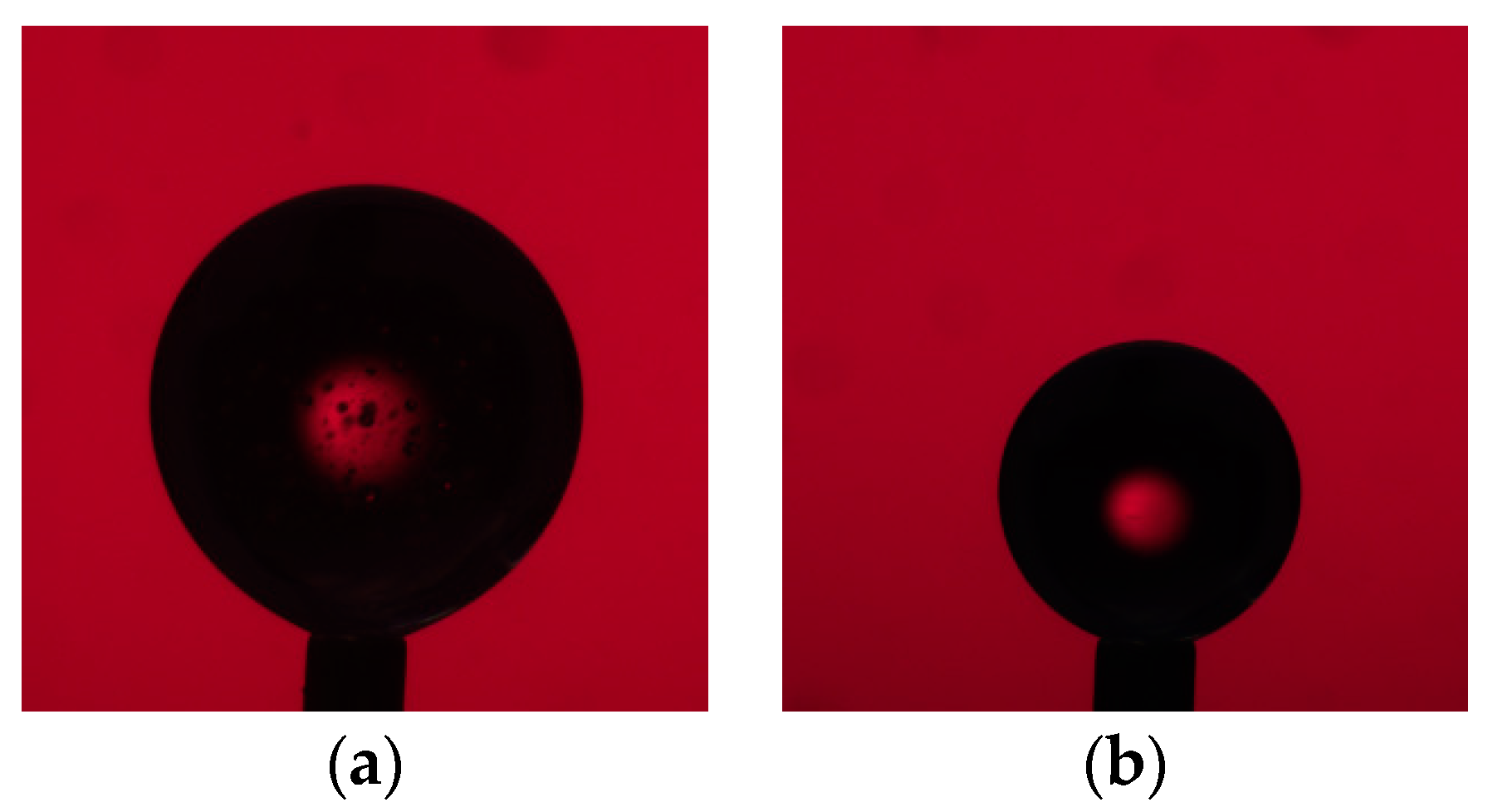
2.6. Axisymmetric Drop Shape Analysis (ADSA)
3. Results
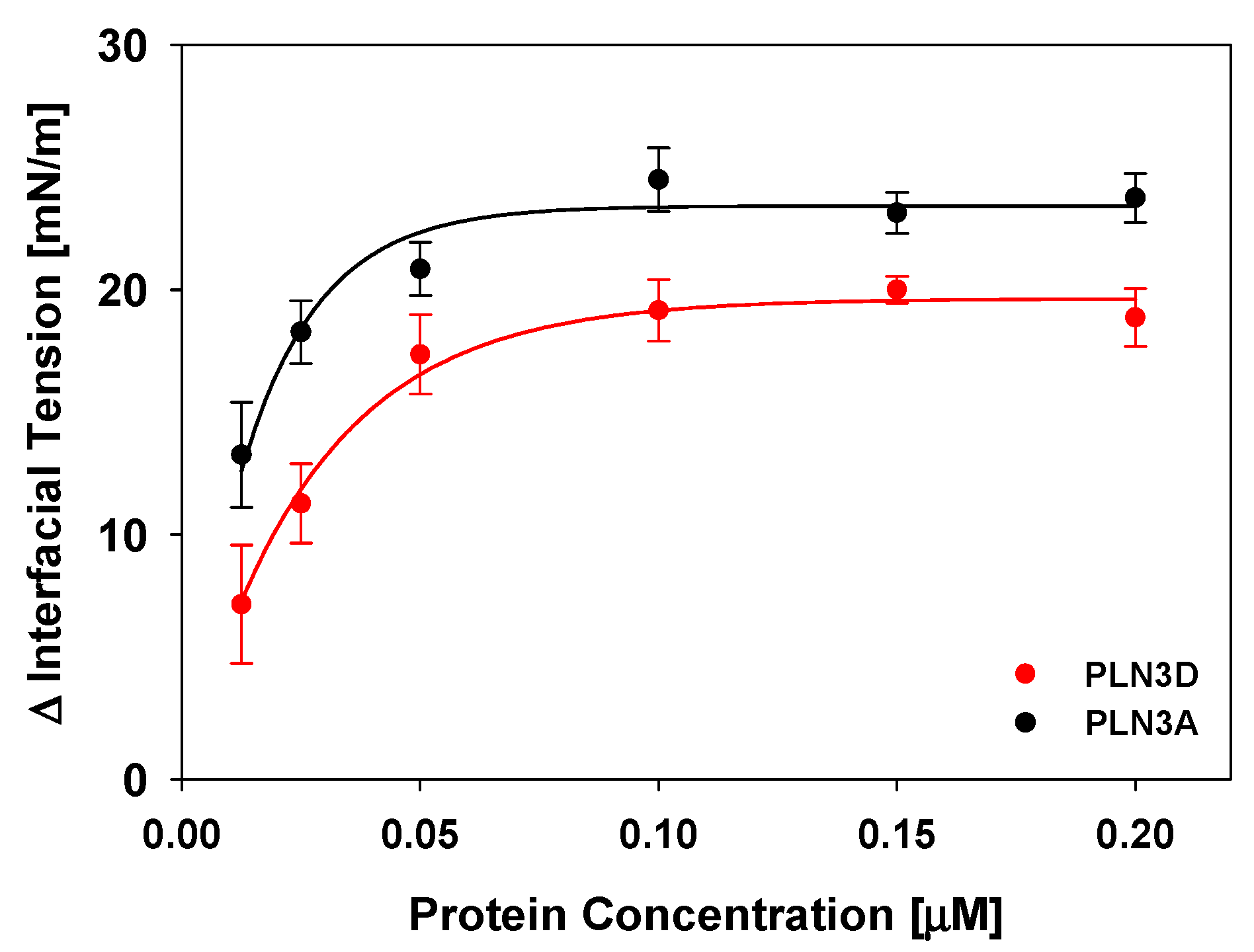
3.1. The C-Terminus of Perilipin 3 Is Surface Active at the Oil-Aqueous Interface
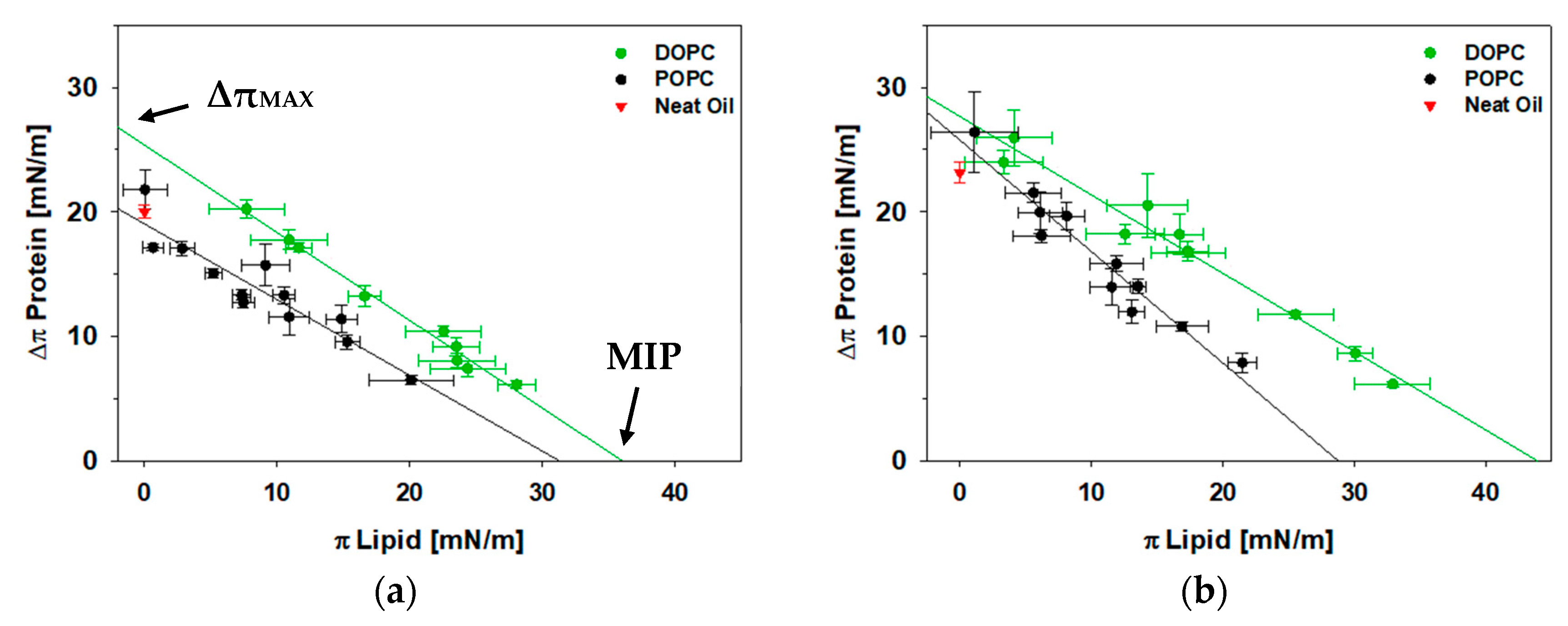
3.2. A Fully Unsaturated PC Monolayer Allows for Greater Protein Insertion for Both Full-Length Perilipin 3 and Its C-Terminus at the Oil-Lipid-Aqueous Interface
3.3. At the Oil-Aqueous Interface, Addition of POPE Increases Insertion of the C-Terminus of Perilipin 3, But Not for the Full-Length Protein
4. Discussion
4.1. Lipid Acyl Chain Unsaturation Assists in Perilipin 3 Binding and Monolayer Insertion at the LD Interface
4.2. PE Facilitates Recruitment of the C-Terminal α-Helix Bundle of Perilipin 3 to LDs, But Not the Full-Length Protein
4.3. Proposed Model of Perilipin 3 Recruitment to Nascent LDs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, D.A. Lipid droplets: Proteins floating on a pool of fat. Curr. Biol. 2001, 11, R446–R449. [Google Scholar] [CrossRef]
- Tauchi-Sato, K.; Ozeki, S.; Houjou, T.; Taguchi, R.; Fujimoto, T. The Surface of Lipid Droplets Is a Phospholipid Monolayer with a Unique Fatty Acid Composition. J. Biol. Chem. 2002, 277, 44507–44512. [Google Scholar] [CrossRef]
- Molenaar, M.R.; Wassenaar, T.A.; Yadav, K.K.; Toulmay, A.; Mari, M.C.; Caillon, L.; Chorlay, A.; Haaker, M.W.; Wubbolts, R.W.; Houweling, M.; et al. Lecithin:Retinol Acyl Transferase (LRAT) induces the formation of lipid droplets. bioRxiv 2019, 733931. [Google Scholar] [CrossRef]
- Thiam, A.R.; Farese, R.V., Jr.; Walther, T.C. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 2013, 14, 775–786. [Google Scholar] [CrossRef]
- Cavonius, L.; Fink, H.; Kiskis, J.; Albers, E.; Undeland, I.; Enejder, A. Imaging of Lipids in Microalgae with Coherent Anti-Stokes Raman Scattering Microscopy. Plant Physiol. 2015, 167, 603–616. [Google Scholar] [CrossRef]
