Liposome Sterile Filtration Characterization via X-ray Computed Tomography and Confocal Microscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Membranes and Filtration
2.2. D Imaging and Analysis
3. Results and Discussion
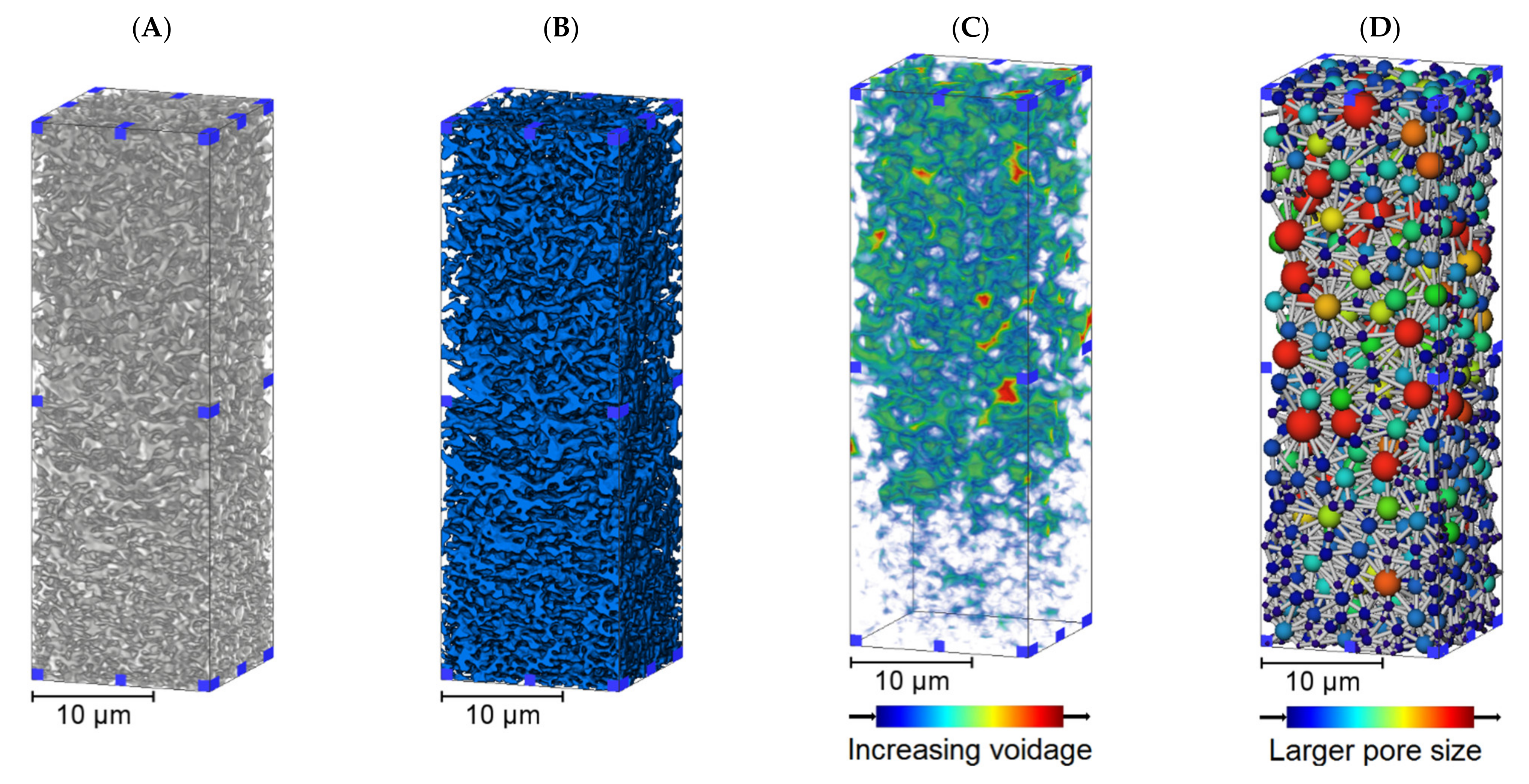
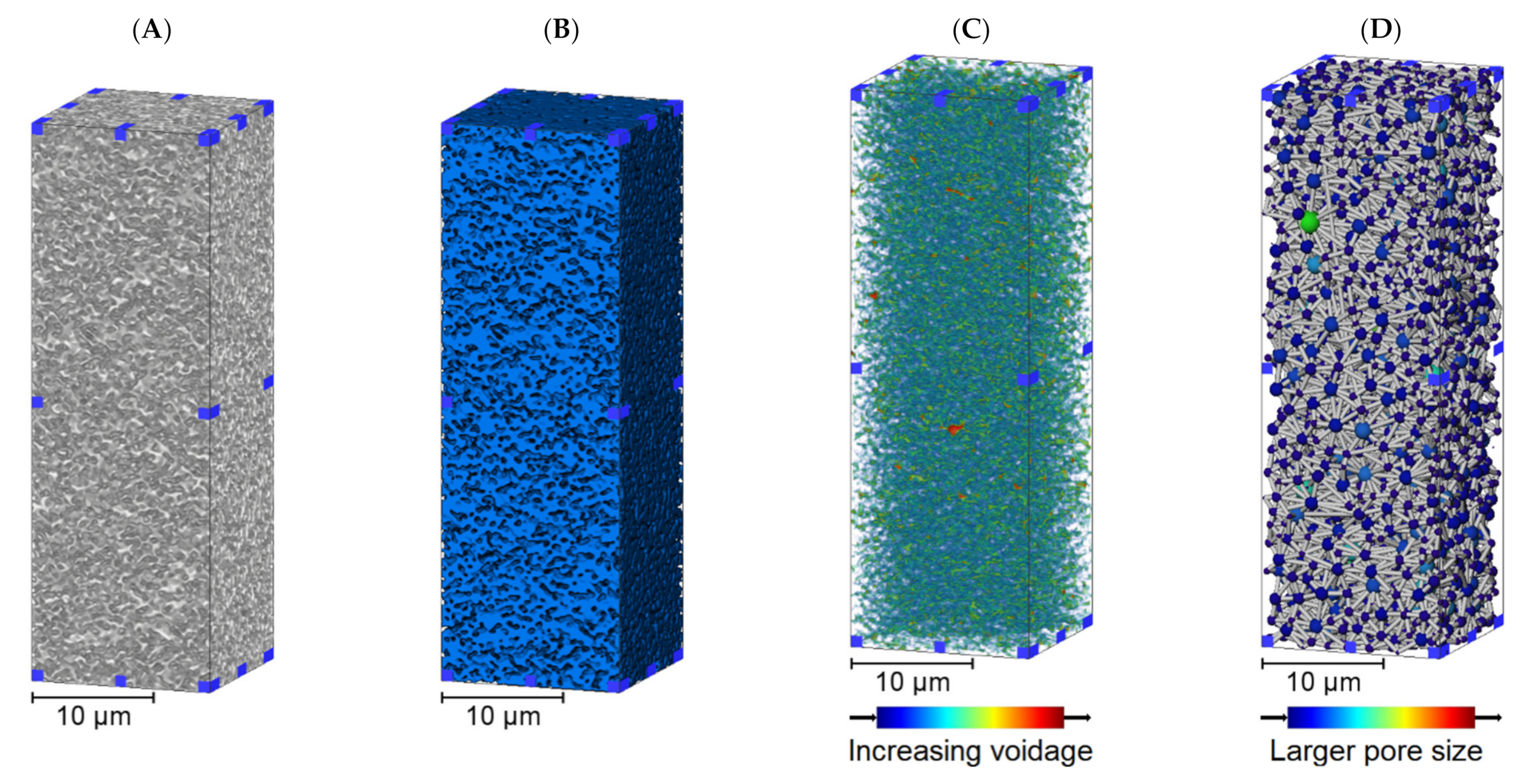
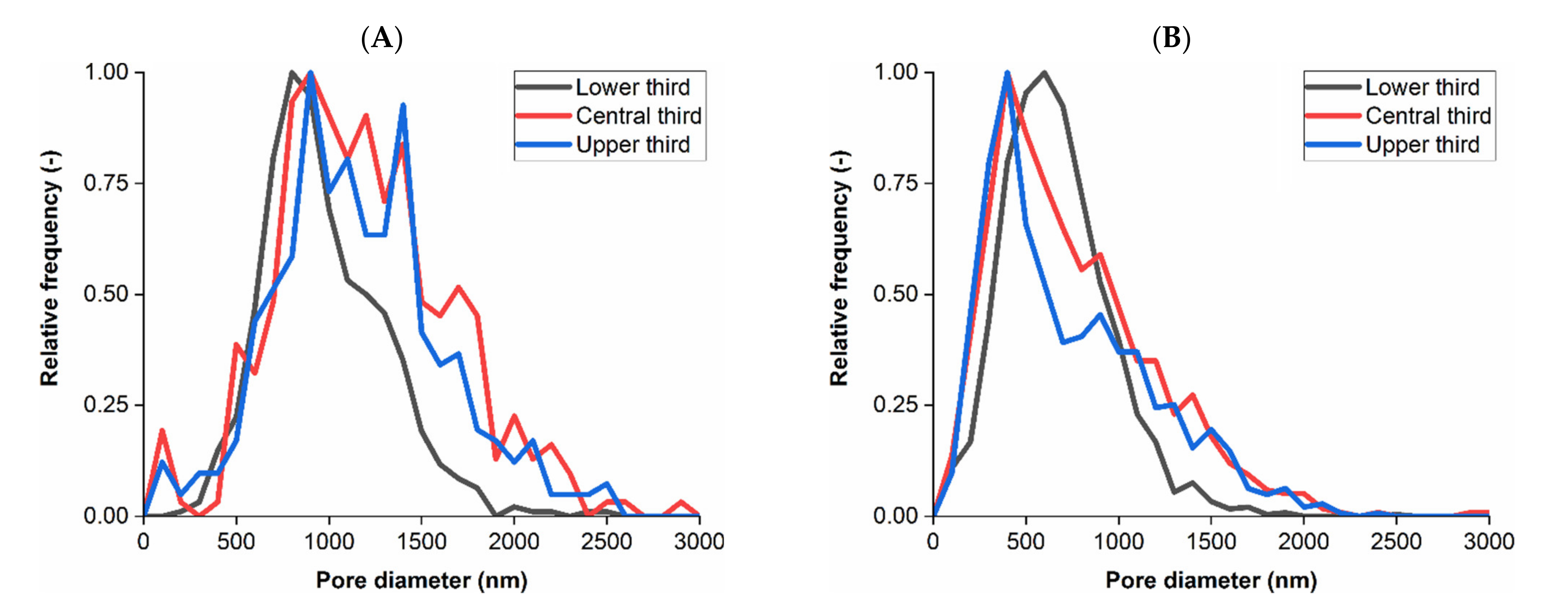
3.1. Membrane Structure Imaging
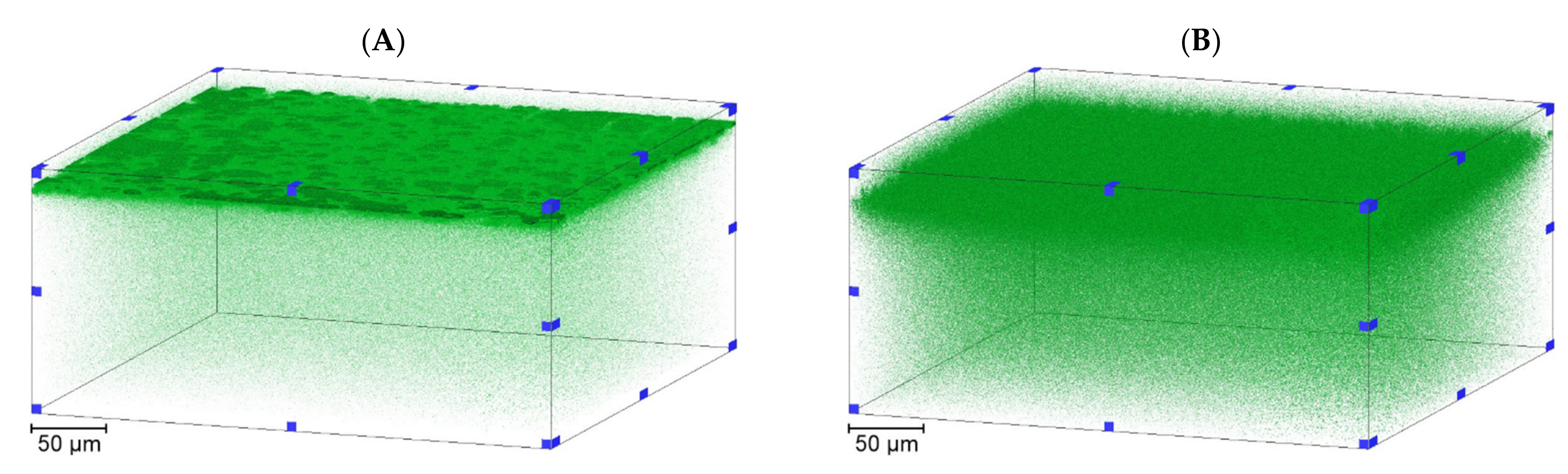
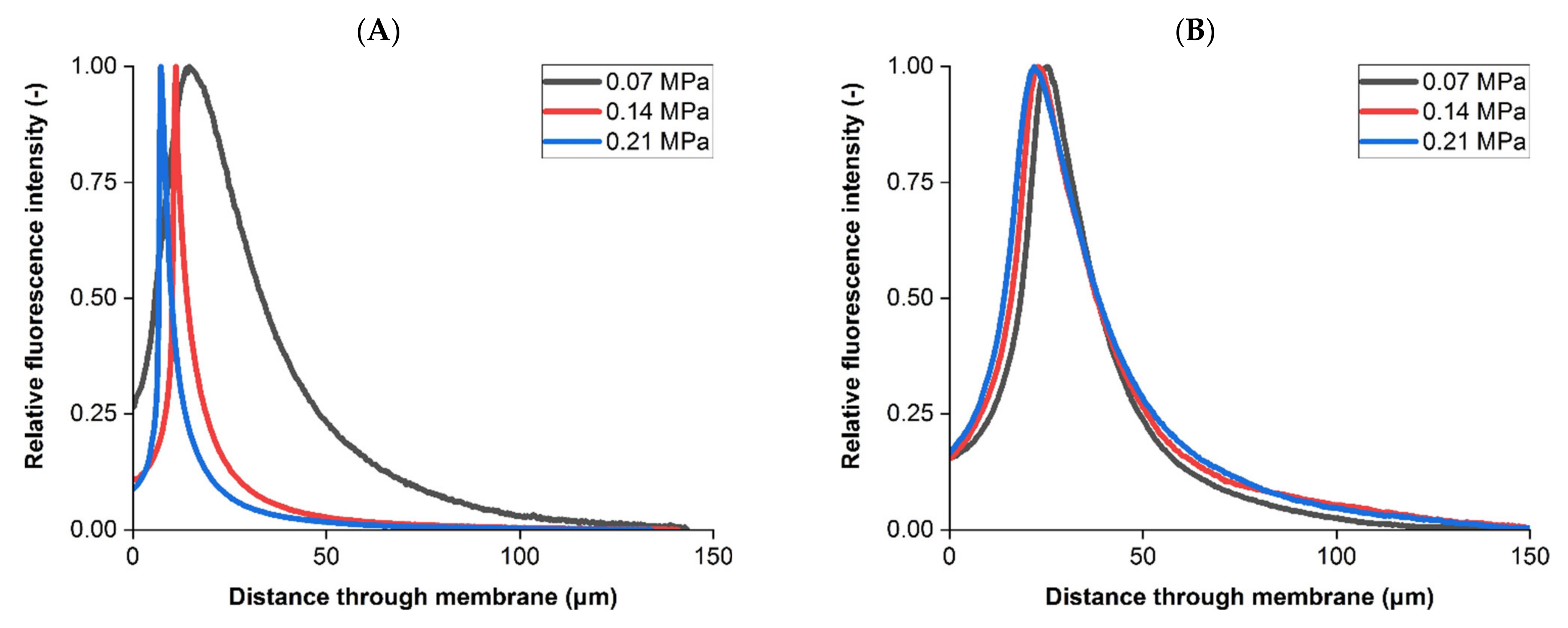
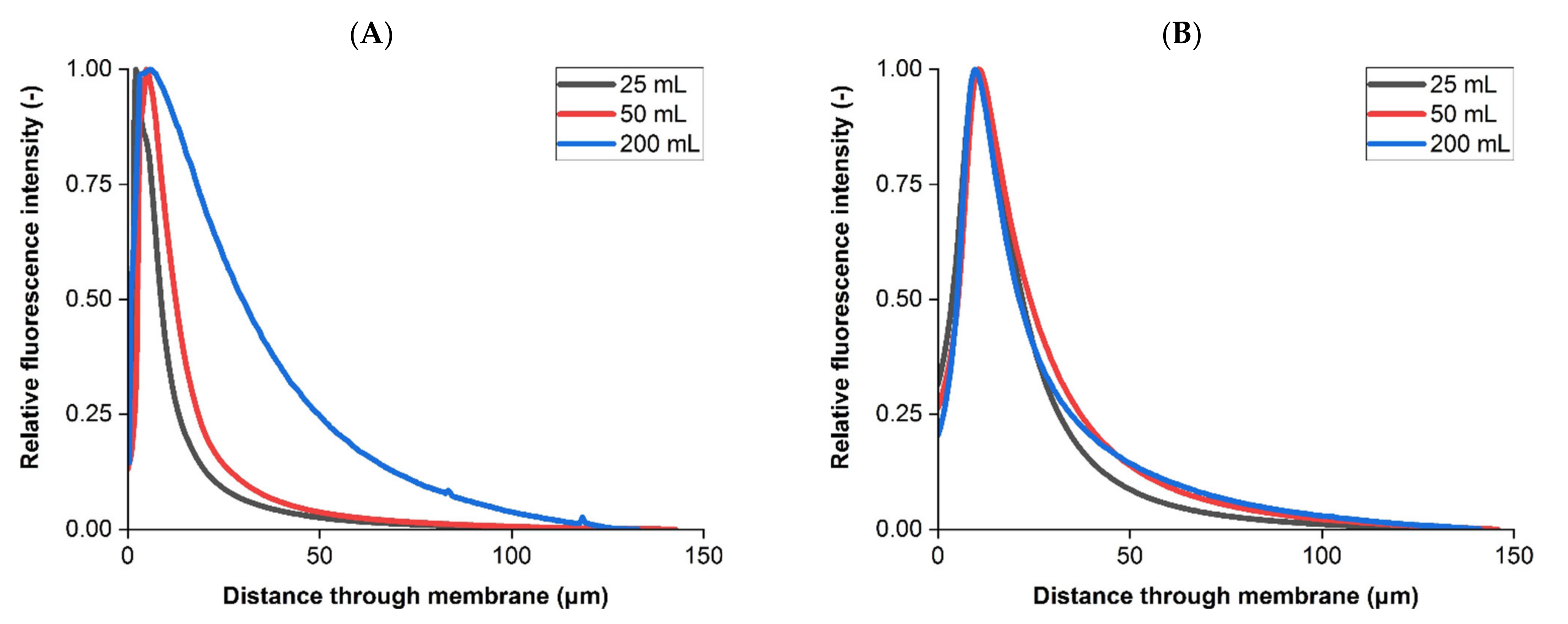
3.2. Liposome Retention Imaging
3.3. Bacterial Challenges
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.L.; Morilla, M.J. Ultradeformable phospholipid vesicles as a drug delivery system: A review. Res. Reports Transdermal Drug Deliv. 2015, 4, 55–69. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-I.; Yeh, M.-K. Clinical development of liposome-based drugs: Formulation, characterization, and therapeutic efficacy. Int. J. Nanomed. 2012, 7, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Bulbake, U.; Doppalapudi, S.; Kommineni, N.; Khan, W. Liposomal formulations in clinical use: An updated review. Pharmaceutics 2017, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Carugo, D.; Bottaro, E.; Owen, J.; Stride, E.; Nastruzzi, C. Liposome production by microfluidics: Potential and limiting factors. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Alavi, M.; Karimi, N.; Safaei, M. Application of various types of liposomes in drug delivery systems. Adv. Pharm. Bull. 2017, 7, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Barenholz, Y. Doxil® The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Pattni, B.S.; Chupin, V.V.; Torchilin, V.P. New Developments in Liposomal Drug Delivery. Chem. Rev. 2015, 115, 10938–10966. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 2015, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ashrafuzzaman, M. The antimicrobial peptide gramicidin S enhances membrane adsorption and ion pore formation potency of chemotherapy drugs in lipid bilayers. Membranes 2021, 11, 247. [Google Scholar] [CrossRef]
- Boca, S.; Gulei, D.; Zimta, A.A.; Onaciu, A.; Magdo, L.; Tigu, A.B.; Ionescu, C.; Irimie, A.; Buiga, R.; Berindan-Neagoe, I. Nanoscale delivery systems for microRNAs in cancer therapy. Cell. Mol. Life Sci. 2020, 77, 1059–1086. [Google Scholar] [CrossRef] [PubMed]
- Batist, G.; Barton, J.; Chaikin, P.; Swenson, C.; Welles, L. Myocet (liposome-encapsulated doxorubicin citrate): A new approach in breast cancer therapy. Expert Opin. Pharmacother. 2002, 3, 1739–1751. [Google Scholar] [CrossRef]
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Soo Park, K.; Sun, X.; Aikins, M.E.; Moon, J.J. Non-viral COVID-19 vaccine delivery systems. Adv. Drug Deliv. Rev. J. 2020, 169, 137–151. [Google Scholar] [CrossRef]
- Wagner, A.; Vorauer-Uhl, K. Liposome Technology for Industrial Purposes. J. Drug Deliv. 2011, 2011, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zylberberg, C.; Gaskill, K.; Pasley, S.; Matosevic, S. Engineering liposomal nanoparticles for targeted gene therapy. Gene Ther. 2017, 24, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Zydney, A.L. New developments in membranes for bioprocessing A review. J. Memb. Sci. 2021, 620, 118804. [Google Scholar] [CrossRef]
