What Can Mushroom Proteins Teach Us about Lipid Rafts?
Abstract
1. Introduction
2. Sphingomyelin/Cholesterol Binding Proteins
2.1. Ostreolysin A
2.2. Ostreolysin A6
2.3. rOlyA
2.4. Pleurotolysin A
2.5. Pleurotolysin A2
2.6. Nakanori
3. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, O.S.; Koeppe, R.E. Bilayer thickness and membrane protein function, an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107–130. [Google Scholar] [CrossRef] [PubMed]
- Ingólfsson, H.I.; Melo, M.N.; van Eerden, F.J.; Arnarez, C.; Lopez, C.A.; Wassenaar, T.A.; Periole, X.; de Vries, A.H.; Tieleman, D.P.; Marrink, S.J. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014, 136, 14554–14559. [Google Scholar] [CrossRef] [PubMed]
- Bagatolli, L.A.; Ipsen, J.H.; Simonsen, A.C.; Mouritsen, O.G. An outlook on organization of lipids in membranes: Searching for a realistic connection with the organization of biological membranes. Prog. Lipid Res. 2010, 49, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Head, B.P.; Patel, H.H.; Insel, P.A. Interaction of membrane/lipid rafts with the cytoskeleton: Impact on signaling and function: Membrane/lipid rafts; mediators of cytoskeletal arrangement and cell signaling. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 532–545. [Google Scholar] [CrossRef]
- Singer, S.J.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef]
- Chichili, G.R.; Rodgers, W. Cytoskeleton-membrane interactions in membrane raft structure. Cell. Mol. Life Sci. 2009, 66, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- Douglass, A.D.; Vale, R.D. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell 2005, 121, 937–950. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A.; Cheng, J.; Fujimoto, T. Segregation of GM1 and GM3 clusters in the cell membrane depends on the intact actin cytoskeleton. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2009, 1791, 388–396. [Google Scholar] [CrossRef]
- Levental, I.; Veatch, S.L. The continuing mystery of lipid rafts. J. Mol. Biol. 2016, 428, 4749–4764. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization, composition; regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef]
- Levental, I.; Levental, K.R.; Heberle, F.A. Lipid rafts, controversies resolved; mysteries remain. Trends Cell Biol. 2020, 30, 341–353. [Google Scholar] [CrossRef]
- Edidin, M. The state of lipid rafts, from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 257–283. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef]
- London, E. Insights into lipid raft structure and formation from experiments in model membranes. Curr. Opin. Struct. Biol. 2002, 12, 480–486. [Google Scholar] [CrossRef]