- Di Napoli, C.; Pope, I.; Masia, F.; Langbein, W.; Watson, P.; Borri, P. Quantitative Spatiotemporal Chemical Profiling of Individual Lipid Droplets by Hyperspectral CARS Microscopy in Living Human Adipose-Derived Stem Cells. Anal. Chem. 2016, 88, 3677–3685. [Google Scholar] [CrossRef]
- Hsieh, K.; Lee, Y.K.; Londos, C.; Raaka, B.M.; Dalen, K.T.; Kimmel, A.R. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 2012, 125, 4067–4076. [Google Scholar] [CrossRef]
- Storey, S.M.; McIntosh, A.L.; Senthivinayagam, S.; Moon, K.C.; Atshaves, B.P. The phospholipid monolayer associated with perilipin-enriched lipid droplets is a highly organized rigid membrane structure. Am. J. Physiol. Metab. 2011, 301, E991–E1003. [Google Scholar] [CrossRef]
- Yoo, H.; Triandafillou, C.; Drummond, D.A. Cellular sensing by phase separation: Using the process, not just the products. J. Biol. Chem. 2019, 294, 7151–7159. [Google Scholar] [CrossRef]
- Kory, N.; Thiam, A.-R.; Farese, R.V.; Walther, T.C. Protein Crowding Is a Determinant of Lipid Droplet Protein Composition. Dev. Cell 2015, 34, 351–363. [Google Scholar] [CrossRef]
- Small, D.M.; Wang, L.; Mitsche, M.A. The adsorption of biological peptides and proteins at the oil/water interface. A potentially important but largely unexplored field. J. Lipid Res. 2009, 50, S329–S334. [Google Scholar] [CrossRef]
- Du, X.; Zhou, L.; Aw, Y.C.; Mak, H.Y.; Xu, Y.; Rae, J.; Wang, W.; Zadoorian, A.; Hancock, S.E.; Osborne, B.; et al. ORP5 localizes to ER–lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J. Cell Biol. 2020, 219, 219. [Google Scholar] [CrossRef]
- Fujimoto, T.; Parton, R.G. Not Just Fat: The Structure and Function of the Lipid Droplet. Cold Spring Harb. Perspect. Biol. 2011, 3, a004838. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, Z.; Wu, Y.; Tsukui, T.; Ma, X.; Zhang, X.; Chiba, H.; Hui, S.-P. Separating and Profiling Phosphatidylcholines and Triglycerides from Single Cellular Lipid Droplet by In-Tip Solvent Microextraction Mass Spectrometry. Anal. Chem. 2019, 91, 4466–4471. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, F.; Thiam, A.R.; Forêt, L. Lipid Droplets Can Spontaneously Bud Off from a Symmetric Bilayer. Biophys. J. 2017, 113, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Wu, X.; Lambert, T.J.; Lai, Z.W.; Walther, T.C.; Farese, R.V. LDAF1 and Seipin Form a Lipid Droplet Assembly Complex. Dev. Cell 2019, 51, 551–563.e7. [Google Scholar] [CrossRef] [PubMed]
- Renne, M.F.; Klug, Y.A.; Carvalho, P. Lipid droplet biogenesis: A mystery “unmixing”? In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Wang, H.; Becuwe, M.; E Housden, B.; Chitraju, C.; Porras, A.J.; Graham, M.M.; Liu, X.N.; Thiam, A.R.; Savage, D.B.; Agarwal, A.K.; et al. Seipin is required for converting nascent to mature lipid droplets. eLife 2016, 5, e16582. [Google Scholar] [CrossRef] [PubMed]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Zoni, V.; Nieto, V.; Endter, L.J.; Risselada, H.J.; Monticelli, L.; Vanni, S. To Bud or Not to Bud: A Perspective on Molecular Simulations of Lipid Droplet Budding. Front. Mol. Biosci. 2019, 6, 124. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Ben M’barek, K.; Ajjaji, D.; Chorlay, A.; Vanni, S.; Foret, L.; Thiam, A.R. ER Membrane Phospholipids and Surface Tension Control Cellular Lipid Droplet Formation. Dev. Cell 2017, 41, 591–604.e597. [Google Scholar] [CrossRef]
- Rowe, E.R.; Mimmack, M.L.; Barbosa, A.D.; Haider, A.; Isaac, I.; Ouberai, M.M.; Thiam, A.R.; Patel, S.; Saudek, V.; Siniossoglou, S.; et al. Conserved Amphipathic Helices Mediate Lipid Droplet Targeting of Perilipins 1–3. J. Biol. Chem. 2016, 291, 6664–6678. [Google Scholar] [CrossRef] [PubMed]
- Kory, N.; Farese, R.V.; Walther, T.C. Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol. 2016, 26, 535–546. [Google Scholar] [CrossRef]
