A Ready-to-Use Metal-Supported Bilayer Lipid Membrane Biosensor for the Detection of Phenol in Water
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
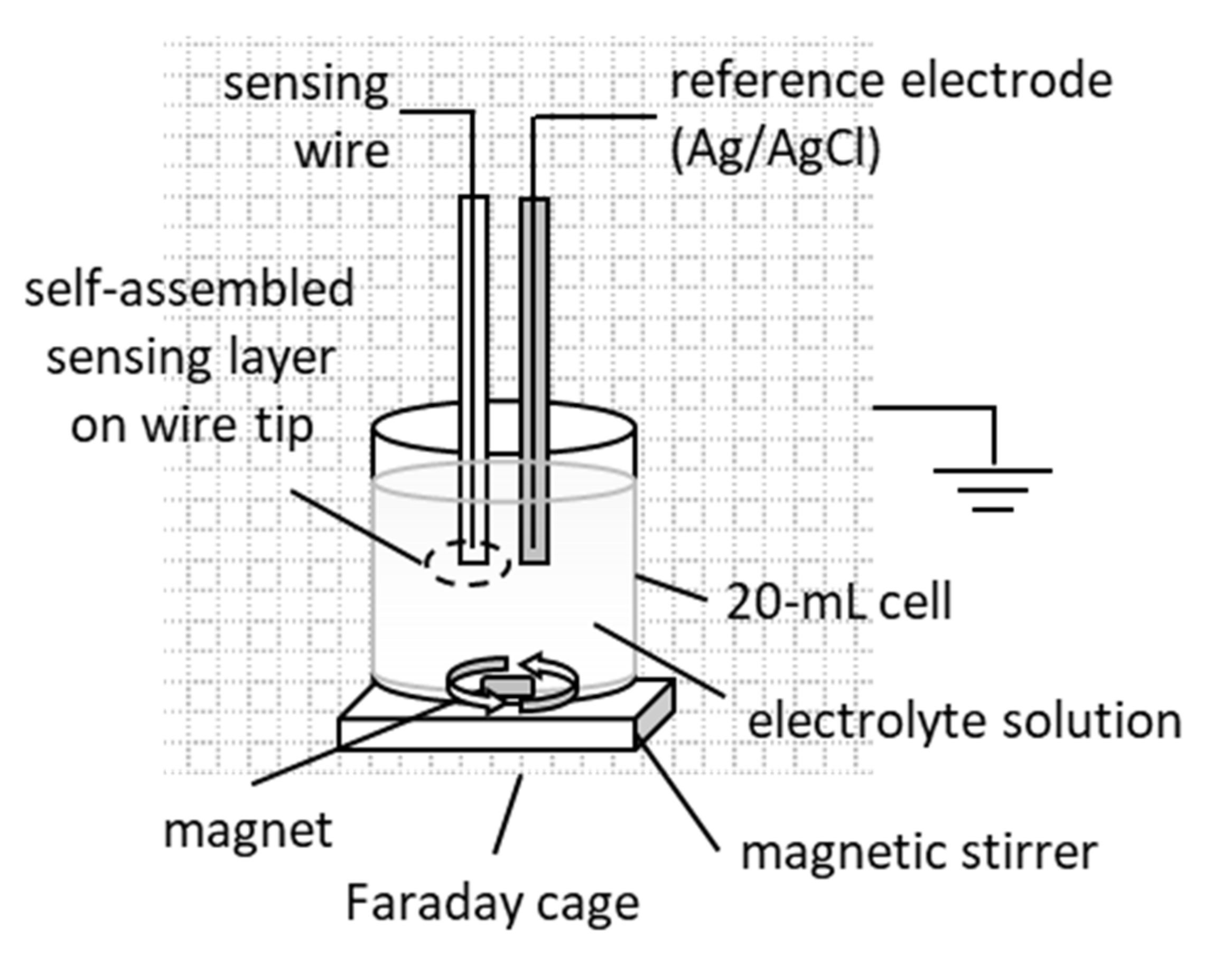
2.2. Apparatus
2.3. Sensor Assembly
2.4. Construction of the Tyrosinase Metal-Supported Lipid Membrane Sensor
2.5. Treatment of Environmental Samples
3. Results and Discussions
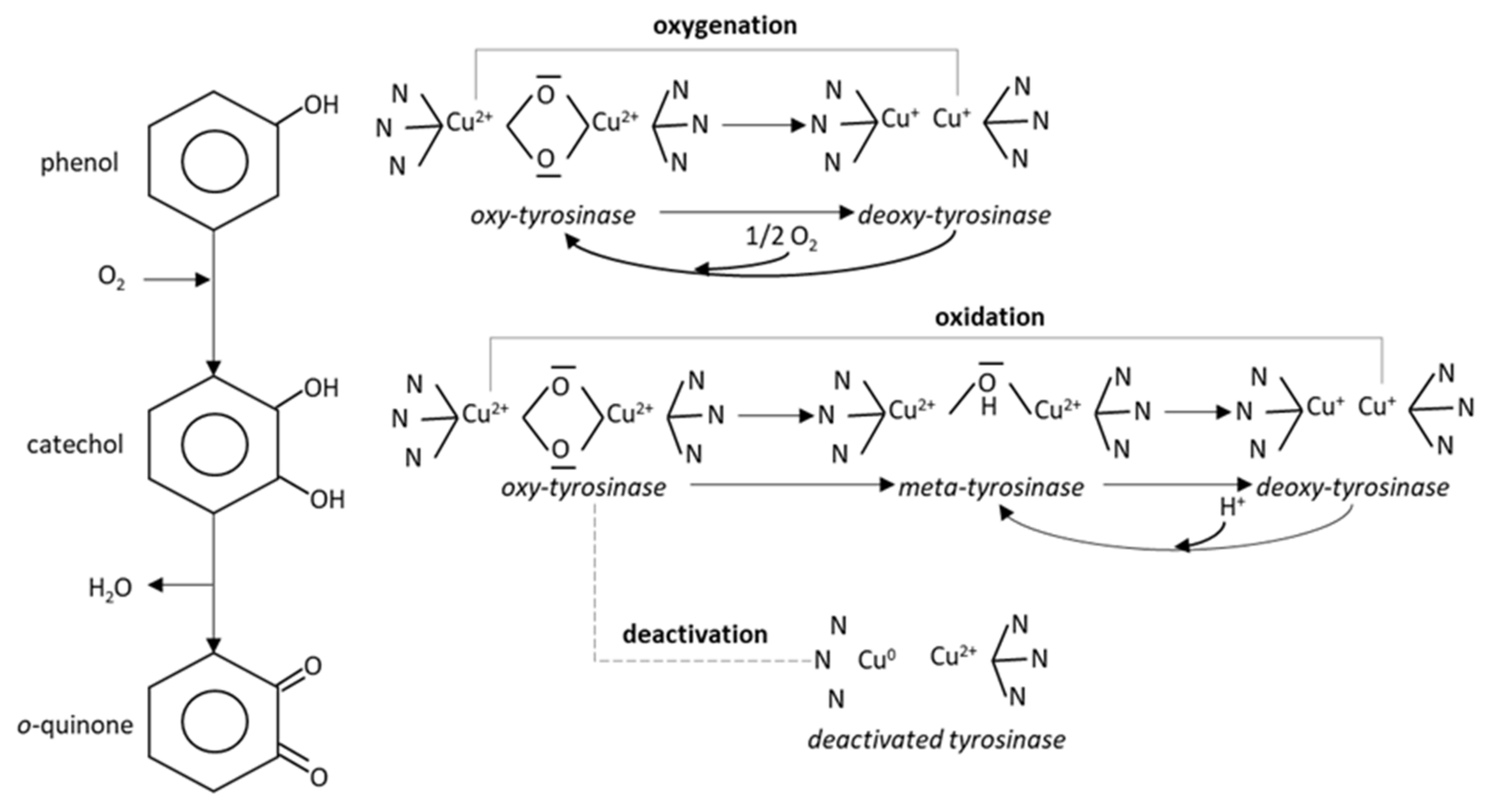
3.1. Biosensor Functioning
3.2. Phenol Detection
3.3. Sensor Reversibility
3.4. Sensor Validation
3.5. Marketability and Miniaturization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anku, W.W.; Mamo, M.A.; Govender, P.P. Phenolic Compounds in Water: Sources, Reactivity, Toxicity and Treatment Methods. In Phenolic Compounds-Natural Sources, Importance and Applications; Soto-Hernandez, M., Palma-Tenango, M., del Rosario Garcia-Mateos, M., Eds.; IntechOpen: London, UK, 2017; pp. 419–443. [Google Scholar] [CrossRef]
- World Health Organization (WHO). A Global Overview of National Regulations and Standards for Drinking-Water Quality; CCBY-NC-SA3.0IGO; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Arfin, T.; Sonawane, K.; Tarannum, A. Review on detection of phenol in water. Adv. Mater. Lett. 2019, 10, 753–785. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z.; Xu, L. Simultaneous determination of phenols (bibenzyl, phenanthrene, and fluorenone) in Dendrobium species by high-performance liquid chromatography with diode array detection. J. Chromatogr. A 2006, 1104, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Vanbeneden, N.; Delvaux, F.; Delvaux, F.R. Determination of hydroxycinnamic acids and volatile phenols in wort and beer by isocratic high-performance liquid chromatography using electrochemical detection. J. Chromatogr. A 2006, 1136, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Z.; Yang, C.; Chen, F.; Mai, B. Simultaneous determination of endocrine-disrupting phenols and steroid estrogens in sediment by gas chromatography–mass spectrometry. J. Chromatogr. A 2006, 1116, 51–56. [Google Scholar] [CrossRef]
- Fiamegos, Y.C.; Nanos, C.G.; Pilidis, G.A.; Stalikas, C.D. Phase-transfer catalytic determination of phenols as methylated derivatives by gas chromatography with flame ionization and mass-selective detection. J. Chromatogr. A 2003, 983, 215–223. [Google Scholar] [CrossRef]
- Rahman, M.M. Selective capturing of phenolic derivative by a binary metal oxide microcubes for its detection. Sci. Rep. 2019, 9, 19234. [Google Scholar] [CrossRef]
- Rahman, M.M.; Alam, M.M.; Asiri, A.M. Development of an efficient phenolic sensor based on facile Ag2O/Sb2O3nanoparticles for environmental safety. Nanoscale Adv. 2019, 1, 696–705. [Google Scholar] [CrossRef]
