Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance
Abstract
1. Introduction
2. Historical Background and Types of Polymyxins
3. Clinically Useful Polymyxins and Their Principle Properties
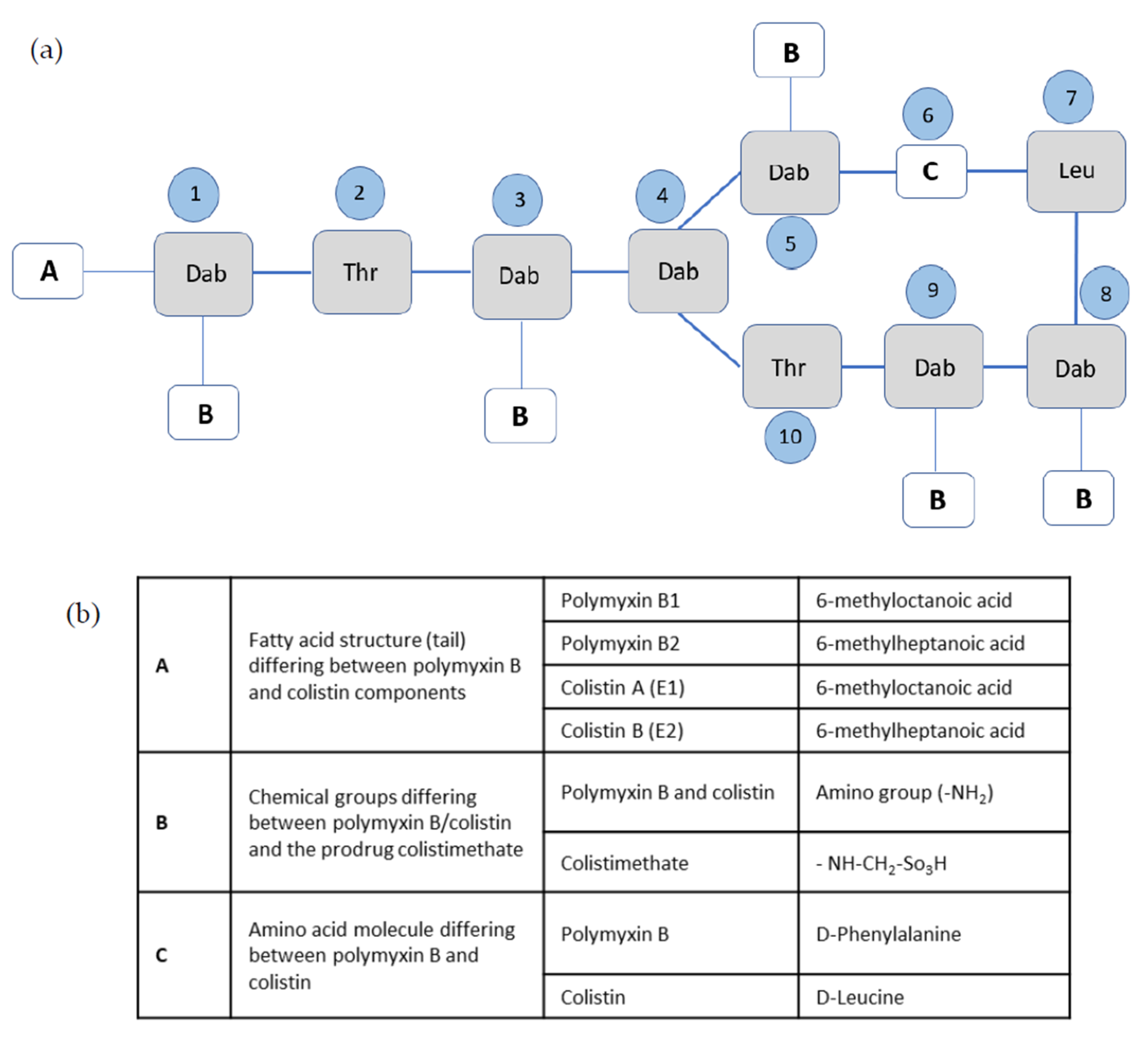
3.1. Chemical Properties and Structure–Activity Relationships
3.2. Spectrum of Antibacterial Activity
3.3. Administration and Clinical Uses
3.3.1. Systemic Use
3.3.2. Topical Use
3.4. Pharmacokinetics
3.5. Toxicity
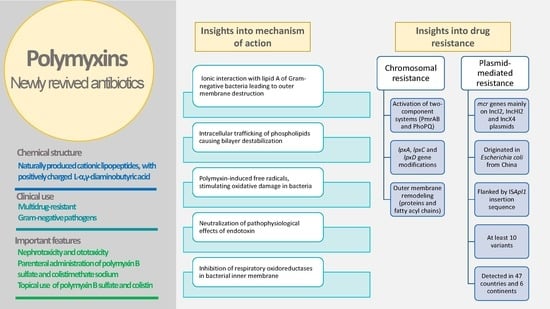
4. Mechanism of Action of Polymyxins and Proposed Interactions with Bacterial Membranes and Other Cellular Structures
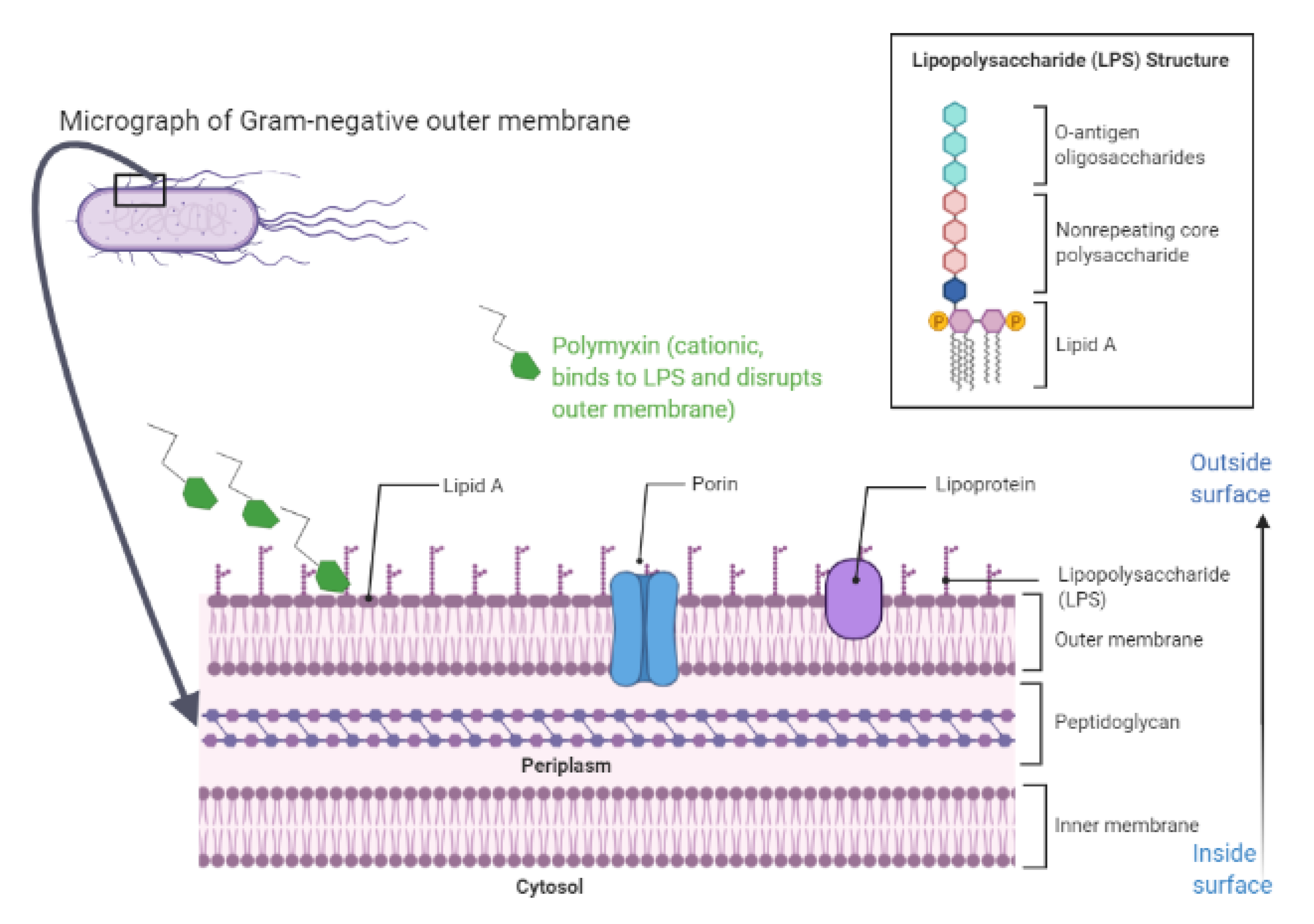
4.1. Overview
4.2. Insights into Models of Polymyxins’ Mechanism of Action
5. Bacterial Resistance to Polymyxins and Changes in Membranes
5.1. Chromosomal Resistance
- The PhoPQ two-component system and its regulatory gene mgrB. This system codes for two proteins, the regulator protein PhoP and the protein kinase PhoQ. While the kinase senses a specific environmental stimulus, the corresponding response regulator mediates the cellular response, mostly through the differential expression of target genes. In the presence of certain environmental stimuli, this system allows the expression of virulence factors, enzymes that modify the LPS to allow resistance to cationic antimicrobial peptides, or enzymes that decrease stress due to acidic pH. The PhoPQ two-component system promotes bacterial survival in low magnesium concentration or in acidic pH or in the presence of cationic antimicrobial peptides. PhoQ is a protein with tyrosine kinase activity that activates PhoP through phosphorylation [102]. Active PhoP drives the transcription of the pmrHFIJKLM operon, involved in the chemical modification of LPS via the addition of L-Ara4N to the LPS. Moreover, PhoP can also activate the pmrA gene, triggering the expression of PmrA protein, causing the addition of pEtN to the LPS [103]. The regulation of the PhoPQ system occurs through the gene mgrB, which acts as a negative regulator. Upon the activation of PhoP, the mgrB gene is upregulated. The translated mgrB protein in turn represses the PhoQ gene. The inactivation of the mgrB gene leads to the overexpression of the phoPQ operon, thus causing pmrHFIJKLM operon activation, leading to the production of L-Ara4N responsible for the acquisition of polymyxin resistance. Studies show that substitutions, insertions, or deletions in the mgrB gene mediate polymyxin resistance [12]. For example, in KPC-producing K. pneumoniae, the transcriptional upregulation of the PhoQ gene was observed in the strains with mgrB alterations, mediating colistin resistance [104]. Although mgrB mutations or inactivation were suggested as major mechanisms for colistin resistance in K. pneumoniae [105,106,107], Borsa et al. reported an overexpression of PhoQ and phoP genes in K. pneumoniae with wild-type mgrB gene, suggesting that other genetic regulations of the PhoPQ system may exist [108]. A very recent report from Korea described the mgrB alteration mediating colistin resistance in E. coli isolated from livestock [109]. It is noteworthy that the mutation of genes other than mgrB may contribute to enhancing PhoPQ system activity, such as ColR/ColS and CprR/CprS regulatory systems in P. aeroginosa [110], and cprR/cprS in C. jejuni [99].