- Simons, K.; Gerl, M.J. Revitalizing membrane rafts, new tools and insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef] [PubMed]
- Michel, V.; Bakovic, M. Lipid rafts in health and disease. Biol. Cell 2007, 99, 129–140. [Google Scholar] [CrossRef] [PubMed]
- McMullen, T.P.W.; Lewis, R.N.A.H.; McElhaney, R.N. Cholesterol-phospholipid interactions; the liquid-ordered phase and lipid rafts in model and biological membranes. Curr. Opin. Colloid Interface Sci. 2004, 8, 459–468. [Google Scholar] [CrossRef]
- Kinoshita, M.; Suzuki, K.G.N.; Matsumori, N.; Takada, M.; Ano, H.; Morigaki, K.; Abe, M.; Makino, A.; Kobayashi, T.; Hirosawa, K.M.; et al. Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs. J. Cell Biol. 2017, 216, 1183–1204. [Google Scholar] [CrossRef] [PubMed]
- Sevcsik, E.; Schütz, G.J. With or without rafts? Alternative views on cell membranes. BioEssays 2016, 38, 129–139. [Google Scholar] [CrossRef]
- Kusumi, A.; Fujiwara, T.K.; Tsunoyama, T.A.; Kasai, R.S.; Liu, A.-A.; Hirosawa, K.M.; Kinoshita, M.; Matsumori, N.; Komura, N.; Ando, H.; et al. Defining raft domains in the plasma membrane. Traffic 2020, 21, 106–137. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberg, D.; Goñi, F.M.; Heerklotz, H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 2005, 30, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Kuerschner, L.; Ejsing, C.S.; Ekroos, K.; Shevchenko, A.; Anderson, K.I.; Thiele, C. Polyene-lipids, a new tool to image lipids. Nat. Methods 2005, 2, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.P.; Mély, Y.; Duportail, G.; Klymchenko, A.S. Monitoring biophysical properties of lipid membranes by environment-sensitive fluorescent probes. Biophys. J. 2009, 96, 3461–3470. [Google Scholar] [CrossRef]
- Orlandi, P.A.; Fishman, P.H. Filipin-dependent inhibition of cholera toxin, evidence for toxin internalization and activation through caveolae-like domains. J. Cell Biol. 1998, 141, 905–915. [Google Scholar] [CrossRef]
- Klymchenko, A.S.; Kreder, R. Fluorescent probes for lipid rafts, from model membranes to living cells. Chem. Biol. 2014, 21, 97–113. [Google Scholar] [CrossRef]
- Bacia, K.; Scherfeld, D.; Kahya, N.; Schwille, P. Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys. J. 2004, 87, 1034–1043. [Google Scholar] [CrossRef]
- Bacia, K.; Schwille, P.; Kurzchalia, T. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc. Natl. Acad. Sci. USA 2005, 102, 3272–3277. [Google Scholar] [CrossRef]
- Kenworthy, A.K.; Petranova, N.; Edidin, M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell 2000, 11, 1645–1655. [Google Scholar] [CrossRef]