- Brasaemle, D.L.; Barber, T.; Kimmel, A.R.; Londos, C. Post-translational Regulation of Perilipin Expression. J. Biol. Chem. 1997, 272, 9378–9387. [Google Scholar] [CrossRef]
- Brasaemle, D.L.; Barber, T.; E Wolins, N.; Serrero, G.; Blanchette-Mackie, E.J.; Londos, C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 1997, 38, 2249–2263. [Google Scholar] [CrossRef]
- Cornell, R.B. Membrane lipid compositional sensing by the inducible amphipathic helix of CCT. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Ingelmo-Torres, M.; Gonzã¡lez-Moreno, E.; Kassan, A.; Hanzal-Bayer, M.; Tebar, F.; Herms, A.; Grewal, T.; Hancock, J.F.; Enrich, C.; Bosch, M.; et al. Hydrophobic and Basic Domains Target Proteins to Lipid Droplets. Traffic 2009, 10, 1785–1801. [Google Scholar] [CrossRef]
- Londos, C.; Brasaemle, D.; Schultz, C.; Segrest, J.; Kimmel, A. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 1999, 10, 51–58. [Google Scholar] [CrossRef]
- Hickenbottom, S.J.; Kimmel, A.R.; Londos, C.; Hurley, J.H. Structure of a lipid droplet protein; the PAT family member TIP47. Structure 2004, 12, 1199–1207. [Google Scholar] [CrossRef]
- Wolins, N.E.; Brasaemle, D.L.; Bickel, P.E. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006, 580, 5484–5491. [Google Scholar] [CrossRef]
- Lee, Y.K.; Sohn, J.H.; Han, J.S.; Park, Y.J.; Jeon, Y.G.; Ji, Y.; Dalen, K.T.; Sztalryd, C.; Kimmel, A.R.; Kim, J.B. Perilipin 3 Deficiency Stimulates Thermogenic Beige Adipocytes Through PPARalpha Activation. Diabetes 2018, 67, 791–804. [Google Scholar] [CrossRef]
- Bickel, P.E.; Tansey, J.T.; Welte, M.A. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2009, 1791, 419–440. [Google Scholar] [CrossRef] [PubMed]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Brasaemle, D.L. Thematic review series: Adipocyte Biology. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 2007, 48, 2547–2559. [Google Scholar] [CrossRef] [PubMed]
- Soulages, J.L.; Arrese, E.L. Dynamics and Hydration of the α-Helices of Apolipophorin III. J. Biol. Chem. 2000, 275, 17501–17509. [Google Scholar] [CrossRef] [PubMed]