- The PmrAB two-component system. Similar to the PhoPQ system, the PmrAB system is a typical two-component system, so it encodes both PmrA and PmrB. PmrB is a protein with tyrosine kinase activity, that activates the transcriptional regulator PmrA by phosphorylation. Environmental stimuli, such as macrophage phagosomes, ferric iron, aluminum ion, and low pH, activate PmrB. PmrA in turn activates the transcription of the pmrCAB operon and the pmrHFIJKLM operon, that are involved in LPS modification by the addition of pEtN and L-Ara4N [111]. Mutations causing constitutive activation in the pmrA and pmrB genes have been described as being responsible for acquired colistin resistance [99]. Reports of such alterations are availble for E. coli [112], Enterobacter cloacae [113], P. aeruginosa [114], and A. baumannii [115,116].
- The lpxA, lpxC and lpxD genes. This unique set of genes exists in A. baumannii, which can become highly resistant to polymyxins via spontaneous mutations in these lipid A biosynthesis genes. If the biosynthetic lipidA genes, lpxA, lpxC, or lpxD, become inactive, LPS is not formed, and interaction with polymyxins is lost [116]. In its attempt to adapt to the antibiotic pressure induced by polymyxins, A. baumannii, through the inactivation of the aforementioned genes, loses LPS, a major virulence factor and structural component. Such adaptation results in a dramatic decrease in the fitness and virulence and major changes in the physiology, thus providing insights into the low prevalence of polymyxin-resistant A. baumannii isolates with LPS loss in the clinical setting [117].
5.2. Plasmid-Mediated Resistance
6. Special Features and Spread of Polymyxin Resistance among Prominent Gram-Negative Pathogens
6.1. Enterobacteriaceae
6.2. Pseudomonas aeruginosa
6.3. Acinetobacter baumannii
7. Future Implications
8. Conclusions
Funding
Conflicts of Interest
Abbreviations
| CLSI | Clinical Laboratory and Standards Institute |
| CMS | colistimethate sodium |
| Dab | L-α,γ-diaminobutyric acid |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| Kdo | 3-deoxy-D-manno-octulosonic acid |
| L-Ara4N | 4-amino-4-deoxy-L-arabinose |
| Leu | leucine |
| LPS | lipopolysaccharide |
| MALDI-TOF/MS | matrix-assisted laser desorption ionization time of flight/mass spectrometry |
| mcr | mobilized colistin resistance |
| MDR | multi-drug resistant |
| MIC | mimimum inhibitory concentration |
| pEtN | phosphoethanolamine |
| ROS | reactive oxygen species |
| SMR | small multidrug resistance |
| Thr | threonine |
References
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve it. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y. Potent Antibiotics Active against Multidrug-Resistant Gram-Negative Bacteria. Chem. Pharm. Bull. 2020, 68, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E.W. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Petrosillo, N.; Taglietti, F.; Granata, G. Treatment Options for Colistin Resistant Klebsiella pneumoniae: Present and Future. J. Clin. Med. 2019, 8, 934. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, M.; Losito, A.R.; Giamarellou, H. Optimizing therapy in carbapenem-resistant Enterobacteriaceae infections. Curr. Opin. Infect. Dis. 2018, 31, 566–577. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.-G.; Zhong, L.-L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.-B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Nang, S.C.; Li, J.; Velkov, T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 2019, 45, 131–161. [Google Scholar] [CrossRef]
- Velkov, T.; Roberts, K.D.; Thompson, P.E.; Li, J. Polymyxins: A new hope in combating Gram-negative superbugs? Future Med. Chem. 2016, 8, 1017–1025. [Google Scholar] [CrossRef]
- Benedict, R.G.; Langlykke, A.F. Antibiotic activity of Bacillus polymyxa. J. Bacteriol. 1947, 54, 24. [Google Scholar]
- Stansly, P.G.; Shepherd, R.G.; White, H.J. Polymyxin: A new chemotherapeutic agent. Bull. Johns Hopkins Hosp. 1947, 81, 43–54. [Google Scholar]
- Stansly, P.G. The polymyxins: A review and assessment. Am. J. Med. 1949, 7, 807–818. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure—Activity Relationships of Polymyxin Antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef] [PubMed]
- Zavascki, A.P.; Goldani, L.Z.; Li, J.; Nation, R.L. Polymyxin B for the treatment of multidrug-resistant pathogens: A critical review. J. Antimicrob. Chemother. 2007, 60, 1206–1215. [Google Scholar] [CrossRef]
- Poudyal, A.; Howden, B.P.; Bell, J.M.; Gao, W.; Owen, R.J.; Turnidge, J.D.; Nation, R.L.; Li, J. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2008, 62, 1311–1318. [Google Scholar] [CrossRef]
- Levin, A.S.; Barone, A.A.; Penço, J.; Santos, M.V.; Marinho, I.S.; Arruda, E.A.; Manrique, E.I.; Costa, S.F. Intravenous colistin as therapy for nosocomial infections caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 1999, 28, 1008–1011. [Google Scholar] [CrossRef]
- Markou, N.; Apostolakos, H.; Koumoudiou, C.; Athanasiou, M.; Koutsoukou, A.; Alamanos, I.; Gregorakos, L. Intravenous colistin in the treatment of sepsis from multiresistant Gram-negative bacilli in critically ill patients. Crit. Care Lond. Engl. 2003, 7, R78–R83. [Google Scholar] [CrossRef]
- Nation, R.L.; Velkov, T.; Li, J. Colistin and polymyxin B: Peas in a pod, or chalk and cheese? Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014, 59, 88–94. [Google Scholar] [CrossRef]
- Vaara, M. Polymyxins and their Potential Next Generation as Therapeutic Antibiotics. Front. Microbiol. 2019, 10, 1689. [Google Scholar] [CrossRef]
- Yu, Z.; Qin, W.; Lin, J.; Fang, S.; Qiu, J. Antibacterial mechanisms of polymyxin and bacterial resistance. BioMed Res. Int. 2015, 2015, 679109. [Google Scholar] [CrossRef]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an “old” class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Clausell, A.; Garcia-Subirats, M.; Pujol, M.; Busquets, M.A.; Rabanal, F.; Cajal, Y. Gram-negative outer and inner membrane models: Insertion of cyclic cationic lipopeptides. J. Phys. Chem. B 2007, 111, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Hadjadj, L.; Rolain, J.-M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Godoy, A.; Hansford, K.A.; Muldoon, C.; Becker, B.; Elliott, A.G.; Huang, J.X.; Pelingon, R.; Butler, M.S.; Blaskovich, M.A.T.; Cooper, M.A. Structure-Function Studies of Polymyxin B Lipononapeptides. Molecules 2019, 24, 553. [Google Scholar] [CrossRef]
- Magee, T.V.; Brown, M.F.; Starr, J.T.; Ackley, D.C.; Abramite, J.A.; Aubrecht, J.; Butler, A.; Crandon, J.L.; Dib-Hajj, F.; Flanagan, M.E.; et al. Discovery of Dap-3 polymyxin analogues for the treatment of multidrug-resistant Gram-negative nosocomial infections. J. Med. Chem. 2013, 56, 5079–5093. [Google Scholar] [CrossRef]
- Pristovsek, P.; Kidric, J. The search for molecular determinants of LPS inhibition by proteins and peptides. Curr. Top. Med. Chem. 2004, 4, 1185–1201. [Google Scholar] [CrossRef]
- Tsubery, H.; Ofek, I.; Cohen, S.; Fridkin, M. Structure-function studies of polymyxin B nonapeptide: Implications to sensitization of gram-negative bacteria. J. Med. Chem. 2000, 43, 3085–3092. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 3 August 2020).