- Rahman, M.M.; Balkhoyor, H.B.; Asiri, A.M. Phenolic sensor development based on chromium oxide-decorated carbon nanotubes for environmental safety. J. Environ. Manag. 2017, 188, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Balkhoyor, H.B.; Rahman, M.M.; Asiri, A.M. Effect of Ce doping into ZnO nanostructures to enhance the phenolic sensor performance. RSC Adv. 2016, 6, 58236–58246. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, S.-Q.; Zhang, K.; Zhou, J.-Q. Cd-doped ZnO quantum dots-based immunoassay for the quantitative determination of bisphenol A. Chemosphere 2014, 95, 105–110. [Google Scholar] [CrossRef]
- Pan, G.; Zhao, G.; Wei, M.; Wang, Y.; Zhao, B. Design of nanogold electrochemical immunosensor for detection of four phenolic estrogens. Chem. Phys. Lett. 2019, 732, 136657. [Google Scholar] [CrossRef]
- Wu, Y.; Lin, J.-M. Determination of phenol in landfill leachate by using microchip capillary electrophoresis with end-channel amperometric detection. J. Sep. Sci. 2006, 29, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Karim, M.R.; Alam, M.; Zaman, M.B.; Alharthi, N.; Alharbi, H.; Asiri, A.M. Facile and efficient 3-chlorophenol sensor development based on photolumenescent core-shell CdSe/ZnS quantum dots. Sci. Rep. 2020, 10, 557. [Google Scholar] [CrossRef]
- Rahman, M.M.; Wahid, A.; Alam, M.; Asiri, A.M. Efficient 4-Nitrophenol sensor development based on facile Ag@Nd2O3 nanoparticles. Mater. Today Commun. 2018, 16, 307–313. [Google Scholar] [CrossRef]
- Wahid, A.; Asiri, A.M.; Rahman, M.M. One-step facile synthesis of Nd2O3/ZnO nanostructures for an efficient selective 2,4-dinitrophenol sensor probe. Appl. Surf. Sci. 2019, 487, 1253–1261. [Google Scholar] [CrossRef]
- Rosatto, S.S.; Neto, G.D.O.; Kubota, L.T. Effect of DNA on the Peroxidase Based Biosensor for Phenol Determination in Waste Waters. Electroanalysis 2001, 13, 445–450. [Google Scholar] [CrossRef]
- Rodríguez-Delgado, M.M.; Alemán-Nava, G.S.; Rodriguez-Delgado, J.M.; Dieck-Assad, G.; Martinez-Chapa, S.O.; Barceló, D.; Parra, R. Laccase-based biosensors for detection of phenolic compounds. TrAC Trends Anal. Chem. 2015, 74, 21–45. [Google Scholar] [CrossRef]
- Bensana, A.; Achi, F. Analytical performance of functional nanostructured biointerfaces for sensing phenolic compounds. Colloids Surf. B 2020, 196, 111344. [Google Scholar] [CrossRef] [PubMed]
- Krainer, F.W.; Glieder, A. An updated view on horseradish peroxidases: Recombinant production and biotechnological applications. Appl. Microbiol. Biotechnol. 2015, 99, 1611–1625. [Google Scholar] [CrossRef] [PubMed]
