Mice Immunized with the Vaccine Candidate HexaPro Spike Produce Neutralizing Antibodies against SARS-CoV-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Expression and Purification of HexaPro Subunit Vaccine
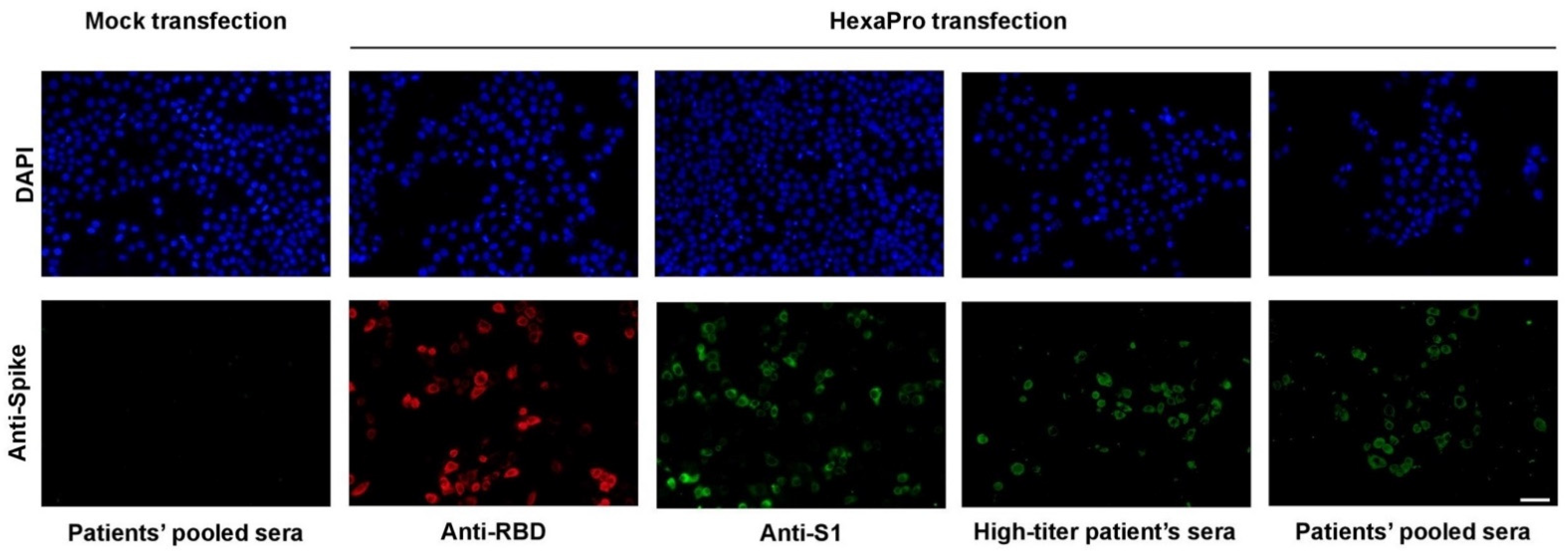
2.3. Immunofluorescence Staining
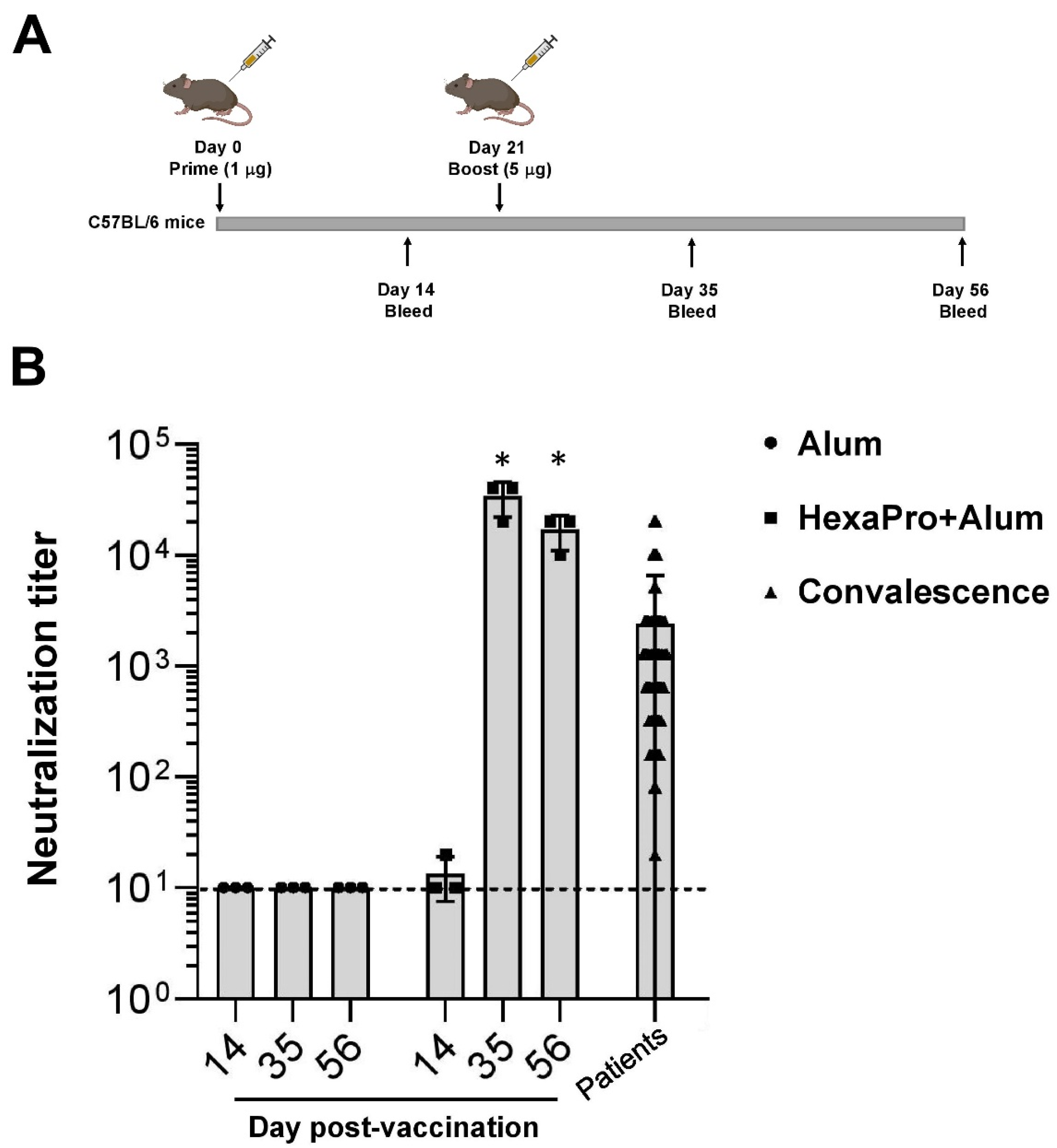
2.4. Mouse Immunization