- Clinical Laboratory and Standards Instiute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; M100 2020; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Niks, M.; Hanzen, J.; Ohlasová, D.; Rovná, D.; Purgelová, A.; Szövényiová, Z.; Vaculíková, A. Multiresistant nosocomial bacterial strains and their “in vitro” susceptibility to chloramphenicol and colistin. Klin. Mikrobiol. Infekcni Lek. 2004, 10, 124–129. [Google Scholar]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2005, 40, 1333–1341. [Google Scholar] [CrossRef]
- Liu, M.-C.; Lin, S.-B.; Chien, H.-F.; Wang, W.-B.; Yuan, Y.-H.; Hsueh, P.-R.; Liaw, S.-J. 10’(Z),13’(E)-heptadecadienylhydroquinone inhibits swarming and virulence factors and increases polymyxin B susceptibility in Proteus mirabilis. PLoS ONE 2012, 7, e45563. [Google Scholar] [CrossRef]
- Jiang, S.-S.; Liu, M.-C.; Teng, L.-J.; Wang, W.-B.; Hsueh, P.-R.; Liaw, S.-J. Proteus mirabilis pmrI, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation, and urothelial cell invasion. Antimicrob. Agents Chemother. 2010, 54, 1564–1571. [Google Scholar] [CrossRef] [PubMed]
- Kerrinnes, T.; Young, B.M.; Leon, C.; Roux, C.M.; Tran, L.; Atluri, V.L.; Winter, M.G.; Tsolis, R.M. Phospholipase A1 modulates the cell envelope phospholipid content of Brucella melitensis, contributing to polymyxin resistance and pathogenicity. Antimicrob. Agents Chemother. 2015, 59, 6717–6724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fage, C.D.; Brown, D.B.; Boll, J.M.; Keatinge-Clay, A.T.; Trent, M.S. Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2730–2739. [Google Scholar] [CrossRef] [PubMed]
- Sorlózano-Puerto, A.; Carrillo-Ávila, J.A.; Gutiérrez-Soto, M.; Navarro-Marí, J.M.; Gutiérrez-Fernández, J. Susceptibility of clinical isolates of Campylobacter jejuni and Campylobacter coli to colistin. New Microbiol. 2018, 41, 235–237. [Google Scholar] [PubMed]
- Bergen, P.J.; Landersdorfer, C.B.; Zhang, J.; Zhao, M.; Lee, H.J.; Nation, R.L.; Li, J. Pharmacokinetics and pharmacodynamics of “old” polymyxins: What is new? Diagn. Microbiol. Infect. Dis. 2012, 74, 213–223. [Google Scholar] [CrossRef]
- Tran, T.B.; Velkov, T.; Nation, R.L.; Forrest, A.; Tsuji, B.T.; Bergen, P.J.; Li, J. Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: Are we there yet? Int. J. Antimicrob. Agents 2016, 48, 592–597. [Google Scholar] [CrossRef]
- Barnett, M.; Bushby, S.R.; Wilkinson, S. Sodium Sulphomethyl Derivatives of Polymyxins. Br. J. Pharmacol. Chemother. 1964, 23, 552–574. [Google Scholar] [CrossRef]
- Bergen, P.J.; Li, J.; Rayner, C.R.; Nation, R.L. Colistin methanesulfonate is an inactive prodrug of colistin against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 1953–1958. [Google Scholar] [CrossRef]
- Beveridge, E.G.; Martin, A.J. Sodium sulphomethyl derivatives of polymyxins. Br. J. Pharmacol. Chemother. 1967, 29, 125–135. [Google Scholar] [CrossRef]
- Li, J.; Nation, R.L.; Milne, R.W.; Turnidge, J.D.; Coulthard, K. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 2005, 25, 11–25. [Google Scholar] [CrossRef]
- Stefaniuk, E.M.; Tyski, S. Colistin Resistance in Enterobacterales Strains—A Current View. Pol. J. Microbiol. 2019, 68, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Nation, R.L.; Li, J.; Cars, O.; Couet, W.; Dudley, M.N.; Kaye, K.S.; Mouton, J.W.; Paterson, D.L.; Tam, V.H.; Theuretzbacher, U.; et al. Framework for optimisation of the clinical use of colistin and polymyxin B: The Prato polymyxin consensus. Lancet Infect. Dis. 2015, 15, 225–234. [Google Scholar] [CrossRef]
- Tsuji, B.T.; Pogue, J.M.; Zavascki, A.P.; Paul, M.; Daikos, G.L.; Forrest, A.; Giacobbe, D.R.; Viscoli, C.; Giamarellou, H.; Karaiskos, I.; et al. International Consensus Guidelines for the Optimal Use of the Polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-Infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 2019, 39, 10–39. [Google Scholar] [CrossRef]
- Hewer, S.C.L.; Smyth, A.R. Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis. Cochrane Database Syst. Rev. 2017, 4, CD004197. [Google Scholar] [CrossRef]
- Bargiacchi, O.; De Rosa, F.G. Intrathecal or intraventricular colistin: A review. Infez. Med. 2016, 24, 3–11. [Google Scholar] [PubMed]
- Robert, P.Y.; Adenis, J.P. Comparative review of topical ophthalmic antibacterial preparations. Drugs 2001, 61, 175–185. [Google Scholar] [CrossRef]
- Kumar, N.; Singh, Y.; Yadav, G.; Mathur, S.K.; Bhadani, U.K. Role of neomycin polymyxin sulfate solution bladder wash for prevention of catheter associated urinary tract infection in traumatic brain injury patient admitted to Intensive Care Unit: A prospective randomized study. Int. J. Crit. Illn. Inj. Sci. 2018, 8, 17–21. [Google Scholar] [CrossRef]
- Amani, S.; Moeini, M. Comparison of Boric Acid and Combination Drug of Polymyxin, Neomycin and Hydrocortisone (Polymyxin NH) in the Treatment of Acute Otitis Externa. J. Clin. Diagn. Res. 2016, 10, MC01–MC04. [Google Scholar] [CrossRef]
- Marchand, S.; Lamarche, I.; Gobin, P.; Couet, W. Dose-ranging pharmacokinetics of colistin methanesulphonate (CMS) and colistin in rats following single intravenous CMS doses. J. Antimicrob. Chemother. 2010, 65, 1753–1758. [Google Scholar] [CrossRef]
- Zavascki, A.P.; Nation, R.L. Nephrotoxicity of Polymyxins: Is there any Difference between Colistimethate and Polymyxin B? Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Kelesidis, T.; Falagas, M.E. The safety of polymyxin antibiotics. Expert Opin. Drug Saf. 2015, 14, 1687–1701. [Google Scholar] [CrossRef] [PubMed]
- Justo, J.A.; Bosso, J.A. Adverse reactions associated with systemic polymyxin therapy. Pharmacotherapy 2015, 35, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Nation, R.L.; Garonzik, S.M.; Li, J.; Thamlikitkul, V.; Giamarellos-Bourboulis, E.J.; Paterson, D.L.; Turnidge, J.D.; Forrest, A.; Silveira, F.P. Updated US and European dose Recommendations for Intravenous Colistin: How do they Perform? Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2016, 62, 552–558. [Google Scholar] [CrossRef]
- Manchandani, P.; Zhou, J.; Ledesma, K.R.; Truong, L.D.; Chow, D.S.-L.; Eriksen, J.L.; Tam, V.H. Characterization of Polymyxin B Biodistribution and Disposition in an Animal Model. Antimicrob. Agents Chemother. 2016, 60, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Li, J.; Tang, S.; Li, J.; Xiao, X. Colistin-induced nephrotoxicity in mice involves the mitochondrial, death receptor, and endoplasmic reticulum pathways. Antimicrob. Agents Chemother. 2014, 58, 4075–4085. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K. Toxicity of polymyxins: A systematic review of the evidence from old and recent studies. Crit. Care Lond. Engl. 2006, 10, R27. [Google Scholar] [CrossRef]
- Dai, C.; Xiao, X.; Li, J.; Ciccotosto, G.D.; Cappai, R.; Tang, S.; Schneider-Futschik, E.K.; Hoyer, D.; Velkov, T.; Shen, J. Molecular Mechanisms of Neurotoxicity Induced by Polymyxins and Chemoprevention. ACS Chem. Neurosci. 2019, 10, 120–131. [Google Scholar] [CrossRef]
- Dai, C.; Tang, S.; Velkov, T.; Xiao, X. Colistin-Induced Apoptosis of Neuroblastoma-2a Cells Involves the Generation of Reactive Oxygen Species, Mitochondrial Dysfunction, and Autophagy. Mol. Neurobiol. 2016, 53, 4685–4700. [Google Scholar] [CrossRef]
- Dai, C.; Ciccotosto, G.D.; Cappai, R.; Wang, Y.; Tang, S.; Xiao, X.; Velkov, T. Minocycline attenuates colistin-induced neurotoxicity via suppression of apoptosis, mitochondrial dysfunction and oxidative stress. J. Antimicrob. Chemother. 2017, 72, 1635–1645. [Google Scholar] [CrossRef]