- Putzbach, W.; Ronkainen, N.J. Immobilization Techniques in the Fabrication of Nanomaterial-Based Electrochemical Biosensors: A Review. Sensors 2013, 13, 4811–4840. [Google Scholar] [CrossRef] [PubMed]
- Olivares, C.; Solano, F. New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins. Pigment. Cell Melanoma Res. 2009, 22, 750–760. [Google Scholar] [CrossRef]
- Ramsden, C.A.; Riley, P.A. Tyrosinase: The four oxidation states of the active site and their relevance to enzymatic activation, oxidation and inactivation. Bioorganic Med. Chem. 2014, 22, 2388–2395. [Google Scholar] [CrossRef] [PubMed]
- Adamski, J.; Nowak, P.; Kochana, J. Simple sensor for the determination of phenol and its derivatives in water based on enzyme tyrosinase. Electrochim. Acta 2010, 55, 2363–2367. [Google Scholar] [CrossRef]
- Guan, H.; Liu, X.; Wang, W. Encapsulation of tyrosinase within liposome bioreactors for developing an amperometric phenolic compounds biosensor. J. Solid State Electrochem. 2013, 17, 2887–2893. [Google Scholar] [CrossRef]
- Yildiz, H.B.; Castillo, J.; Guschin, D.A.; Toppare, L.; Schuhmann, W.; Castillo-León, J. Phenol biosensor based on electrochemically controlled integration of tyrosinase in a redox polymer. Microchim. Acta 2007, 159, 27–34. [Google Scholar] [CrossRef]
- Fartas, F.M.; Abdullah, J.; Yusof, N.A.; Sulaiman, Y.; Saiman, M.I. Biosensor Based on Tyrosinase Immobilized on Graphene-Decorated Gold Nanoparticle/Chitosan for Phenolic Detection in Aqueous. Sensors 2017, 17, 1132. [Google Scholar] [CrossRef]
- Zheng, H.; Yan, Z.; Wang, M.; Chen, J.; Zhang, X. Biosensor based on polyaniline-polyacrylonitrile-graphene hybrid assemblies for the determination of phenolic compounds in water samples. J. Hazard. Mater. 2019, 378, 120714. [Google Scholar] [CrossRef]
- Wee, Y.; Park, S.; Kwon, Y.H.; Ju, Y.; Yeon, K.-M.; Kim, J. Tyrosinase-immobilized CNT based biosensor for highly-sensitive detection of phenolic compounds. Biosens. Bioelectron. 2019, 132, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Manan, F.A.A.; Hong, W.W.; Abdullah, J.; Yusof, N.A.; Ahmad, I. Nanocrystalline cellulose decorated quantum dots based tyrosinase biosensor for phenol determination. Mater. Sci. Eng. C 2019, 99, 37–46. [Google Scholar] [CrossRef]
- Siontorou, C.G. Bilayer lipid membrane constructs: A strategic technology evaluation approach. In Advanced Bioelectronic Materials; Tiwari, A., Patra, H.K., Turner, A.P., Eds.; Wiley: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Salva, M.L.; Rocca, M.; Niemeyer, C.M.; Delamarche, E. Methods for immobilizing receptors in microfluidic devices: A review. Micro Nano Eng. 2021, 11, 100085. [Google Scholar] [CrossRef]