- Joseph, A.; Kenty, B.; Mollet, M.; Hwang, K.; Rose, S.; Goldrick, S.; Bender, J.; Farid, S.S.; Titchener-Hooker, N. A scale-down mimic for mapping the process performance of centrifugation, depth and sterile filtration. Biotechnol. Bioeng. 2016, 113, 1934–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moleirinho, M.G.; Silva, R.J.S.; Alves, P.M.; Carrondo, M.J.T.; Peixoto, C. Current challenges in biotherapeutic particles manufacturing. Expert Opin. Biol. Ther. 2019, 20, 451–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delma, K.L.; Lechanteur, A.; Evrard, B.; Semdé, R.; Piel, G. Sterilization methods of liposomes: Drawbacks of conventional methods and perspectives. Int. J. Pharm. 2021, 597, 120271. [Google Scholar] [CrossRef]
- Vetten, M.A.; Yah, C.S.; Singh, T.; Gulumian, M. Challenges facing sterilization and depyrogenation of nanoparticles: Effects on structural stability and biomedical applications. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Zourna, K.; Iwaniec, A.; Turner, S.; Jackson, N.B.; Welsh, J.H. Optimizing the Filtration of Liposomes Using Sterilizing-Grade Filters. PDA J. Pharm. Sci. Technol. 2021, 75, 128–140. [Google Scholar] [CrossRef]
- Loewe, D.; Grein, T.A.; Dieken, H.; Weidner, T.; Salzig, D.; Czermak, P. Tangential flow filtration for the concentration of oncolytic measles virus: The influence of filter properties and the cell culture medium. Membranes 2019, 9, 160. [Google Scholar] [CrossRef] [Green Version]
- Humpert, D.; Ebrahimi, M.; Czermak, P. Membrane technology for the recovery of lignin: A review. Membranes 2016, 6, 42. [Google Scholar] [CrossRef]