- Dai, C.; Ciccotosto, G.D.; Cappai, R.; Wang, Y.; Tang, S.; Hoyer, D.; Schneider, E.K.; Velkov, T.; Xiao, X. Rapamycin Confers Neuroprotection against Colistin-Induced Oxidative Stress, Mitochondria Dysfunction, and Apoptosis through the Activation of Autophagy and mTOR/Akt/CREB Signaling Pathways. ACS Chem. Neurosci. 2018, 9, 824–837. [Google Scholar] [CrossRef]
- Li, Y.M.; Milikowski, C.; Selvaggi, G.; Abbo, L.M.; Skiada, D.; Galimberti, F. Polymyxin B-induced skin hyperpigmentation. Transpl. Infect. Dis. Off. J. Transplant. Soc. 2020, e13312. [Google Scholar] [CrossRef]
- Mattos, K.P.H.; Cintra, M.L.; Gouvêa, I.R.; Ferreira, L.Á.; Velho, P.E.N.F.; Moriel, P. Skin hyperpigmentation following intravenous polymyxin B treatment associated with melanocyte activation and inflammatory process. J. Clin. Pharm. Ther. 2017, 42, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Elbediwi, M.; Li, Y.; Paudyal, N.; Pan, H.; Li, X.; Xie, S.; Rajkovic, A.; Feng, Y.; Fang, W.; Rankin, S.C.; et al. Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Resistance Genes Study (1980–2018). Microorganisms 2019, 7, 461. [Google Scholar] [CrossRef]
- Evans, M.E.; Feola, D.J.; Rapp, R.P. Polymyxin B sulfate and colistin: Old antibiotics for emerging multiresistant gram-negative bacteria. Ann. Pharmacother. 1999, 33, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Choi, U.; Lee, C.-R. Antimicrobial Agents that Inhibit the Outer Membrane Assembly Machines of Gram-Negative Bacteria. J. Microbiol. Biotechnol. 2019, 29, 1–10. [Google Scholar] [CrossRef]
- Patel, D.S.; Qi, Y.; Im, W. Modeling and simulation of bacterial outer membranes and interactions with membrane proteins. Curr. Opin. Struct. Biol. 2017, 43, 131–140. [Google Scholar] [CrossRef]
- May, K.L.; Silhavy, T.J. Making a membrane on the other side of the wall. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1386–1393. [Google Scholar] [CrossRef]
- Grabowicz, M. Lipoprotein Transport: Greasing the Machines of Outer Membrane Biogenesis: Re-Examining Lipoprotein Transport Mechanisms Among Diverse Gram-Negative Bacteria While Exploring New Discoveries and Questions. BioEssays News Rev. Mol. Cell. Dev. Biol. 2018, 40, e1700187. [Google Scholar] [CrossRef]
- Zeth, K.; Thein, M. Porins in prokaryotes and eukaryotes: Common themes and variations. Biochem. J. 2010, 431, 13–22. [Google Scholar] [CrossRef]
- Doerrler, W.T. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol. Microbiol. 2006, 60, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Riedemann, N.C.; Guo, R.-F.; Ward, P.A. The enigma of sepsis. J. Clin. Investig. 2003, 112, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Holst, O.; Di Lorenzo, F.; Callaghan, M.; Nurisso, A.; D’Errico, G.; Zamyatina, A.; Peri, F.; Berisio, R.; Jerala, R.; et al. Chemistry of lipid A: At the heart of innate immunity. Chem. Weinh. Bergstr. Ger. 2015, 21, 500–519. [Google Scholar] [CrossRef]
- Kawahara, K. Synthetic chemistry with friendships that unveiled the long-lasting mystery of lipid A. Innate Immun. 2019, 25, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Laloux, G.; Collet, J.-F. Major Tom to Ground Control: How Lipoproteins Communicate Extracytoplasmic Stress to the Decision Center of the Cell. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef]
- Re, H.; Ds, C. Peptide Antibiotics. Available online: https://pubmed.ncbi.nlm.nih.gov/10348745/?from_single_result=peptide+antibiotics+chapple+1999 (accessed on 10 June 2020).
- Clausell, A.; Rabanal, F.; Garcia-Subirats, M.; Alsina, M.A.; Cajal, Y. Membrane association and contact formation by a synthetic analogue of polymyxin B and its fluorescent derivatives. J. Phys. Chem. B 2006, 110, 4465–4471. [Google Scholar] [CrossRef]
- Cajal, Y.; Rogers, J.; Berg, O.G.; Jain, M.K. Intermembrane molecular contacts by polymyxin B mediate exchange of phospholipids. Biochemistry 1996, 35, 299–308. [Google Scholar] [CrossRef]
- Deris, Z.Z.; Swarbrick, J.D.; Roberts, K.D.; Azad, M.A.K.; Akter, J.; Horne, A.S.; Nation, R.L.; Rogers, K.L.; Thompson, P.E.; Velkov, T.; et al. Probing the penetration of antimicrobial polymyxin lipopeptides into gram-negative bacteria. Bioconjug. Chem. 2014, 25, 750–760. [Google Scholar] [CrossRef]
- Berglund, N.A.; Piggot, T.J.; Jefferies, D.; Sessions, R.B.; Bond, P.J.; Khalid, S. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: A molecular dynamics study. PLoS Comput. Biol. 2015, 11, e1004180. [Google Scholar] [CrossRef]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.R.; Ferreira, G.F.; Neto, W.R.N.; De Melo Monteiro, J.; Santos, Á.R.C.; Tavares, P.B.; Denadai, Â.M.L.; Bomfim, M.R.Q.; Dos Santos, V.L.; Marques, S.G.; et al. Evaluation of the interaction between polymyxin B and Pseudomonas aeruginosa biofilm and planktonic cells: Reactive oxygen species induction and zeta potential. BMC Microbiol. 2019, 19, 115. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Liu, X.; Schroeder, M.R.; Kraft, C.S.; Burd, E.M.; Weiss, D.S. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 2012, 56, 5642–5649. [Google Scholar] [CrossRef]
- Dong, T.G.; Dong, S.; Catalano, C.; Moore, R.; Liang, X.; Mekalanos, J.J. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc. Natl. Acad. Sci. USA 2015, 112, 2181–2186. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, Y.; Qin, W.; Yin, J.; Qiu, J. Oxidative Stress Induced by Polymyxin E is Involved in Rapid Killing of Paenibacillus Polymyxa. BioMed Res. Int. 2017, 2017, 5437139. [Google Scholar] [CrossRef]
- Yu, Z.; Cai, Y.; Qin, W.; Lin, J.; Qiu, J. Polymyxin E Induces Rapid Paenibacillus Polymyxa Death by Damaging Cell Membrane while Ca2+ can Protect Cells from Damage. PLoS ONE 2015, 10, e0135198. [Google Scholar] [CrossRef]
- Deris, Z.Z.; Akter, J.; Sivanesan, S.; Roberts, K.D.; Thompson, P.E.; Nation, R.L.; Li, J.; Velkov, T. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J. Antibiot. 2014, 67, 147–151. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, Y.; Fu, J.; Qiu, J.; Yin, J. Enhanced NADH Metabolism Involves Colistin-Induced Killing of Bacillus Subtilis and Paenibacillus Polymyxa. Molecules 2019, 24, 387. [Google Scholar] [CrossRef]
- Mortensen, N.P.; Fowlkes, J.D.; Sullivan, C.J.; Allison, D.P.; Larsen, N.B.; Molin, S.; Doktycz, M.J. Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir ACS J. Surf. Colloids 2009, 25, 3728–3733. [Google Scholar] [CrossRef]
- McCoy, L.S.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Velkov, T.; Li, J.; Tor, Y. Polymyxins and analogues bind to ribosomal RNA and interfere with eukaryotic translation in vitro. Chembiochem Eur. J. Chem. Biol. 2013, 14, 2083–2086. [Google Scholar] [CrossRef] [PubMed]
- Koen, N.; Van Breda, S.V.; Loots, D.T. Elucidating the antimicrobial mechanisms of colistin sulfate on Mycobacterium tuberculosis using metabolomics. Tuberc. Edinb. Scotl. 2018, 111, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Mogi, T.; Murase, Y.; Mori, M.; Shiomi, K.; Omura, S.; Paranagama, M.P.; Kita, K. Polymyxin B identified as an inhibitor of alternative NADH dehydrogenase and malate: Quinone oxidoreductase from the Gram-positive bacterium Mycobacterium smegmatis. J. Biochem. 2009, 146, 491–499. [Google Scholar] [CrossRef]
- Bax, H.I.; De Steenwinkel, J.E.M.; Kate, M.T.T.; Van der Meijden, A.; Verbon, A.; Bakker-Woudenberg, I.A.J.M. Colistin as a potentiator of anti-TB drug activity against Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2015, 70, 2828–2837. [Google Scholar] [CrossRef]