- Hianik, T. Mechanical properties of bilayer lipid membranes and protein–lipid interactions. In Advances in Planar Lipid Bilayers and Liposomes; Iglič, A., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 13, pp. 33–72. [Google Scholar] [CrossRef]
- Siontorou, C.G.; Nikolelis, D.P.; Krull, U.J. Flow Injection Monitoring and Analysis of Mixtures of Hydrazine Compounds Using Filter-Supported Bilayer Lipid Membranes with Incorporated DNA. Anal. Chem. 2000, 72, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Zakharian, E. Recording of Ion Channel Activity in Planar Lipid Bilayer Experiments. Methods Mol. Biol. 2013, 998, 109–118. [Google Scholar] [CrossRef]
- Miguel, V.; Villarreal, M.A.; García, D.A. Effects of gabergic phenols on the dynamic and structure of lipid bilayers: A molecular dynamic simulation approach. PLoS ONE 2019, 14, e0218042. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-M.; Liu, X.-C.; Li, Y.-Q.; Wang, P.; Han, R.-M.; Zhang, J.-P.; Skibsted, L.H. Synergy between plant phenols and carotenoids in stabilizing lipid-bilayer membranes of giant unilamellar vesicles against oxidative destruction. Soft Matter 2020, 16, 1792–1800. [Google Scholar] [CrossRef]
- Malekar, S.A.; Sarode, A.L.; Bach, A.C.; Worthen, D.R.; Ii, A.C.B. The Localization of Phenolic Compounds in Liposomal Bilayers and Their Effects on Surface Characteristics and Colloidal Stability. AAPS PharmSciTech 2016, 17, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Tien, H.T.; Salamon, Z. Self-assembling bilayer lipid membranes on solid support. Biotechnol. Appl. Biochem. 1990, 12, 478–484. [Google Scholar]
- Castellana, E.T.; Cremer, P.S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006, 61, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Becucci, L.; Moncelli, M.R.; Guidelli, R. Ion Carriers and Channels in Metal-Supported Lipid Bilayers as Probes of Transmembrane and Dipole Potentials. Langmuir 2003, 19, 3386–3392. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, F.; Liu, J. Headgroup-Inversed Liposomes: Biointerfaces, Supported Bilayers and Applications. Langmuir 2018, 34, 9337–9348. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Nikolelis, D.P.; Krull, U.J. A carbon dioxide biosensor based on hemoglobin incorporated in metal supported bilayer lipid membranes (BLMs): Investigations for enhancement of response characteristics by using platelet-activating factor. Electroanalysis 1997, 9, 1043–1048. [Google Scholar] [CrossRef]
- Siontorou, C.; Nikolelis, D.; Tarus, B.; Dumbrava, J.; Krull, U.J. DNA Biosensor Based on Self-Assembled Bilayer Lipid Membranes for the Detection of Hydrazines. Electroanalysis 1998, 10, 691–694. [Google Scholar] [CrossRef]