- Chinnapen, D.J.-F.; Chinnapen, H.; Saslowsky, D.; Lencer, W.I. Rafting with cholera toxin, endocytosis and trafficking from plasma membrane to ER. FEMS Microbiol. Lett. 2007, 266, 129–137. [Google Scholar] [CrossRef]
- Blank, N.; Schiller, M.; Krienke, S.; Wabnitz, G.; Ho, A.D.; Lorenz, H.-M. Cholera toxin binds to lipid rafts but has a limited specificity for ganglioside GM1. Immunol. Cell Biol. 2007, 85, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Day, C.A.; Kenworthy, A.K. Functions of cholera toxin B-subunit as a raft cross-linker. Essays Biochem. 2015, 57, 135–145. [Google Scholar] [CrossRef]
- Harder, T.; Simons, K. Clusters of glycolipid and glycosylphosphatidylinositol-anchored proteins in lymphoid cells, accumulation of actin regulated by local tyrosine phosphorylation. Eur. J. Immunol. 1999, 29, 556–562. [Google Scholar] [CrossRef]
- Wang, R.; Shi, J.; Parikh, A.N.; Shreve, A.P.; Chen, L.; Swanson, B.I. Evidence for cholera aggregation on GM1-decorated lipid bilayers. Colloids Surf. B Biointerfaces 2004, 33, 45–51. [Google Scholar] [CrossRef]
- Sepčić, K.; Berne, S.; Rebolj, K.; Batista, U.; Plemenitaš, A.; Šentjurc, M.; Macek, P. Ostreolysin; a pore-forming protein from the oyster mushroom; interacts specifically with membrane cholesterol-rich lipid domains. FEBS Lett. 2004, 575, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Skočaj, M.; Resnik, N.; Grundner, M.; Ota, K.; Rojko, N.; Hodnik, V.; Anderluh, G.; Sobota, A.; Maček, P.; Veranič, P.; et al. Tracking cholesterol/sphingomyelin-rich membrane domains with the ostreolysin A-mCherry protein. PLoS ONE 2014, 9, e92783. [Google Scholar] [CrossRef] [PubMed]
- Bhat, H.B.; Kishimoto, T.; Abe, M.; Makino, A.; Inaba, T.; Murate, M.; Dohmae, N.; Kurahashi, A.; Nishibori, K.; Fujimori, F.; et al. Binding of a pleurotolysin ortholog from Pleurotus eryngii to sphingomyelin and cholesterol-rich membrane domains. J. Lipid Res. 2013, 54, 2933–2943. [Google Scholar] [CrossRef]
- Ohno-Iwashita, Y.; Shimada, Y.; Waheed, A.A.; Hayashi, M.; Inomata, M.; Nakamura, M.; Maruya, M.; Iwashita, S. Perfringolysin O; a cholesterol-binding cytolysin; as a probe for lipid rafts. Anaerobe 2004, 10, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Makino, A.; Abe, M.; Ishitsuka, R.; Murate, M.; Kishimoto, T.; Sakai, S.; Hullin-Matsuda, F.; Shimada, Y.; Inaba, T.; Miyatake, H.; et al. A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C. FASEB J. 2016, 31, 1301–1322. [Google Scholar] [CrossRef]