- Lim, L.M.; Ly, N.; Anderson, D.; Yang, J.C.; Macander, L.; Jarkowski, A.; Forrest, A.; Bulitta, J.B.; Tsuji, B.T. Resurgence of Colistin: A Review of Resistance, Toxicity, Pharmacodynamics, and Dosing. Pharmacotherapy 2010, 30, 1279–1291. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, C.; Song, J.; Velkov, T.; Wang, L.; Zhu, Y.; Li, J. Regulating polymyxin resistance in Gram-negative bacteria: Roles of two-component systems PhoPQ and PmrAB. Future Microbiol. 2020, 15, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, E.L.; Kwasnicka, A.; Ochs, M.M.; Hancock, R.E. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 1999, 34, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Groisman, E.A. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol. Microbiol. 2014, 91, 135–144. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; Brink, A.; Poirel, L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 2015, 59, 2780–2784. [Google Scholar] [CrossRef]
- Cannatelli, A.; Giani, T.; D’Andrea, M.M.; Di Pilato, V.; Arena, F.; Conte, V.; Tryfinopoulou, K.; Vatopoulos, A.; Rossolini, G.M. COLGRIT Study Group mgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 2014, 58, 5696–5703. [Google Scholar] [CrossRef] [PubMed]
- Haeili, M.; Javani, A.; Moradi, J.; Jafari, Z.; Feizabadi, M.M.; Babaei, E. mgrB Alterations Mediate Colistin Resistance in Klebsiella pneumoniae Isolates from Iran. Front. Microbiol. 2017, 8, 2470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, D.; Shi, Q.; Quan, J.; Li, X.; Yu, Y. mcr-1 Gene has no Effect on Colistin Resistance when it Coexists with Inactivated mgrB Gene in Klebsiella pneumoniae. Microb. Drug Resist. 2018, 24, 1117–1120. [Google Scholar] [CrossRef] [PubMed]
- Zafer, M.M.; El-Mahallawy, H.A.; Abdulhak, A.; Amin, M.A.; Al-Agamy, M.H.; Radwan, H.H. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 40. [Google Scholar] [CrossRef] [PubMed]
- Borsa, B.A.; Demirci, M.; Gungordu-Dalar, Z.; Karabiyik, G.; Aygun, G.; Kucukbasmaci, O. Molecular Mechanisms of Colistin Resistance among Klebsiella pneumoniae Strains. Clin. Lab. 2019, 65. [Google Scholar] [CrossRef]
- Kim, S.; Woo, J.H.; Kim, N.; Kim, M.H.; Kim, S.Y.; Son, J.H.; Moon, D.C.; Lim, S.-K.; Shin, M.; Lee, J.C. Characterization of Chromosome-Mediated Colistin Resistance in Escherichia coli Isolates from Livestock in Korea. Infect. Drug Resist. 2019, 12, 3291–3299. [Google Scholar] [CrossRef]
- Gutu, A.D.; Sgambati, N.; Strasbourger, P.; Brannon, M.K.; Jacobs, M.A.; Haugen, E.; Kaul, R.K.; Johansen, H.K.; Høiby, N.; Moskowitz, S.M. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob. Agents Chemother. 2013, 57, 2204–2215. [Google Scholar] [CrossRef]
- Gunn, J.S. The Salmonella PmrAB regulon: Lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008, 16, 284–290. [Google Scholar] [CrossRef]
- Sato, T.; Shiraishi, T.; Hiyama, Y.; Honda, H.; Shinagawa, M.; Usui, M.; Kuronuma, K.; Masumori, N.; Takahashi, S.; Tamura, Y.; et al. Contribution of Novel Amino Acid Alterations in PmrA or PmrB to Colistin Resistance in mcr-Negative Escherichia coli Clinical Isolates, Including Major Multidrug-Resistant Lineages O25b:H4-ST131-H30Rx and Non-x. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Hong, Y.-K.; Ko, K.S. PmrAB and PhoPQ Variants in Colistin-Resistant Enterobacter spp. Isolates in Korea. Curr. Microbiol. 2019, 76, 644–649. [Google Scholar] [CrossRef]
- Lin, J.; Xu, C.; Fang, R.; Cao, J.; Zhang, X.; Zhao, Y.; Dong, G.; Sun, Y.; Zhou, T. Resistance and Heteroresistance to Colistin in Pseudomonas aeruginosa Isolates from Wenzhou, China. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Gerson, S.; Betts, J.W.; Lucaßen, K.; Nodari, C.S.; Wille, J.; Josten, M.; Göttig, S.; Nowak, J.; Stefanik, D.; Roca, I.; et al. Investigation of Novel pmrB and eptA Mutations in Isogenic Acinetobacter baumannii Isolates Associated with Colistin Resistance and Increased Virulence In Vivo. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.A.; Halat, D.H. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Ledesma, M.; García-Quintanilla, M.; Martín-Peña, R.; Pulido, M.R.; Pachón, J.; McConnell, M.J. Phenotypic changes associated with Colistin resistance due to Lipopolysaccharide loss in Acinetobacter baumannii. Virulence 2018, 9, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Jasim, R.; Baker, M.A.; Zhu, Y.; Han, M.; Schneider-Futschik, E.K.; Hussein, M.; Hoyer, D.; Li, J.; Velkov, T. A Comparative Study of Outer Membrane Proteome between Paired Colistin-Susceptible and Extremely Colistin-Resistant Klebsiella pneumoniae Strains. ACS Infect. Dis. 2018, 4, 1692–1704. [Google Scholar] [CrossRef]
- Kádár, B.; Kocsis, B.; Tóth, Á.; Kristóf, K.; Felső, P.; Kocsis, B.; Böddi, K.; Szabó, D. Colistin resistance associated with outer membrane protein change in Klebsiella pneumoniae and Enterobacter asburiae. Acta Microbiol. Immunol. Hung. 2017, 64, 217–227. [Google Scholar] [CrossRef][Green Version]
- Velkov, T.; Soon, R.L.; Chong, P.L.; Huang, J.X.; Cooper, M.A.; Azad, M.A.K.; Baker, M.A.; Thompson, P.E.; Roberts, K.; Nation, R.L.; et al. Molecular basis for the increased polymyxin susceptibility of Klebsiella pneumoniae strains with under-acylated lipid A. Innate Immun. 2013, 19, 265–277. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C. Occurrence and Characteristics of Mobile Colistin Resistance (mcr) Gene-Containing Isolates from the Environment: A Review. Int. J. Environ. Res. Public Health 2020, 17, 1028. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm animals and aquaculture: Significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 2020. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Schwarz, S.; Johnson, A.P. Transferable resistance to colistin: A new but old threat. J. Antimicrob. Chemother. 2016, 71, 2066–2070. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Hu, Y.; Li, Z.; Sun, J.; Wang, Q.; Lin, J.; Ye, H.; Liu, F.; Srinivas, S.; Li, D.; et al. Dissemination and Mechanism for the mcr-1 Colistin Resistance. PLoS Pathog. 2016, 12, e1005957. [Google Scholar] [CrossRef] [PubMed]
- Khondker, A.; Dhaliwal, A.K.; Saem, S.; Mahmood, A.; Fradin, C.; Moran-Mirabal, J.; Rheinstädter, M.C. Membrane charge and lipid packing determine polymyxin-induced membrane damage. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. Bull. Eur. Sur. Mal. Transm. Eur. Commun. Dis. Bull. 2016, 21. [Google Scholar] [CrossRef]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. Bull. Eur. Sur. Mal. Transm. Eur. Commun. Dis. Bull. 2017, 22. [Google Scholar] [CrossRef]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi, B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef]
- AbuOun, M.; Stubberfield, E.J.; Duggett, N.A.; Kirchner, M.; Dormer, L.; Nunez-Garcia, J.; Randall, L.P.; Lemma, F.; Crook, D.W.; Teale, C.; et al. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2018, 73, 2904. [Google Scholar] [CrossRef]
- Yang, Y.-Q.; Li, Y.-X.; Lei, C.-W.; Zhang, A.-Y.; Wang, H.-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Halat, D.H.; Moubareck, C.A. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-T.; Song, F.-J.; Zou, M.; Hao, Z.-H.; Shan, H. Emergence of Colistin Resistance Gene mcr-1 in Cronobacter sakazakii Producing NDM-9 and in Escherichia coli from the Same Animal. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Snesrud, E.; He, S.; Chandler, M.; Dekker, J.P.; Hickman, A.B.; McGann, P.; Dyda, F. A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 2016, 60, 6973–6976. [Google Scholar] [CrossRef]