- Passechnik, V.I.; Hianik, T.; Ivanov, S.A.; Sivak, B. Specific capacitance of metal supported lipid membranes. Electroanalysis 1998, 10, 295–302. [Google Scholar] [CrossRef]
- An, H.H.; Lee, S.J.; Kim, H.-S.; Han, W.B.; Yoon, C.S. Structure of solid-supported lipid membrane probed by noble metal nanoparticle deposition. Biochim. Biophys. Acta Biomembr. 2012, 1818, 2884–2891. [Google Scholar] [CrossRef][Green Version]
- Siontorou, C.G.; Georgopoulos, K.N. A biosensor platform for soil management: The case of nitrites. J. Clean. Prod. 2016, 111, 133–142. [Google Scholar] [CrossRef]
- Dimitrov, D.S.; Jain, R.K. Membrane stability. Biochim. Biophys. Acta Rev. Biomembr. 1984, 779, 437–468. [Google Scholar] [CrossRef]
- Zhou, Y.; Raphael, R.M. Solution pH Alters Mechanical and Electrical Properties of Phosphatidylcholine Membranes: Relation between Interfacial Electrostatics, Intramembrane Potential, and Bending Elasticity. Biophys. J. 2007, 92, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Petelska, A.D.; Figaszewski, Z.A. Effect of pH on the interfacial tension of bilayer lipid membrane formed from phosphatidylcholine or phosphatidylserine. Biochim. et Biophys. Acta-Biomembr. 2002, 1561, 135–146. [Google Scholar] [CrossRef]
- Naumowicz, M.; Figaszewski, Z.A.; Poltorak, L. Electrochemical impedance spectroscopy as a useful method for examination of the acid–base equilibria at interface separating electrolyte solution and phosphatidylcholine bilayer. Electrochim. Acta 2013, 91, 367–372. [Google Scholar] [CrossRef]
- Naumowicz, M.; Kotyńska, J.; Petelska, A.; Figaszewski, Z. Impedance analysis of phosphatidylcholine membranes modified with valinomycin. Eur. Biophys. J. 2005, 35, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Dimitriou, E.; Karaouzas, I.; Sarantakos, K.; Zacharias, I.; Bogdanos, K.; Diapoulis, A. Groundwater risk assessment at a heavily industrialised catchment and the associated impacts on a peri-urban wetland. J. Environ. Manag. 2008, 88, 526–538. [Google Scholar] [CrossRef] [PubMed]
- Battle, A.; Ridone, P.; Bavi, N.; Nakayama, Y.; Nikolaev, Y.; Martinac, B. Lipid–protein interactions: Lessons learned from stress. Biochim. Biophys. Acta Biomembr. 2015, 1848, 1744–1756. [Google Scholar] [CrossRef] [PubMed]
- Van Dael, H.; Ceuterickx, P. The interaction of phenol with lipid bilayers. Chem. Phys. Lipids 1984, 35, 171–181. [Google Scholar] [CrossRef]