- Berne, S.; Sepčić, K.; Anderluh, G.; Turk, T.; Maček, P.; Ulrih, N.P. Effect of pH on the pore forming activity and conformational stability of ostreolysin; a lipid raft-binding protein from the edible mushroom Pleurotus ostreatus. Biochemistry 2005, 44, 11137–11147. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Leonardi, A.; Mikelj, M.; Skočaj, M.; Wohlschlager, T.; Künzler, M.; Aebi, M.; Narat, M.; Križaj, I.; Anderluh, G.; et al. Membrane cholesterol and sphingomyelin; and ostreolysin A are obligatory for pore-formation by a MACPF/CDC-like pore-forming protein; pleurotolysin B. Biochimie 2013, 95, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Nimri, L.; Spivak, O.; Tal, D.; Schälling, D.; Peri, I.; Graeve, L.; Salame, T.M.; Yarden, O.; Hadar, Y.; Schwartz, B. A recombinant fungal compound induces anti-proliferative and pro-apoptotic effects on colon cancer cells. Oncotarget 2017, 8, 28854–28864. [Google Scholar] [CrossRef]
- Sakurai, N.; Kaneko, J.; Kamio, Y.; Tomita, T. Cloning; expression; and pore-forming properties of mature and precursor forms of pleurotolysin; a sphingomyelin-specific two-component cytolysin from the edible mushroom Pleurotus ostreatus. Biochim. Biophys. Acta BBA Gene Struct. Expr. 2004, 1679, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, H.H.; Rebolj, K.; Kreft, M.; Zorec, R.; Maček, P.; Sepčić, K. Lysophospholipids prevent binding of a cytolytic protein ostreolysin to cholesterol-enriched membrane domains. Toxicon 2008, 51, 1345–1356. [Google Scholar] [CrossRef]
- Skočaj, M.; Yu, Y.; Grundner, M.; Resnik, N.; Bedina Zavec, A.; Leonardi, A.; Križaj, I.; Guella, G.; Maček, P.; Kreft, M.E.; et al. Characterisation of plasmalemmal shedding of vesicles induced by the cholesterol/sphingomyelin binding protein; ostreolysin A-mCherry. Biochim. Biophys. Acta BBA Biomembr. 2016, 1858, 2882–2893. [Google Scholar] [CrossRef]
- Resnik, N.; Repnik, U.; Kreft, M.E.; Sepčić, K.; Maček, P.; Turk, B.; Veranič, P. Highly selective anti-cancer activity of cholesterol-interacting agents methyl-β-cyclodextrin and ostreolysin A/pleurotolysin B protein complex on urothelial cancer cells. PLoS ONE 2015, 10, e0137878. [Google Scholar] [CrossRef]
- Oren, T.; Nimri, L.; Yehuda-Shnaidman, E.; Staikin, K.; Hadar, Y.; Friedler, A.; Amartely, H.; Slutzki, M.; Di Pizio, A.; Niv, M.Y.; et al. Recombinant ostreolysin induces brown fat-like phenotype in HIB-1B cells. Mol. Nutr. Food Res. 2017, 61, 1700057. [Google Scholar] [CrossRef]
- Nimri, L.; Staikin, K.; Peri, I.; Yehuda-Shnaidman, E.; Schwartz, B. Ostreolysin induces browning of adipocytes and ameliorates hepatic steatosis. J. Gastroenterol. Hepatol. 2018, 33, 1990–2000. [Google Scholar] [CrossRef]