- Emami, P.; Fallahianbijan, F.; Dinse, E.; Motevalian, S.P.; Conde, B.C.; Reilly, K.; Zydney, A.L. Modified intermediate pore blockage model describes fouling behavior during sterile filtration of glycoconjugate vaccines. J. Memb. Sci. 2020, 613, 118495. [Google Scholar] [CrossRef]
- Shah, S.; Dhawan, V.; Holm, R.; Nagarsenker, M.S.; Perrie, Y. Liposomes: Advancements and innovation in the manufacturing process. Adv. Drug Deliv. Rev. 2020, 154–155, 102–122. [Google Scholar] [CrossRef] [PubMed]
- Affandy, A.; Keshavarz-Moore, E.; Versteeg, H.K. Application of filtration blocking models to describe fouling and transmission of large plasmids DNA in sterile filtration. J. Memb. Sci. 2013, 437, 150–159. [Google Scholar] [CrossRef] [Green Version]
- Laska, M.E.; Brooks, R.P.; Gayton, M.; Pujar, N.S. Robust scale-up of dead end filtration: Impact of filter fouling mechanisms and flow distribution. Biotechnol. Bioeng. 2005, 92, 308–320. [Google Scholar] [CrossRef]
- Folmsbee, M. Evaluation of the Effect of the Volume Throughput and Maximum Flux of Low-Surface-Tension Fluids on Bacterial Penetration of 0.2 Micron-Rated Filters during Process-Specific Filter Validation Testing. PDA J. Pharm. Sci. Technol. 2015, 69, 307–316. [Google Scholar] [CrossRef]
- Taylor, N.; Ma, W.; Kristopeit, A.; Wang, S.C.; Zydney, A.L. Evaluation of a sterile filtration process for viral vaccines using a model nanoparticle suspension. Biotechnol. Bioeng. 2021, 118, 106–115. [Google Scholar] [CrossRef]