- Poirel, L.; Kieffer, N.; Brink, A.; Coetze, J.; Jayol, A.; Nordmann, P. Genetic Features of mcr-1-Producing Colistin-Resistant Escherichia coli Isolates in South Africa. Antimicrob. Agents Chemother. 2016, 60, 4394–4397. [Google Scholar] [CrossRef]
- Li, R.; Xie, M.; Zhang, J.; Yang, Z.; Liu, L.; Liu, X.; Zheng, Z.; Chan, E.W.-C.; Chen, S. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 2017, 72, 393–401. [Google Scholar] [CrossRef]
- Snesrud, E.; McGann, P.; Chandler, M. The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: The Vehicle for Transferable Colistin Resistance. mBio 2018, 9. [Google Scholar] [CrossRef]
- Zurfluh, K.; Tasara, T.; Poirel, L.; Nordmann, P.; Stephan, R. Draft Genome Sequence of Escherichia coli S51, a Chicken Isolate Harboring a Chromosomally Encoded mcr-1 Gene. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, H.; Liu, Y.-H.; Feng, Y. Towards Understanding mcr-Like Colistin Resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef]
- Sun, J.; Xu, Y.; Gao, R.; Lin, J.; Wei, W.; Srinivas, S.; Li, D.; Yang, R.-S.; Li, X.-P.; Liao, X.-P.; et al. Deciphering mcr-2 Colistin Resistance. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Prim, N.; Turbau, M.; Rivera, A.; Rodríguez-Navarro, J.; Coll, P.; Mirelis, B. Prevalence of colistin resistance in clinical isolates of Enterobacteriaceae: A four-year cross-sectional study. J. Infect. 2017, 75, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Peter, S.; Bezdan, D.; Oberhettinger, P.; Vogel, W.; Dörfel, D.; Dick, J.; Marschal, M.; Liese, J.; Weidenmaier, C.; Autenrieth, I.; et al. Whole-genome sequencing enabling the detection of a colistin-resistant hypermutating Citrobacter werkmanii strain harbouring a novel metallo-β-lactamase VIM-48. Int. J. Antimicrob. Agents 2018, 51, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-Y.; Chen, Y.-F.; Peng, H.-L. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 2010, 17, 60. [Google Scholar] [CrossRef]
- Formosa, C.; Herold, M.; Vidaillac, C.; Duval, R.E.; Dague, E. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J. Antimicrob. Chemother. 2015, 70, 2261–2270. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462. [Google Scholar] [CrossRef]
- Moosavian, M.; Emam, N. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect. Drug Resist. 2019, 12, 1001–1010. [Google Scholar] [CrossRef]
- Jo, H.; Jeong, E.Y.; Jeon, J.; Ban, C. Structural insights into Escherichia coli polymyxin B resistance protein D with X-ray crystallography and small-angle X-ray scattering. BMC Struct. Biol. 2014, 14, 24. [Google Scholar] [CrossRef][Green Version]
- Moon, K.; Gottesman, S. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 2009, 74, 1314–1330. [Google Scholar] [CrossRef]
- Vounba, P.; Rhouma, M.; Arsenault, J.; Alambédji, R.B.; Fravalo, P.; Fairbrother, J.M. Prevalence of colistin resistance and mcr-1/mcr-2 genes in extended-spectrum β-lactamase/AmpC-producing Escherichia coli isolated from chickens in Canada, Senegal and Vietnam. J. Glob. Antimicrob. Resist. 2019, 19, 222–227. [Google Scholar] [CrossRef]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Miller, S.I. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 1996, 178, 6857–6864. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.; Porrero, M.C.; Téllez, S.; Palomo, G.; García, M.; Domínguez, L. Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J. Antimicrob. Chemother. 2015, 70, 71–74. [Google Scholar] [CrossRef]
- Sheng, X.; Wang, W.; Chen, L.; Zhang, H.; Zhang, Y.; Xu, S.; Xu, H.; Huang, X. Mig-14 may contribute to Salmonella enterica serovar Typhi resistance to polymyxin B by decreasing the permeability of the outer-membrane and promoting the formation of biofilm. Int. J. Med. Microbiol. 2019, 309, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Hammerl, J.A.; Deneke, C.; Fischer, J.; Szabo, I.; Malorny, B. Characterization of mcr-5-Harboring Salmonella enterica subsp. enterica Serovar Typhimurium Isolates from Animal and Food Origin in Germany. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Miller, A.K.; Brannon, M.K.; Stevens, L.; Johansen, H.K.; Selgrade, S.E.; Miller, S.I.; Høiby, N.; Moskowitz, S.M. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2011, 55, 5761–5769. [Google Scholar] [CrossRef]
- Barrow, K.; Kwon, D.H. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 5150–5154. [Google Scholar] [CrossRef]
- Fernández, L.; Jenssen, H.; Bains, M.; Wiegand, I.; Gooderham, W.J.; Hancock, R.E.W. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob. Agents Chemother. 2012, 56, 6212–6222. [Google Scholar] [CrossRef]
- Goli, H.R.; Nahaei, M.R.; Rezaee, M.A.; Hasani, A.; Kafil, H.S.; Aghazadeh, M. Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iran. J. Microbiol. 2016, 8, 62–69. [Google Scholar]
- Puja, H.; Bolard, A.; Noguès, A.; Plésiat, P.; Jeannot, K. The Efflux Pump MexXY/OprM Contributes to the Tolerance and Acquired Resistance of Pseudomonas aeruginosa to Colistin. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Han, M.-L.; Velkov, T.; Zhu, Y.; Roberts, K.D.; Le Brun, A.P.; Chow, S.H.; Gutu, A.D.; Moskowitz, S.M.; Shen, H.-H.; Li, J. Polymyxin-Induced Lipid A Deacylation in Pseudomonas aeruginosa Perturbs Polymyxin Penetration and Confers High-Level Resistance. ACS Chem. Biol. 2018, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Snesrud, E.; Maybank, R.; Kwak, Y.I.; Jones, A.R.; Hinkle, M.K.; McGann, P. Chromosomally Encoded mcr-5 in Colistin-Nonsusceptible Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: First report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-Y.; Gregg, K.A.; Napier, B.A.; Ernst, R.K.; Weiss, D.S. A PmrB-Regulated Deacetylase Required for Lipid A Modification and Polymyxin Resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015, 59, 7911–7914. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-F.; Lin, Y.-Y.; Lan, C.-Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. Seoul Korea 2017, 55, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Shen, C.; Zheng, X.; Liu, Y.; Chen, H.; Zhong, L.; Liang, Y.; Liao, K.; Xia, Y.; Tian, G.-B.; et al. Identification of a Novel Plasmid Carrying mcr-4.3 in an Acinetobacter baumannii Strain in China. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Mularski, A.; Wilksch, J.; Hanssen, E.; Li, J.; Tomita, T.; Pidot, S.J.; Stinear, T.; Separovic, F.; Strugnell, D. A nanomechanical study of the effects of colistin on the Klebsiella pneumoniae AJ218 capsule. Eur. Biophys. J. 2017, 46, 351–361. [Google Scholar] [CrossRef]
- Llobet, E.; Tomás, J.M.; Bengoechea, J.A. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiol. Read. Engl. 2008, 154, 3877–3886. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Emerging plasmid-encoded colistin resistance: The animal world as the culprit? J. Antimicrob. Chemother. 2016, 71, 2326–2327. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, G.-B.; Zhang, R.; Shen, Y.; Tyrrell, J.M.; Huang, X.; Zhou, H.; Lei, L.; Li, H.-Y.; Doi, Y.; et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: An epidemiological and clinical study. Lancet Infect. Dis. 2017, 17, 390–399. [Google Scholar] [CrossRef]