- Tomita, T.; Noguchi, K.; Mimuro, H.; Ukaji, F.; Ito, K.; Sugawara-Tomita, N.; Hashimoto, Y. Pleurotolysin; a novel sphingomyelin-specific two-component cytolysin from the edible mushroom Pleurotus ostreatus, assembles into a transmembrane pore complex. J. Biol. Chem. 2004, 279, 26975–26982. [Google Scholar] [CrossRef]
- Berne, S.; Križaj, I.; Pohleven, F.; Turk, T.; Maček, P.; Sepčić, K. Pleurotus and Agrocybe hemolysins; new proteins hypothetically involved in fungal fruiting. Biochim Biophys Acta BBA Gen. Subj. 2002, 1570, 153–159. [Google Scholar] [CrossRef]
- Butala, M.; Novak, M.; Kraševec, N.; Skočaj, M.; Veranič, P.; Maček, P.; Sepčić, K. Aegerolysins, lipid-binding proteins with versatile functions. Semin. Cell Dev. Biol. 2017, 72, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Sepčić, K.; Berne, S.; Potrich, C.; Turk, T.; Maček, P.; Menestrina, G. Interaction of ostreolysin; a cytolytic protein from the edible mushroom Pleurotus ostreatus; with lipid membranes and modulation by lysophospholipids. Eur. J. Biochem. 2003, 270, 1199–1210. [Google Scholar] [CrossRef]
- Maličev, E.; Chowdhury, H.H.; Maček, P.; Sepčić, K. Effect of ostreolysin; an Asp-hemolysin isoform; on human chondrocytes and osteoblasts; and possible role of Asp-hemolysin in pathogenesis. Med. Mycol. 2007, 45, 123–130. [Google Scholar] [CrossRef][Green Version]
- Rebolj, K.; Batista, U.; Sepčić, K.; Cestnik, V.; Maček, P.; Frangež, R. Ostreolysin affects rat aorta ring tension and endothelial cell viability in vitro. Toxicon 2007, 49, 1211–1213. [Google Scholar] [CrossRef]
- Endapally, S.; Frias, D.; Grzemska, M.; Gay, A.; Tomchick, D.R.; Radhakrishnan, A. Molecular discrimination between two conformations of sphingomyelin in plasma membranes. Cell 2019, 176, 1040–1053.e17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Endapally, S.; Vazquez, D.C.; Infante, R.E.; Radhakrishnan, A. Ostreolysin A and anthrolysin O use different mechanisms to control movement of cholesterol from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 2019, 294, 17289–17300. [Google Scholar] [CrossRef]
- Vrecl, M.; Babnik, M.; Diacci, U.; Benoit, E.; Frangež, R. Effect of the ostreolysin A/pleurotolysin B pore-forming complex on neuroblastoma cell morphology and intracellular Ca2+ activity. Toxicol. Sci. Off. J. Soc. Toxicol. 2015, 144, 276–283. [Google Scholar] [CrossRef]
- Keyel, P.A.; Loultcheva, L.; Roth, R.; Salter, R.D.; Watkins, S.C.; Yokoyama, W.M.; Heuser, J.E. Streptolysin O clearance through sequestration into blebs that bud passively from the plasma membrane. J. Cell Sci. 2011, 124, 2414–2423. [Google Scholar] [CrossRef] [PubMed]