- Mertens, J.C.E.; Henderson, K.; Cordes, N.L.; Pacheco, R.; Xiao, X.; Williams, J.J.; Chawla, N.; Patterson, B.M. Analysis of thermal history effects on mechanical anisotropy of 3D-printed polymer matrix composites via in situ X-ray tomography. J. Mater. Sci. 2017, 52, 12185–12206. [Google Scholar] [CrossRef]
- Tariq, F.; Yufit, V.; Kishimoto, M.; Shearing, P.R.; Menkin, S.; Golodnitsky, D.; Gelb, J.; Peled, E.; Brandon, N.P. Three-dimensional high resolution X-ray imaging and quantification of lithium ion battery mesocarbon microbead anodes. J. Power Sources 2014, 248, 1014–1020. [Google Scholar] [CrossRef]
- Bhartiya, A.; Madi, K.; Disney, C.M.; Courtois, L.; Jupe, A.; Zhang, F.; Bodey, A.J.; Lee, P.; Rau, C.; Robinsona, I.K.; et al. Phase-contrast 3D tomography of HeLa cells grown in PLLA polymer electrospun scaffolds using synchrotron X-rays Bhartiya A. J. Synchrotron Radiat. 2020, 27, 158–163. [Google Scholar] [CrossRef]
- Zankel, A.; Wagner, J.; Poelt, P. Serial sectioning methods for 3D investigations in materials science. Micron 2014, 62, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Jungreuthmayer, C.; Steppert, P.; Sekot, G.; Zankel, A.; Reingruber, H.; Zanghellini, J.; Jungbauer, A. The 3D pore structure and fluid dynamics simulation of macroporous monoliths: High permeability due to alternating channel width. J. Chromatogr. A 2015, 1425, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Denk, W.; Horstmann, H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004, 2, 1900–1909. [Google Scholar] [CrossRef]