- Forsberg, M.M.; Huotari, M.; Savolainen, J.; Männistö, P.T. The role of physicochemical properties of entacapone and tolcapone on their efficacy during local intrastriatal administration. Eur. J. Pharm. Sci. 2005, 24, 503–511. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors-Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Stoytcheva, M.; Zlatev, R.; Velkova, Z.; Valdez, B.; Ovalle, M. Analytical Characteristics of Electrochemical Biosensors. Port. Electrochim. Acta 2009, 27, 353–362. [Google Scholar] [CrossRef]
- Agarwal, P.; Gupta, R.; Agarwal, N. A Review on Enzymatic Treatment of Phenols in Wastewater. J. Biotechnol. Biomater. 2016, 6, 249. [Google Scholar] [CrossRef]
- Aloraefy, M.; Pfefer, T.J.; Ramella-Roman, J.C.; Sapsford, K.E. In Vitro Evaluation of Fluorescence Glucose Biosensor Response. Sensors 2014, 14, 12127–12148. [Google Scholar] [CrossRef]
- European Medicines Agency. ICH Validation of Analytical Procedures: Text and Methodology Q2(R1); European Medicines Agency: Amsterdam, The Netherlands, 2005. [Google Scholar]




| Day | Phenol (pg/mL) | Signal 1/Analyst 1 | Signal 2/Analyst 2 | Signal 3/Analyst 3 | Mean | SD | Estimated Concentration | Error of Measurement |
|---|---|---|---|---|---|---|---|---|
| 1 | 2.48 | 90 | 88 | 95 | 91.0 | 3.61 | 2.601 | +4.89% |
| 3.72 | 120 | 128 | 119 | 122.3 | 4.93 | 3.538 | −4.89% | |
| 6.20 | 202 | 199 | 210 | 203.7 | 5.69 | 5.970 | −3.71% | |
| 2 | 2.48 | 78 | 85 | 90 | 84.3 | 6.03 | 2.402 | −3.15% |
| 3.72 | 128 | 125 | 130 | 127.7 | 2.52 | 3.698 | −0.60% | |
| 6.20 | 200 | 195 | 220 | 205.0 | 13.23 | 6.010 | −3.07% | |
| 3 | 2.48 | 86 | 89 | 85 | 86.7 | 2.08 | 2.472 | −0.33% |
| 3.72 | 125 | 125 | 122 | 124.0 | 1.73 | 3.588 | −3.55% | |
| 6.20 | 225 | 210 | 218 | 217.7 | 7.51 | 6.388 | +3.04% | |
| 4 | 2.48 | 80 | 94 | 87 | 87.0 | 7.00 | 2.482 | +0.07% |
| 3.72 | 127 | 122 | 130 | 126.3 | 4.04 | 3.658 | −1.68% | |
| 6.20 | 225 | 212 | 198 | 211.7 | 13.50 | 6.209 | +0.14% | |
| 5 | 2.48 | 85 | 90 | 92 | 89.0 | 3.61 | 2.541 | +2.48% |
| 3.72 | 129 | 133 | 125 | 129.0 | 4.00 | 3.737 | +0.47% | |
| 6.20 | 215 | 200 | 199 | 204.7 | 8.96 | 6.000 | −3.23% |
| 2.48 ng/mL | Sum of Squares | Degrees of Freedom | Mean Square | F (DFn, DFd) | p-Value |
|---|---|---|---|---|---|
| analyst | 109.2 | 2 | 54.60 | (2, 8) = 3.576 | 0.077 |
| day | 76.27 | 4 | 19.07 | (4, 8) = 1.249 | 0.3644 |
| residual | 122.1 | 8 | 15.27 | ||
| Source of variation | % of total variation | p-value | |||
| analyst | 35.50 | 0.077 | |||
| day | 24.79 | 0.3644 | |||
| 3.72 ng/mL | Sum of squares | Degrees of freedom | Mean square | F (DFn, DFd) | p-value |
| analyst | 4.933 | 2 | 2.467 | (2, 8) = 0.1553 | 0.8587 |
| day | 87.73 | 4 | 21.93 | (4, 8) = 1.381 | 0.3225 |
| residual | 127.1 | 8 | 15.88 | ||
| Source of variation | % of total variation | p-value | |||