- Lencer, W.I.; Saslowsky, D. Raft trafficking of AB5 subunit bacterial toxins. Biochim. Biophys. Acta 2005, 1746, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Schlumberger, S.; Kristan, K.Č.; Ota, K.; Frangež, R.; Molgό, J.; Sepčić, K.; Benoit, E.; Maček, P. Permeability characteristics of cell-membrane pores induced by ostreolysin A/pleurotolysin B; binary pore-forming proteins from the oyster mushroom. FEBS Lett. 2014, 588, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Panevska, A.; Skočaj, M.; Križaj, I.; Maček, P.; Sepčić, K. Ceramide phosphoethanolamine; an enigmatic cellular membrane sphingolipid. Biochim. Biophys. Acta BBA Biomembr. 2019, 1861, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Bhat, H.B.; Ishitsuka, R.; Inaba, T.; Murate, M.; Abe, M.; Makino, A.; Kohyama-Koganeya, A.; Nagao, K.; Kurahashi, A.; Kishimoto, T.; et al. Evaluation of aegerolysins as novel tools to detect and visualize ceramide phosphoethanolamine; a major sphingolipid in invertebrates. FASEB J. 2015, 29, 3920–3934. [Google Scholar] [CrossRef] [PubMed]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-forming protein complexes from Pleurotus mushrooms kill western corn rootworm and Colorado potato beetle through targeting membrane ceramide phosphoethanolamine. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Krpan, T.; Panevska, A.; Shewell, L.K.; Day, C.J.; Jennings, M.P.; Guella, G.; Sepčić, K. Binding specificity of ostreolysin A6 towards Sf9 insect cell lipids. Biochim. Biophys. Acta BBA Biomembr. 2020, 1862, 183307. [Google Scholar] [CrossRef]
- Panevska, A.; Skočaj, M.; Modic, Š.; Razinger, J.; Sepčić, K. Aegerolysins from the fungal genus Pleurotus Bioinsecticidal proteins with multiple potential applications. J. Invertebr. Pathol. 2020, 107474. [Google Scholar] [CrossRef]
- Younis, A.M.; Wu, F.-S.; Shikh, H.H.E. Antimicrobial activity of extracts of the oyster culinary medicinal mushroom Pleurotus ostreatus (Higher Basidiomycetes) and identification of a new antimicrobial compound. Int. J. Med. Mushrooms 2015, 17, 579–590. [Google Scholar] [CrossRef]
- Pilch, P.F.; Liu, L. Fat caves, caveolae; lipid trafficking and lipid metabolism in adipocytes. Trends Endocrinol. Metab. 2011, 22, 318–324. [Google Scholar] [CrossRef]
- Bernheimer, A.W.; Avigad, L.S. A cytolytic protein from the edible mushroom, Pleurotus ostreatus. Biochim. Biophys. Acta BBA Gen. Subj. 1979, 585, 451–461. [Google Scholar] [CrossRef]
- Lukoyanova, N.; Kondos, S.C.; Farabella, I.; Law, R.H.P.; Reboul, C.F.; Caradoc-Davies, T.T.; Spicer, B.A.; Kleifeld, O.; Traore, D.A.K.; Ekkel, S.M.; et al. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 2015, 13, e1002049. [Google Scholar] [CrossRef]