- Bailey, J.J.; Heenan, T.M.M.; Finegan, D.P.; Lu, X.; Daemi, S.R.; Iacoviello, F.; Backeberg, N.R.; Taiwo, O.O.; Brett, D.J.L.; Atkinson, A.; et al. Laser-preparation of geometrically optimised samples for X-ray nano-CT. J. Microsc. 2017, 267, 384–396. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Taiwo, O.O.; Bertei, A.; Li, T.; Li, K.; Brett, D.J.L.; Shearing, P.R. Multi-length scale tomography for the determination and optimization of the effective microstructural properties in novel hierarchical solid oxide fuel cell anodes. J. Power Sources 2017, 367, 177–186. [Google Scholar] [CrossRef]
- Johnson, T.F.; Bailey, J.J.; Iacoviello, F.; Welsh, J.H.; Levison, P.R.; Shearing, P.R.; Bracewell, D.G. Three dimensional characterisation of chromatography bead internal structure using X-ray computed tomography and focused ion beam microscopy. J. Chromatogr. A 2018, 1566, 79–88. [Google Scholar] [CrossRef]
- Johnson, T.F.; Iacoviello, F.; Hayden, D.J.; Welsh, J.H.; Levison, P.R.; Shearing, P.R.; Bracewell, D.G. Packed bed compression visualisation and flow simulation using an erosion-dilation approach. J. Chromatogr. A 2020, 1611, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jackson, N.B.; Bakhshayeshi, M.; Zydney, A.L.; Mehta, A.; van Reis, R.; Kuriyel, R. Internal virus polarization model for virus retention by the Ultipor® VF Grade DV20 membrane. Biotechnol. Prog. 2014, 30, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Dishari, S.K.; Venkiteshwaran, A.; Zydney, A.L. Probing effects of pressure release on virus capture during virus filtration using confocal microscopy. Biotechnol. Bioeng. 2015, 112, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Fallahianbijan, F.; Giglia, S.; Carbrello, C.; Zydney, A.L. Use of fluorescently-labeled nanoparticles to study pore morphology and virus capture in virus filtration membranes. J. Memb. Sci. 2017, 536, 52–58. [Google Scholar] [CrossRef]