| analyst | 2.245 | 0.8587 | |||
| day | 39.93 | 0.3225 | |||
| 6.20 ng/mL | Sum of squares | Degrees of freedom | Mean square | F (DFn, DFd) | p-value |
| analyst | 261.7 | 2 | 130.9 | (2, 8) = 1.324 | 0.3187 |
| day | 433.1 | 4 | 108.3 | (4, 8) = 1.095 | 0.4211 |
| residual | 790.9 | 8 | 98.87 | ||
| Source of variation | % of total variation | p-value | |||
| analyst | 17.62 | 0.3187 | |||
| day | 29.15 | 0.4211 | |||
| Methodology | Detection Limit | Sensitivity | Refs |
|---|---|---|---|
| Liquid chromatography combined with UV | 0.13–1.83 μg/mL | 524–5593 mAU per μg/mL of phenol concentration | [4] |
| Liquid chromatography combined with electrochemistry | 0.017–0.126 μg/mL | 0.0167–0.2650 nA/min per mg/L of phenol concentration | [5] |
| Gas chromatography combined with mass spectrometry | 0.1–0.5 μg/mL | not mentioned | [6] |
| Gas chromatography combined with flame ionization | 0.005–0.120 μg/mL | not mentioned | [7] |
| Binary metal oxide microcube-based glass carbon electrode | 0.018 pg/mL | 7.12 µA/µM cm2 | [8] |
| Ag2O/Sb2O3 nanoparticles deposited on a glassy carbon electrode | 0.009 pg/mL | 11.67 μA/μM cm2 | [9] |
| Chromium (III) oxide nanomaterials-decorated carbon nanotubes | 0.008 pg/mL | 1.4768 μA/mM cm2 | [10] |
| Ce-doped ZnO nanostructures | 1.43 pg/mL | 94.937 μA/μM cm2 | [11] |
| ELISA/quantum dots conjugated with bisphenol A | 13.1 ng/mL | not mentioned | [12] |
| Gold nanoparticles on glassy carbon immunoassay | 0.25 ng/mL | not mentioned | [13] |
| Microchip capillary electrophoresis | 37.6 ng/mL | not mentioned | [14] |
| CdSe/ZnS core/shell type quantum dots on glass carbon electrode | 3.355 pg/mL | 3.6392 µA/µM cm2 | [15] |
| Silver-doped neodymium oxide aggregated nanoparticles | 0.06 pg/mL | 0.2215 μA/μM cm2 | [16] |
| Neodymium oxide co-doped zinc oxide nanostructures | 0.061 pg/mL | 28.481 nA/nM cm2 | [17] |
| Tyrosinase glass carbon sensor | 1.29 ng/mL | 0.256 mC/μM | [25] |
| Liposome bioreactor and chitosan nanocomposite tyrosinase sensor | 1.02 ng/mL | not mentioned | [26] |
| Tyrosinase/redox polymer composite sensor | 9.4 ng/mL | 0.15 nA per µM of analyte concentration | [27] |
| Graphene–Au nanoparticle platforms with chitosan-bound tyrosinase | 4.67 ng/mL | 0.624 μA/μM | [28] |
| Hybrid assemblies of polyaniline, polyacrylonitrile and nanostructured graphene | 24.9 ng/mL | 6.46 μA/μM cm2 | [29] |
| Tyrosinase screen-printed dispersed graphene electrode | 3.25 ng/mL | 1170 µA/mM cm2 | [30] |
| Tyrosinase immobilized on nanocrystalline cellulose quantum dots nanocomposites | 7.7 ng/mL | 0.078 μA/μM | [31] |
| Metal-supported lipid membrane with incorporated tyrosinase | 1.24 pg/mL | 33.45 nA per pg/mL of analyte concentration | This work |