- Mancheño, J.M.; Martín-Benito, J.; Martínez-Ripoll, M.; Gavilanes, J.G.; Hermoso, J.A. Crystal and electron microscopy structures of sticholysin II actinoporin reveal insights into the mechanism of membrane pore formation. Structure 2003, 11, 1319–1328. [Google Scholar] [CrossRef]
- Ebina, K.; Sakagami, H.; Yokota, K.; Kondo, H. Cloning and nucleotide sequence of cDNA encoding Asp-hemolysin from Aspergillus fumigatus. Biochim. Biophys. Acta 1994, 1219, 148–150. [Google Scholar] [CrossRef]
- Kurahashi, A.; Sato, M.; Kobayashi, T.; Nishibori, K.; Fujimori, F. Homologous genes; Pe.pleurotolysin A and Pe.ostreolysin; are both specifically and highly expressed in primordia and young fruiting bodies of Pleurotus eryngii. Mycoscience 2014, 55, 113–117. [Google Scholar] [CrossRef]
- Rebolj, K.; Ulrih, N.P.; Maček, P.; Sepčić, K. Steroid structural requirements for interaction of ostreolysin; a lipid-raft binding cytolysin; with lipid monolayers and bilayers. Biochim. Biophys. Acta BBA Biomembr. 2006, 1758, 1662–1670. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kobayashi, T.; Beuchat, M.-H.; Lindsay, M.; Frias, S.; Palmiter, R.D.; Sakuraba, H.; Parton, R.G.; Gruenberg, J. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1999, 1, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Beuchat, M.-H.; Chevallier, J.; Makino, A.; Mayran, N.; Escola, J.-M.; Lebrand, C.; Cosson, P.; Kobayashi, T.; Gruenberg, J. Separation and characterization of late endosomal membrane domains. J. Biol. Chem. 2002, 277, 32157–32164. [Google Scholar] [CrossRef] [PubMed]
- Kraft, M.L. Plasma membrane organization and function, moving past lipid rafts. Mol. Biol. Cell 2013, 24, 2765–2768. [Google Scholar] [CrossRef] [PubMed]
- Skočaj, M.; Bakrač, B.; Križaj, I.; Maček, P.; Anderluh, G.; Sepčić, K. The sensing of membrane microdomains based on pore-forming toxins. Curr. Med. Chem. 2013, 20, 491–501. [Google Scholar] [CrossRef]
- Yamaji-Hasegawa, A.; Hullin-Matsuda, F.; Greimel, P.; Kobayashi, T. Pore-forming toxins: Properties, diversity; and uses as tools to image sphingomyelin and ceramide phosphoethanolamine. Biochim. Biophys. Acta 2016, 1858, 576–592. [Google Scholar] [CrossRef]
- Watkins, E.B.; Miller, E.C.; Majewski, J.; Kuhl, T.L. Membrane texture induced by specific protein binding and receptor clustering: Active roles for lipids in cellular function. Proc. Natl. Acad. Sci. USA 2011, 108, 6975–6980. [Google Scholar] [CrossRef]