- Weers, P.M.; Narayanaswami, V.; Choy, N.; Luty, R.; Hicks, L.; Kay, C.M.; O Ryan, R. Lipid binding ability of human apolipoprotein E N-terminal domain isoforms: Correlation with protein stability? Biophys. Chem. 2002, 100, 481–492. [Google Scholar] [CrossRef]
- Weers, P.M.; Narayanaswami, V.; Kay, C.M.; Ryan, R.O. Interaction of an Exchangeable Apolipoprotein with Phospholipid Vesicles and Lipoprotein Particles: Role of leucines 32, 34, and 95 in Locusta migratoria apolipophorin III. J. Biol. Chem. 1999, 274, 21804–21810. [Google Scholar] [CrossRef] [PubMed]
- Weers, P.; Ryan, R. Apolipophorin III: Role model apolipoprotein. Insect Biochem. Mol. Biol. 2006, 36, 231–240. [Google Scholar] [CrossRef]
- Wolins, N.E.; Rubin, B.; Brasaemle, D.L. TIP47 Associates with Lipid Droplets. J. Biol. Chem. 2001, 276, 5101–5108. [Google Scholar] [CrossRef]
- Ajjaji, D.; Ben M’barek, K.; Mimmack, M.L.; England, C.; Herscovitz, H.; Dong, L.; Kay, R.G.; Patel, S.; Saudek, V.; Small, D.M.; et al. Dual binding motifs underpin the hierarchical association of perilipins1-3 with lipid droplets. Mol. Biol. Cell 2019, 30, 703–716. [Google Scholar] [CrossRef]
- Bulankina, A.V.; Deggerich, A.; Wenzel, D.; Mutenda, K.; Wittmann, J.G.; Rudolph, M.G.; Burger, K.N.; Höning, S. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 2009, 185, 641–655. [Google Scholar] [CrossRef]
- Orlicky, D.J.; Degala, G.; Greenwood, C.; Bales, E.S.; Russell, T.D.; McManaman, J.L. Multiple functions encoded by the N-terminal PAT domain of adipophilin. J. Cell Sci. 2008, 121, 2921–2929. [Google Scholar] [CrossRef] [PubMed]
- Mirheydari, M.; Rathnayake, S.S.; Frederick, H.; Arhar, T.; Mann, E.K.; Cocklin, S.; Kooijman, E.E. Insertion of perilipin 3 into a glycero(phospho)lipid monolayer depends on lipid headgroup and acyl chain species. J. Lipid Res. 2016, 57, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Mirheydari, M.; Mann, E.K.; Kooijman, E.E. Interaction of a model apolipoprotein, apoLp-III, with an oil-phospholipid interface. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Mirheydari, M.S.; Putta, P.; Mann, E.K.; Kooijman, E.E. Interaction of two amphipathic α-helix bundle proteins, apoLp-III and apoE 3, with the oil-aqueous interface. J. Phys. Chem. B 2020, submitted. [Google Scholar]
- Thiam, A.R.; Dugail, I. Lipid droplet-membrane contact sites—From protein binding to function. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Saad, S.M.; Neumann, A.W. Axisymmetric Drop Shape Analysis (ADSA): An Outline. Adv. Colloid Interface Sci. 2016, 238, 62–87. [Google Scholar] [CrossRef] [PubMed]
- Canny, J. A Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI-8, 679–698. [Google Scholar] [CrossRef]
- Hoorfar, M.; Neumann, A.W. Axisymmetric drop shape analysis (ADSA) for the determination of surface tension and contact angle. J. Adhes. 2004, 80, 727–743. [Google Scholar] [CrossRef]
- Titus, A.R.; Ferreira, L.A.; Belgovskiy, A.I.; Kooijman, E.E.; Mann, E.K.; Mann, J.A.; Meyer, W.V.; Smart, A.E.; Uversky, V.N.; Zaslavsky, B.Y. Interfacial tension and mechanism of liquid–liquid phase separation in aqueous media. Phys. Chem. Chem. Phys. 2020, 22, 4574–4580. [Google Scholar] [CrossRef]
- Yang, J.; Yu, K.; Zuo, Y.Y. Accuracy of Axisymmetric Drop Shape Analysis in Determining Surface and Interfacial Tensions. Langmuir 2017, 33, 8914–8923. [Google Scholar] [CrossRef]
- Calvez, P.; Bussières, S.; Demers, É.; Salesse, C. Parameters modulating the maximum insertion pressure of proteins and peptides in lipid monolayers. Biochimie 2009, 91, 718–733. [Google Scholar] [CrossRef] [PubMed]
- de Kruijff, B. Lipid polymorphism and biomembrane function. Curr. Opin. Chem. Biol. 1997, 1, 564–569. [Google Scholar] [CrossRef]
- Laan, E.V.D.B.-V.D.; Killian, J.A.; De Kruijff, B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta (BBA) Biomembr. 2004, 1666, 275–288. [Google Scholar] [CrossRef]