- Close, E.J.; Salm, J.R.; Iskra, T.; Sørensen, E.; Bracewell, D.G. Fouling of an anion exchange chromatography operation in a monoclonal antibody process: Visualization and kinetic studies. Biotechnol. Bioeng. 2013, 110, 2425–2435. [Google Scholar] [CrossRef] [Green Version]
- Cooper, S.J.; Bertei, A.; Shearing, P.R.; Kilner, J.A.; Brandon, N.P. TauFactor: An open-source application for calculating tortuosity factors from tomographic data. SoftwareX 2016, 5, 203–210. [Google Scholar] [CrossRef]
- Tjaden, B.; Cooper, S.J.; Brett, D.J.; Kramer, D.; Shearing, P.R. On the origin and application of the Bruggeman correlation for analysing transport phenomena in electrochemical systems. Curr. Opin. Chem. Eng. 2016, 12, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Manickam, S.S.; McCutcheon, J.R. Understanding mass transfer through asymmetric membranes during forward osmosis: A historical perspective and critical review on measuring structural parameter with semi-empirical models and characterization approaches. Desalination 2017, 421, 110–126. [Google Scholar] [CrossRef]
- Sark, J.F.; Jullok, N.; Lau, W.J. Improving the structural parameter of the membrane sublayer for enhanced forward osmosis. Membranes 2020, 11, 448. [Google Scholar] [CrossRef]
- Tjaden, B.; Brett, D.J.L.; Shearing, P.R. Tortuosity in electrochemical devices: A review of calculation approaches. Int. Mater. Rev. 2018, 63, 47–67. [Google Scholar] [CrossRef] [Green Version]

| Membrane | Location | Permeability (µm2) | Tortuosity Factor |
|---|---|---|---|
| Upstream | Upper third | 0.0279 ± 0.0015 | 1.21 ± 0.02 |