- Caselli, E.; D’Accolti, M.; Soffritti, I.; Piffanelli, M.; Mazzacane, S. Spread of mcr-1-Driven Colistin Resistance on Hospital Surfaces, Italy. Emerg. Infect. Dis. 2018, 24, 1752–1753. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.J.; Hong, J.S.; Yang, J.W.; Lee, K.J.; Lee, H.; Jeong, S.H. Detection of mcr-1 Plasmids in Enterobacteriaceae Isolates from Human Specimens: Comparison with those in Escherichia coli Isolates from Livestock in Korea. Ann. Lab. Med. 2018, 38, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-T.; Li, X.; Zhang, Q.; Shan, H.; Zou, M.; Song, F.-J. Colistin-Resistant mcr-Positive Enterobacteriaceae in Fresh Vegetables, an Increasing Infectious Threat in China. Int. J. Antimicrob. Agents 2019, 54, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.S.; Elshafiee, E.A.; Khalefa, H.S.; Kadry, M.; Hamza, D.A. Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob. Resist. Infect. Control 2019, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Feng, S.; Chen, H.; Dai, M.; Paterson, D.L.; Zheng, X.; Wu, X.; Zhong, L.-L.; Liu, Y.; Xia, Y.; et al. Transmission of mcr-1-Producing Multidrug-resistant Enterobacteriaceae in Public Transportation in Guangzhou, China. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 67, S217–S224. [Google Scholar] [CrossRef]
- Hassen, B.; Saloua, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Hammami, S.; Torres, C. mcr-1 encoding colistin resistance in CTX-M-1/CTX-M-15- producing Escherichia coli isolates of bovine and caprine origins in Tunisia. First report of CTX-M-15-ST394/D E. coli from goats. Comp. Immunol. Microbiol. Infect. Dis. 2019, 67, 101366. [Google Scholar] [CrossRef]
- Dalmolin, T.V.; Martins, A.F.; Zavascki, A.P.; De Lima-Morales, D.; Barth, A.L. Acquisition of the mcr-1 gene by a high-risk clone of KPC-2-producing Klebsiella pneumoniae ST437/CC258, Brazil. Diagn. Microbiol. Infect. Dis. 2018, 90, 132–133. [Google Scholar] [CrossRef]
- Caltagirone, M.; Nucleo, E.; Spalla, M.; Zara, F.; Novazzi, F.; Marchetti, V.M.; Piazza, A.; Bitar, I.; De Cicco, M.; Paolucci, S.; et al. Occurrence of Extended Spectrum β-Lactamases, KPC-Type, and mcr-1.2-Producing Enterobacteriaceae from Wells, River Water, and Wastewater Treatment Plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 2017, 8, 2232. [Google Scholar] [CrossRef]
- Moubareck, C.A.; Mouftah, S.F.; Pál, T.; Ghazawi, A.; Halat, D.H.; Nabi, A.; AlSharhan, M.A.; AlDeesi, Z.O.; Peters, C.C.; Celiloglu, H.; et al. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int. J. Antimicrob. Agents 2018, 52, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Orsi, T.D.; Neto, L.V.P.; Martins, R.C.R.; Levin, A.S.; Costa, S.F. Polymyxin-resistant Pseudomonas aeruginosa assigned as ST245: First report in an intensive care unit in São Paulo, Brazil. J. Glob. Antimicrob. Resist. 2019, 16, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Ko, K.S. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn. Microbiol. Infect. Dis. 2014, 78, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Abraham, N.; Kwon, D.H. A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009, 298, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Owusu-Anim, D.; Kwon, D.H. Differential Role of Two-Component Regulatory Systems (phoPQ and pmrAB) in Polymyxin B Susceptibility of Pseudomonas aeruginosa. Adv. Microbiol. 2012, 2. [Google Scholar] [CrossRef]
- Muller, C.; Plésiat, P.; Jeannot, K. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.-I.; Hakamada, H.; Yamamoto, S.; Sato, T.; Shiraishi, T.; Shinagawa, M.; Takahashi, S. Release of large amounts of lipopolysaccharides from Pseudomonas aeruginosa cells reduces their susceptibility to colistin. Int. J. Antimicrob. Agents 2018, 51, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-L.; Lu, M.-C.; Shao, P.-L.; Lu, P.-L.; Chen, Y.-H.; Cheng, S.-H.; Ko, W.-C.; Lin, C.-Y.; Wu, T.-S.; Yen, M.-Y.; et al. Nationwide surveillance of antimicrobial resistance among clinically important Gram-negative bacteria, with an emphasis on carbapenems and colistin: Results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) in 2018. Int. J. Antimicrob. Agents 2019, 54, 318–328. [Google Scholar] [CrossRef]
- Katchanov, J.; Asar, L.; Klupp, E.-M.; Both, A.; Rothe, C.; König, C.; Rohde, H.; Kluge, S.; Maurer, F.P. Carbapenem-resistant Gram-negative pathogens in a German university medical center: Prevalence, clinical implications and the role of novel β-lactam/β-lactamase inhibitor combinations. PLoS ONE 2018, 13, e0195757. [Google Scholar] [CrossRef]
- Del Barrio-Tofiño, E.; Zamorano, L.; Cortes-Lara, S.; López-Causapé, C.; Sánchez-Diener, I.; Cabot, G.; Bou, G.; Martínez-Martínez, L.; Oliver, A. GEMARA-SEIMC/REIPI Pseudomonas study Group Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J. Antimicrob. Chemother. 2019, 74, 1825–1835. [Google Scholar] [CrossRef]
- Wi, Y.M.; Choi, J.-Y.; Lee, J.-Y.; Kang, C.-I.; Chung, D.R.; Peck, K.R.; Song, J.-H.; Ko, K.S. Emergence of colistin resistance in Pseudomonas aeruginosa ST235 clone in South Korea. Int. J. Antimicrob. Agents 2017, 49, 767–769. [Google Scholar] [CrossRef]
- Tunyapanit, W.; Pruekprasert, P.; Laoprasopwattana, K.; Chelae, S. In vitro activity of colistin against multidrug-resistant Pseudomonas aeruginosa isolates from patients in Songklanagarind Hospital, Thailand. Southeast Asian J. Trop. Med. Public Health 2013, 44, 273–280. [Google Scholar]
- Moubareck, C.A.; Halat, D.H.; Akkawi, C.; Nabi, A.; AlSharhan, M.A.; AlDeesi, Z.O.; Peters, C.C.; Celiloglu, H.; Sarkis, D.K. Role of outer membrane permeability, efflux mechanism, and carbapenemases in carbapenem-nonsusceptible Pseudomonas aeruginosa from Dubai hospitals: Results of the first cross-sectional survey. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2019, 84, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Garg, J.; Kumar, S.; Bhattacharya, A.; Agarwal, S.; Upadhyay, G.C. Molecular epidemiology & therapeutic options of carbapenem-resistant Gram-negative bacteria. Indian J. Med. Res. 2019, 149, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Giani, T.; Arena, F.; Pollini, S.; Di Pilato, V.; D’Andrea, M.M.; De Angelis, L.H.; Bassetti, M.; Rossolini, G.M. Pseudomonas aeruginosa Working Group Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: Activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J. Antimicrob. Chemother. 2018, 73, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Trebosc, V.; Gartenmann, S.; Tötzl, M.; Lucchini, V.; Schellhorn, B.; Pieren, M.; Lociuro, S.; Gitzinger, M.; Tigges, M.; Bumann, D.; et al. Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Lima, W.G.; Alves, M.C.; Cruz, W.S.; Paiva, M.C. Chromosomally encoded and plasmid-mediated polymyxins resistance in Acinetobacter baumannii: A huge public health threat. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018, 37, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Adler, B.; Nation, R.L.; Li, J.; Boyce, J.D. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3022–3024. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.F.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef]
- Charretier, Y.; Diene, S.M.; Baud, D.; Chatellier, S.; Santiago-Allexant, E.; Van Belkum, A.; Guigon, G.; Schrenzel, J. Colistin Heteroresistance and Involvement of the PmrAB Regulatory System in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Whitfield, C.; Trent, M.S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. [Google Scholar] [CrossRef]