- Čopič, A.; Antoine-Bally, S.; Giménez-Andrés, M.; Garay, C.L.T.; Antonny, B.; Manni, M.M.; Pagnotta, S.; Guihot, J.; Jackson, C.L. A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bacle, A.; Gautier, R.; Jackson, C.L.; Fuchs, P.F.; Vanni, S. Interdigitation between Triglycerides and Lipids Modulates Surface Properties of Lipid Droplets. Biophys. J. 2017, 112, 1417–1430. [Google Scholar] [CrossRef]
- Prévost, C.; Sharp, M.E.; Kory, N.; Lin, Q.; Voth, G.A.; Farese, R.V.; Walther, T.C. Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev. Cell 2018, 44, 73–86.e4. [Google Scholar] [CrossRef]
- Putta, P.; Rankenberg, J.; Korver, R.A.; van Wijk, R.; Munnik, T.; Testerink, C.; Kooijman, E.E. Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 2709–2716. [Google Scholar] [CrossRef]
- Bigay, J.; Casella, J.-F.; Drin, G.; Mesmin, B.; Antonny, B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005, 24, 2244–2253. [Google Scholar] [CrossRef]
- Listenberger, L.; Townsend, E.; Rickertsen, C.; Hains, A.; Brown, E.; Inwards, E.G.; Stoeckman, A.K.; Matis, M.P.; Sampathkumar, R.S.; Osna, N.A.; et al. Decreasing Phosphatidylcholine on the Surface of the Lipid Droplet Correlates with Altered Protein Binding and Steatosis. Cells 2018, 7, 230. [Google Scholar] [CrossRef] [PubMed]
- Leikin, S.; Kozlov, M.; Fuller, N.; Rand, R. Measured effects of diacylglycerol on structural and elastic properties of phospholipid membranes. Biophys. J. 1996, 71, 2623–2632. [Google Scholar] [CrossRef]
- Szule, J.A.; Fuller, N.L.; Rand, R.P. The effects of acyl chain length and saturation of diacylglycerols and phosphatidylcholines on membrane monolayer curvature. Biophys. J. 2002, 83, 977–984. [Google Scholar] [CrossRef]