- Hullin-Matsuda, F.; Makino, A.; Murate, M.; Kobayashi, T. Probing phosphoethanolamine-containing lipids in membranes with duramycin/cinnamycin and aegerolysin proteins. Biochimie 2016, 130, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef] [PubMed]
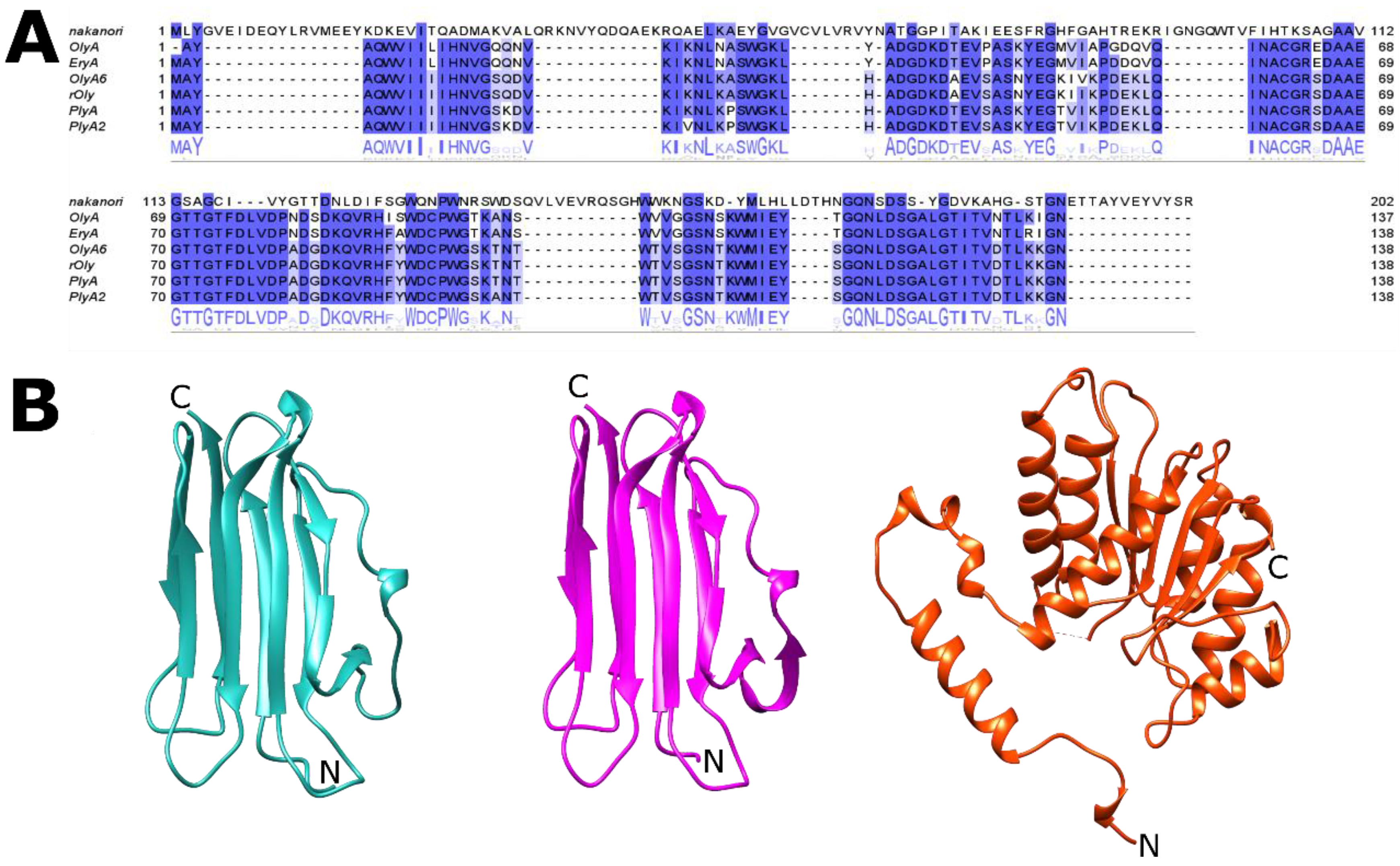
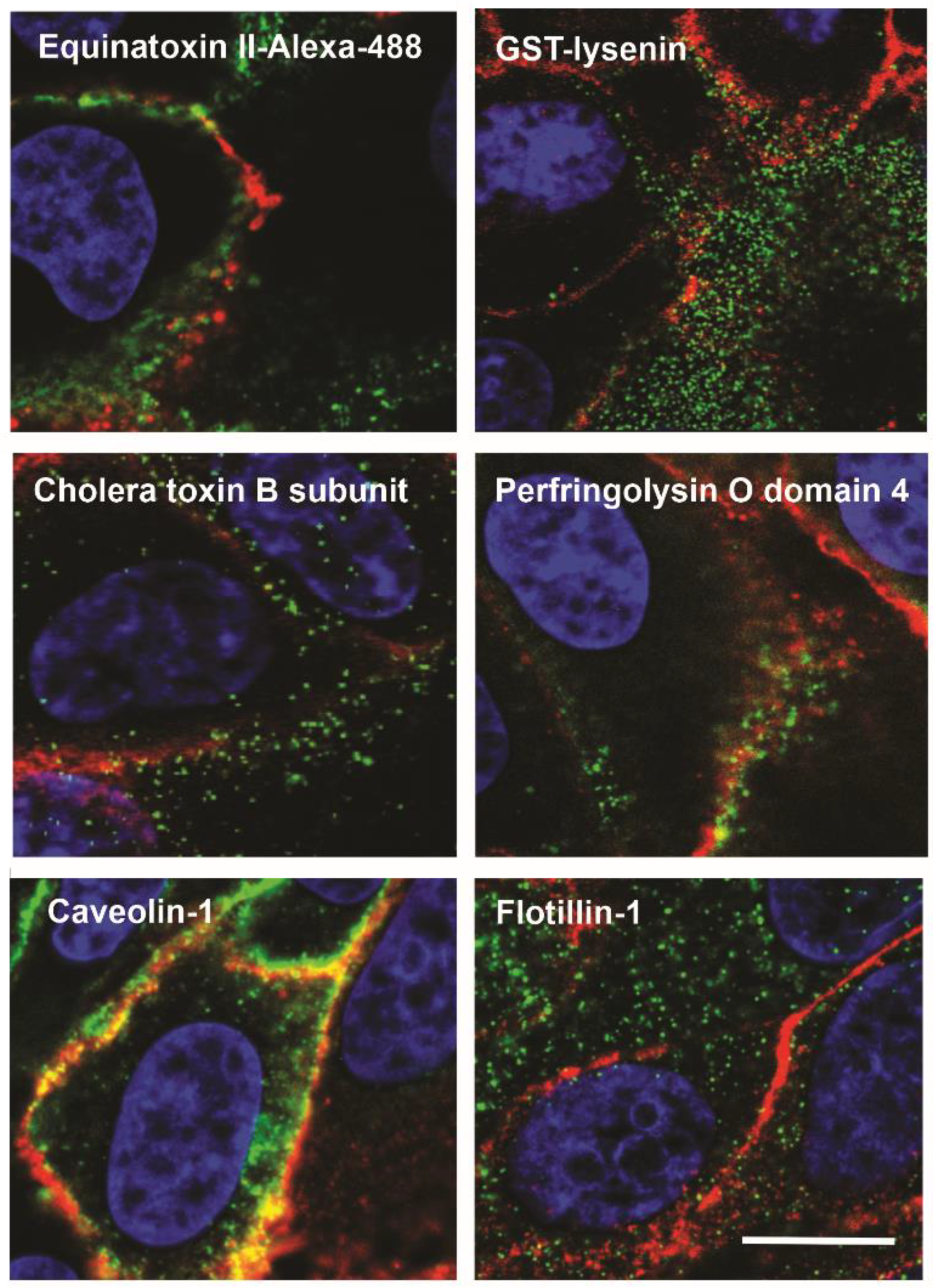
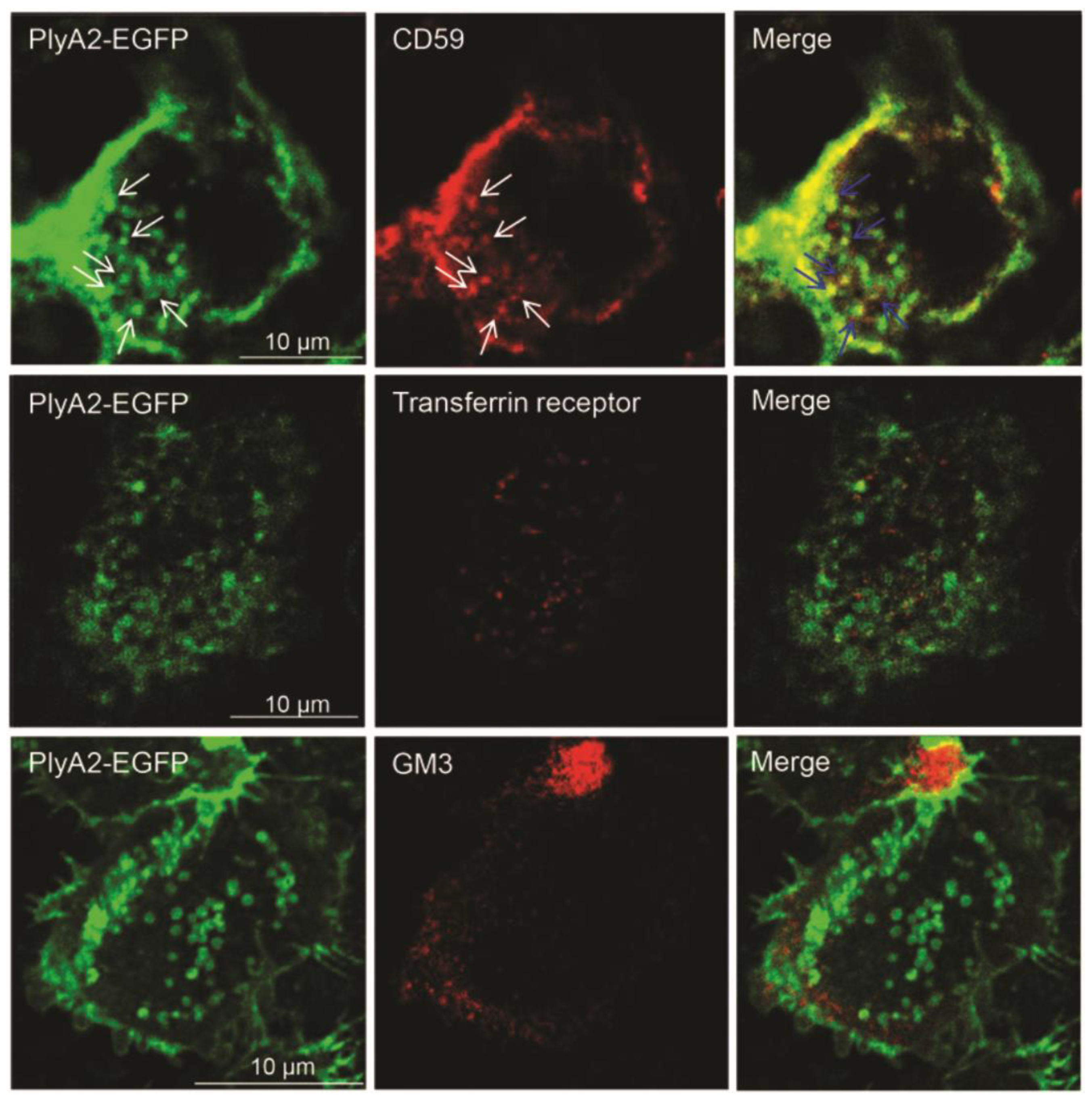
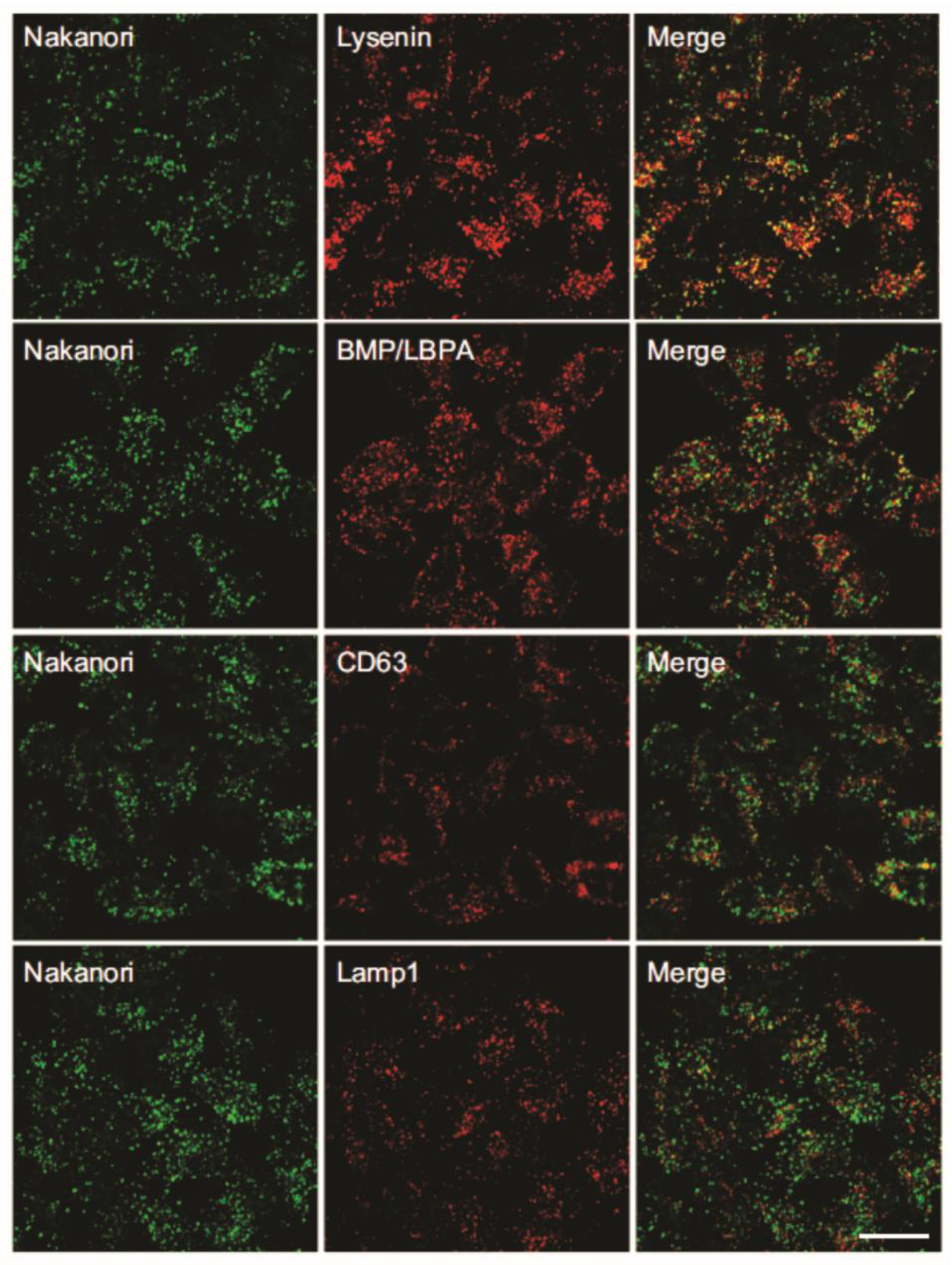
| Protein | NCBI Accession Code | Source Organism | MW a (kDa) | pI b | Cytolytic Protein Partner | Reference |
|---|---|---|---|---|---|---|
| OlyA c | AAX21097.1 | P. ostreatus | 14.9 | 4.8 | PlyB h | [41] |
| OlyA6 d | AGH25589.1 | P. ostreatus | 15.0 | 5.5 | Ply B | [37,42] |
| rOly e | KDQ25828.1 | P. ostreatus | 15.0 | 5.5 | Ply B | [43] |
| PlyA f | BAD66668.1 | P. ostreatus | 14.9 | 5.9 | Ply B | [44] |
| PlyA2 g | BAN83906.1 | P. eryngii | 15.0 | 5.5 | Erylysin B | [38] |
| Nakanori | BAO31550.1 | G. frondosa | 22.6 | 6.2 | / | [40] |
| Protein | KD (nM; SM/Chol) | Co-Localization | Cell Model | Biotechnological Applications | Reference |
|---|---|---|---|---|---|
| OlyA | >1000 | / | Lipid raft biomarker | [36,41,45] | |
| OlyA6 | ND | Caveolin 1 | MDCK cells | Lipid raft biomarker; OlyA6-induced vesicles for sampling of cytoplasm or as model plasma membrane vesicles; tool for delivering pharmacologically active substances to specific intracellular compartments; treatment of bladder cancer (with PlyB) | [37,42,46,47] |
| rOly | ND | ND | ND | Treatment of colorectal tumors and metabolic disorders | [43,48,49] |
| PlyA | ND | ND | ND | Lipid raft biomarker | [50] |
| PlyA2 | >1000 | CD59 | Hela cells | Lipid raft biomarker | [38] |
| Nakanori | 141 | Lysenin; BMP/LBPA; CD63; Lamp1 | Hela cells | Lipid raft biomarker; tool for detecting alterations in membrane dynamics (detection of Niemann–Pick disease); inhibition of release of influenza virus from MDCK cells | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grundner, M.; Panevska, A.; Sepčić, K.; Skočaj, M. What Can Mushroom Proteins Teach Us about Lipid Rafts? Membranes 2021, 11, 264. https://doi.org/10.3390/membranes11040264
Grundner M, Panevska A, Sepčić K, Skočaj M. What Can Mushroom Proteins Teach Us about Lipid Rafts? Membranes. 2021; 11(4):264. https://doi.org/10.3390/membranes11040264
Chicago/Turabian StyleGrundner, Maja, Anastasija Panevska, Kristina Sepčić, and Matej Skočaj. 2021. "What Can Mushroom Proteins Teach Us about Lipid Rafts?" Membranes 11, no. 4: 264. https://doi.org/10.3390/membranes11040264
APA StyleGrundner, M., Panevska, A., Sepčić, K., & Skočaj, M. (2021). What Can Mushroom Proteins Teach Us about Lipid Rafts? Membranes, 11(4), 264. https://doi.org/10.3390/membranes11040264