| Matrix Composition | Signal Deviation% (n = 5) |
|---|---|
| Carbonates (32.78 mM as HCO3−) | 0.9 ± 0.3 |
| Nitrates (44.5 mM as NO3) | 3.3 ± 0.2 |
| Phosphates (12.8 mM as PO43−) | 1.5 ± 0.1 |
| Chloride (34.44 mM Cl−) | 3.2 ± 0.2 |
| Sulfates (10 mM as SO42−) | 1.0 ± 0.3 |
| Sulfides (10 mM as (NH4)2S) | 0.5 ± 0.05 |
| Ammonium (10 mM as (NH4)2S) | 1.3 ± 0.6 |
| Calcium (1.05 mM Ca2+) | 1.7 ± 0.1 |
| Magnesium (1.40 mM Mg2+) | 0.4 ± 0.1 |
| HCO3−/NO3/Cl− (at max. concentrations) | 4.2 ± 0.5 |
| PO43−/SO42− (at max. concentrations) | 2.6 ± 0.7 |
| Cl−/(NH4)2S (at max. concentrations) | 4.4 ± 1.1 |
| Ca2+/Mg2+/NO3 (at max. concentrations) | 4.0 ± 0.6 |
| HCO3−/Cl−/SO42−/Ca2+/Mg2+ (at max. concentrations) | 4.5 ± 0.4 |
| Tap Water | River Water | ||||
|---|---|---|---|---|---|
| # | Phenol Detected with the Sensor (ng/mL) | % Relative Error | # | Phenol Detected with the Sensor (ng/mL) | % Relative Error |
| 1 | 9.06 | −3.617 | 1 | 9.39 | −0.126 |
| 2 | 9.36 | −0.426 | 2 | 9.63 | +2.418 |
| 3 | 9.84 | +4.681 | 3 | 9.81 | +4.327 |
| 4 | 9.48 | +0.851 | 4 | 9.03 | −3.943 |
| 5 | 9.66 | +2.766 | 5 | 8.73 | −7.123 |
| 6 | 9.18 | −2.340 | 6 | 9.57 | +1.782 |
| 7 | 9.90 | +5.319 | 7 | 9.33 | −0.762 |
| 8 | 9.84 | +4.681 | 8 | 9.81 | +4.327 |
| 9 | 9.12 | −2.979 | 9 | 8.85 | −5.851 |
| 10 | 8.76 | −6.809 | 10 | 9.15 | −2.670 |
| # | Phenol Detected with the Sensor (ng/mL) | % Relative Error |
|---|---|---|
| 1 | 19.26 | +2.447 |
| 2 | 18.96 | +0.851 |
| 3 | 20.04 | +6.596 |
| 4 | 19.56 | +4.043 |
| 5 | 19.02 | +1.144 |
| 6 | 19.49 | +3.688 |
| 7 | 19.19 | +2.098 |
| 8 | 18.90 | +0.508 |
| 9 | 19.86 | +5.638 |
| 10 | 19.80 | +5.319 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siontorou, C.G.; Georgopoulos, K.N. A Ready-to-Use Metal-Supported Bilayer Lipid Membrane Biosensor for the Detection of Phenol in Water. Membranes 2021, 11, 871. https://doi.org/10.3390/membranes11110871
Siontorou CG, Georgopoulos KN. A Ready-to-Use Metal-Supported Bilayer Lipid Membrane Biosensor for the Detection of Phenol in Water. Membranes. 2021; 11(11):871. https://doi.org/10.3390/membranes11110871
Chicago/Turabian StyleSiontorou, Christina G., and Konstantinos N. Georgopoulos. 2021. "A Ready-to-Use Metal-Supported Bilayer Lipid Membrane Biosensor for the Detection of Phenol in Water" Membranes 11, no. 11: 871. https://doi.org/10.3390/membranes11110871
APA StyleSiontorou, C. G., & Georgopoulos, K. N. (2021). A Ready-to-Use Metal-Supported Bilayer Lipid Membrane Biosensor for the Detection of Phenol in Water. Membranes, 11(11), 871. https://doi.org/10.3390/membranes11110871