- Zoni, V.; Khaddaj, R.; Campomanes, P.; Thiam, R.; Schneiter, R.; Vanni, S. Lipid Droplet Biogenesis is Driven by Liquid-Liquid Phase Separation. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Hynson, R.M.G.; Jeffries, C.M.; Trewhella, J.; Cocklin, S. Solution structure studies of monomeric human TIP47/perilipin-3 reveal a highly extended conformation. Proteins: Struct. Funct. Bioinform. 2012, 80, 2046–2055. [Google Scholar] [CrossRef] [PubMed]


| Triolein Drop | 15 μL [mN/m] | 10 μL [mN/m} | 5 μL [mN/m] |
|---|---|---|---|
| Neat Oil | 37.9 ± 0.5 | 39.6 ± 0.3 | 64 ± 9 |
| POPC | 31.6 ± 0.4 | 30.4 ± 0.4 | 31.2 ± 1.5 |
| PLN3D | ΔπMAX (mN/m) | MIP (mN/m) |
|---|---|---|
| POPC | 19.1 ± 2.0 | 31 ± 4 |
| POPC/POPA | 22.2 ± 2.5 | 29 ± 4 |
| POPC/POPE | 27 ± 3 | 37 ± 4 |
| POPC/POG | 22.1 ± 2.2 | 35 ± 4 |
| PLN3A | ΔπMAX (mN/m) | MIP (mN/m) |
|---|---|---|
| POPC | 25.8 ± 1.2 | 30 ± 3 |
| POPC/POPA | 26 ± 3 | 31 ± 4 |
| POPC/POPE | 27.3 ± 1.2 | 33 ± 4 |
| POPC/POG | 22.9 ± 2.3 | 26 ± 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Titus, A.R.; Ridgway, E.N.; Douglas, R.; Brenes, E.S.; Mann, E.K.; Kooijman, E.E. The C-Terminus of Perilipin 3 Shows Distinct Lipid Binding at Phospholipid-Oil-Aqueous Interfaces. Membranes 2021, 11, 265. https://doi.org/10.3390/membranes11040265
Titus AR, Ridgway EN, Douglas R, Brenes ES, Mann EK, Kooijman EE. The C-Terminus of Perilipin 3 Shows Distinct Lipid Binding at Phospholipid-Oil-Aqueous Interfaces. Membranes. 2021; 11(4):265. https://doi.org/10.3390/membranes11040265
Chicago/Turabian StyleTitus, Amber R., Ellyse N. Ridgway, Rebecca Douglas, Elena Sánchez Brenes, Elizabeth K. Mann, and Edgar E. Kooijman. 2021. "The C-Terminus of Perilipin 3 Shows Distinct Lipid Binding at Phospholipid-Oil-Aqueous Interfaces" Membranes 11, no. 4: 265. https://doi.org/10.3390/membranes11040265
APA StyleTitus, A. R., Ridgway, E. N., Douglas, R., Brenes, E. S., Mann, E. K., & Kooijman, E. E. (2021). The C-Terminus of Perilipin 3 Shows Distinct Lipid Binding at Phospholipid-Oil-Aqueous Interfaces. Membranes, 11(4), 265. https://doi.org/10.3390/membranes11040265