- Lucas, D.D.; Crane, B.; Wright, A.; Han, M.-L.; Moffatt, J.; Bulach, D.; Gladman, S.L.; Powell, D.; Aranda, J.; Seemann, T.; et al. Emergence of High-Level Colistin Resistance in an Acinetobacter baumannii Clinical Isolate Mediated by Inactivation of the Global Regulator H-NS. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Gelbíčová, T.; Baráková, A.; Florianová, M.; Karpíšková, R. Detection of colistin-resistant Acinetobacter baumannii with the mcr-4 gene. Klin. Mikrobiol. Infekcni Lek. 2019, 25, 4–6. [Google Scholar]
- Martins-Sorenson, N.; Snesrud, E.; Xavier, D.E.; Cacci, L.C.; Iavarone, A.T.; McGann, P.; Riley, L.W.; Moreira, B.M. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020, 75, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Wang, J.; Thompson, P.E.; Li, J. Teaching “old” polymyxins new tricks: New-generation lipopeptides targeting gram-negative “superbugs”. ACS Chem. Biol. 2014, 9, 1172–1177. [Google Scholar] [CrossRef]
- Yu, S.; Wang, S.; Zou, P.; Chai, G.; Lin, Y.-W.; Velkov, T.; Li, J.; Pan, W.; Zhou, Q.T. Inhalable liposomal powder formulations for co-delivery of synergistic ciprofloxacin and colistin against multi-drug resistant gram-negative lung infections. Int. J. Pharm. 2020, 575, 118915. [Google Scholar] [CrossRef]
- Dubashynskaya, N.V.; Skorik, Y.A. Polymyxin Delivery Systems: Recent Advances and Challenges. Pharmaceuticals 2020, 13, 83. [Google Scholar] [CrossRef] [PubMed]
- Obuobi, S.; Voo, Z.X.; Low, M.W.; Czarny, B.; Selvarajan, V.; Ibrahim, N.L.; Yang, Y.Y.; Ee, P.L.R. Phenylboronic Acid Functionalized Polycarbonate Hydrogels for Controlled Release of Polymyxin B in Pseudomonas aeruginosa Infected Burn Wounds. Adv. Healthc. Mater. 2018, 7, 1701388. [Google Scholar] [CrossRef]
- Dillon, C.; Hughes, H.; O’Reilly, N.J.; McLoughlin, P. Formulation and characterisation of dissolving microneedles for the transdermal delivery of therapeutic peptides. Int. J. Pharm. 2017, 526, 125–136. [Google Scholar] [CrossRef]
- Dillon, C.; Hughes, H.; O’Reilly, N.J.; Allender, C.J.; Barrow, D.A.; McLoughlin, P. Dissolving microneedle based transdermal delivery of therapeutic peptide analogues. Int. J. Pharm. 2019, 565, 9–19. [Google Scholar] [CrossRef]
- Bartelds, R.; Nematollahi, M.H.; Pols, T.; Stuart, M.C.A.; Pardakhty, A.; Asadikaram, G.; Poolman, B. Niosomes, an alternative for liposomal delivery. PLoS ONE 2018, 13, e0194179. [Google Scholar] [CrossRef]
- Chauhan, M.K.; Bhatt, N. Bioavailability Enhancement of Polymyxin B with Novel Drug Delivery: Development and Optimization Using Quality-by-Design Approach. J. Pharm. Sci. 2019, 108, 1521–1528. [Google Scholar] [CrossRef]
- Hussein, M.; Schneider-Futschik, E.K.; Paulin, O.K.A.; Allobawi, R.; Crawford, S.; Zhou, Q.T.; Hanif, A.; Baker, M.; Zhu, Y.; Li, J.; et al. Effective Strategy Targeting Polymyxin-Resistant Gram-Negative Pathogens: Polymyxin B in Combination with the Selective Serotonin Reuptake Inhibitor Sertraline. ACS Infect. Dis. 2020, 6, 1436–1450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Li, X.; Dong, L.; Hu, X.; Nie, T.; Lu, Y.; Lu, X.; Pang, J.; Li, G.; et al. Synergistic Effect of Colistin Combined with PFK-158 against Colistin-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Loose, M.; Naber, K.G.; Hu, Y.; Coates, A.; Wagenlehner, F.M.E. Urinary bactericidal activity of colistin and azidothymidine combinations against mcr-1-positive colistin-resistant Escherichia coli. Int. J. Antimicrob. Agents 2019, 54, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.; Han, M.-L.; Zhu, Y.; Schneider-Futschik, E.K.; Hu, X.; Zhou, Q.T.; Lin, Y.-W.; Anderson, D.; Creek, D.J.; Hoyer, D.; et al. Mechanistic Insights from Global Metabolomics Studies into Synergistic Bactericidal Effect of a Polymyxin B Combination with Tamoxifen against Cystic Fibrosis MDR Pseudomonas aeruginosa. Comput. Struct. Biotechnol. J. 2018, 16, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Geitani, R.; Moubareck, C.A.; Touqui, L.; Sarkis, D.K. Cationic antimicrobial peptides: Alternatives and/or adjuvants to antibiotics active against methicillin-resistant Staphylococcus aureus and multidrug-resistant Pseudomonas aeruginosa. BMC Microbiol. 2019, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Dosler, S.; Karaaslan, E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef]
- Frecer, V.; Ho, B.; Ding, J.L. De novo design of potent antimicrobial peptides. Antimicrob. Agents Chemother. 2004, 48, 3349–3357. [Google Scholar] [CrossRef]
- Brown, P.; Abbott, E.; Abdulle, O.; Boakes, S.; Coleman, S.; Divall, N.; Duperchy, E.; Moss, S.; Rivers, D.; Simonovic, M.; et al. Design of next Generation Polymyxins with Lower Toxicity: The Discovery of SPR206. ACS Infect. Dis. 2019, 5, 1645–1656. [Google Scholar] [CrossRef]
| EUCAST Breakpoints (mg/L) | CLSI Breakpoints | |||
|---|---|---|---|---|
| S≤ | R> | I≤ | R≥ | |
| Enterobacteriaceae | 2 | 2 | 2 | 4 |
| Pseudomonas | 2 | 2 | 2 | 4 |
| Acinetobacter | 2 | 2 | 2 | 4 |
| mcr Gene Type | Species of First Detection | Country of First Detection | Sequence Homology to mcr-1 (%) | Strain Information | Reference |
|---|---|---|---|---|---|
| mcr-1 | Escherichia coli | China | 100 | SHP45 | [123] |
| mcr-2 | E. coli | Belgium | 76.7 | CP011374 | [128] |
| mcr-3 | E. coli | China | 45 | WJ1 | [129] |
| mcr-4 | Salmonella enterica serovar Typhimurium | Italy | 34 | R3445 | [130] |
| mcr-5 | S. enterica subsp. enterica | Germany | 63.89 | 11-00422 | [131] |
| mcr-6 | Moraxella pluranimalium | Great Britain | 62 | 248-01T/DSM-22804) | [132] |
| mcr-7 | Klebsiella pneumoniae | China | 65 | SC20141012 | [133] |
| mcr-8 | K. pneumoniae | China | 31.08 | KP91 | [134] |
| mcr-9 | S. enterica subsp. enterica | New York | 63 | GCF_002091095.1 | [135] |
| mcr-10 | Enterobacter roggenkampii | China | 29.31 | 090065 (WCHER090065) | [125] |
| Chromosomal Resistance | Plasmid-Encoded Resistance | ||||
|---|---|---|---|---|---|
| Two-component systems [ref] | Additional mechanisms [ref] | mcr type | Ref | ||
| Enterobacteriaceae | Klebsiella pneumoniae | PhoPQ [103] PmrAB [147] | Shedding of capsular polysaccharide capable of trapping polymyxins [148] Overexpression of the efflux pump kpnEF [149] | mcr-1 | [108] |
| mcr-2 | [108,150] | ||||
| mcr-7 | [133] | ||||
| mcr-8 | [134] | ||||
| E. coli | PhoPQ [151] PmrAB [112] | Modification of Kdo (3-deoxy-D-manno-octulosonic acid) [152] | mcr-1 | [123] | |
| mcr-2 | [153] | ||||
| mcr-3 | [129] | ||||
| mcr-4 | [154] | ||||
| mcr-5 | [154] | ||||
| Salmonella | PhoPQ [155] PmrAB [156] | Inhibition of expression of outer membrane proteins OmpF and/or OmpC [157] | mcr-4 | [130] | |
| mcr-5 | [158] | ||||
| Pseudomonas | PhoPQ [159] PmrAB [160] CprRS [161] ColRS [110] ParRS [162] | Efflux pump MexXY/OprM [163] Lipid A diacylation [164] Chromosomal mcr-5 [165] | mcr-1 | [166] | |
| Acinetobacter | PmrAB [167] | Expression of the efflux pump EmrAB [168] | mcr-1 | [166] | |
| mcr-4.3 | [169] | ||||
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayoub Moubareck, C. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 2020, 10, 181. https://doi.org/10.3390/membranes10080181
Ayoub Moubareck C. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes. 2020; 10(8):181. https://doi.org/10.3390/membranes10080181
Chicago/Turabian StyleAyoub Moubareck, Carole. 2020. "Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance" Membranes 10, no. 8: 181. https://doi.org/10.3390/membranes10080181
APA StyleAyoub Moubareck, C. (2020). Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes, 10(8), 181. https://doi.org/10.3390/membranes10080181