| Central third | 0.0257 ± 0.0013 | 1.24 ± 0.02 | |
| Lower third | 0.0139 ± 0.0011 | 1.32 ± 0.05 | |
| Downstream | Upper third | 0.0013 ± 0.0004 | 2.47 ± 0.58 |
| Central third | 0.0013 ± 0.0004 | 2.47 ± 0.59 | |
| Lower third | 0.0013 ± 0.0004 | 2.51 ± 0.61 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johnson, T.F.; Jones, K.; Iacoviello, F.; Turner, S.; Jackson, N.B.; Zourna, K.; Welsh, J.H.; Shearing, P.R.; Hoare, M.; Bracewell, D.G. Liposome Sterile Filtration Characterization via X-ray Computed Tomography and Confocal Microscopy. Membranes 2021, 11, 905. https://doi.org/10.3390/membranes11110905
Johnson TF, Jones K, Iacoviello F, Turner S, Jackson NB, Zourna K, Welsh JH, Shearing PR, Hoare M, Bracewell DG. Liposome Sterile Filtration Characterization via X-ray Computed Tomography and Confocal Microscopy. Membranes. 2021; 11(11):905. https://doi.org/10.3390/membranes11110905
Chicago/Turabian StyleJohnson, Thomas F., Kyle Jones, Francesco Iacoviello, Stephen Turner, Nigel B. Jackson, Kalliopi Zourna, John H. Welsh, Paul R. Shearing, Mike Hoare, and Daniel G. Bracewell. 2021. "Liposome Sterile Filtration Characterization via X-ray Computed Tomography and Confocal Microscopy" Membranes 11, no. 11: 905. https://doi.org/10.3390/membranes11110905
APA StyleJohnson, T. F., Jones, K., Iacoviello, F., Turner, S., Jackson, N. B., Zourna, K., Welsh, J. H., Shearing, P. R., Hoare, M., & Bracewell, D. G. (2021). Liposome Sterile Filtration Characterization via X-ray Computed Tomography and Confocal Microscopy. Membranes, 11(11), 905. https://doi.org/10.3390/membranes11110905