2.5. Microneutralization Assay
3. Results
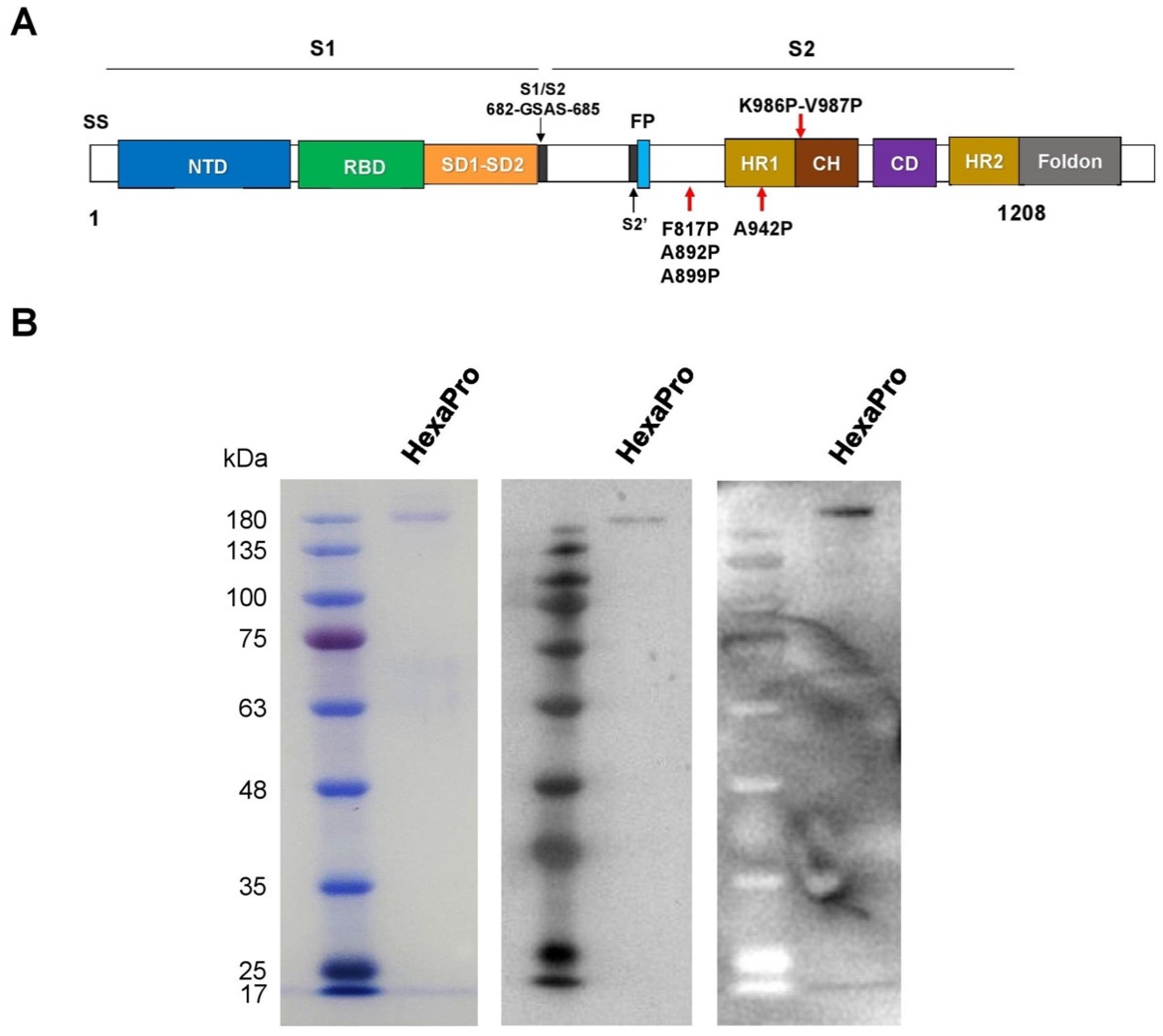
3.1. Expression and Purification of Recombinant SARS-CoV-2 HexaPro Spike Protein
3.2. Neutralization of SARS-CoV-2 by Sera Collected from HexaPro-Immunized Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292 e286. [Google Scholar] [CrossRef]
- Ozono, S.; Zhang, Y.; Ode, H.; Sano, K.; Tan, T.S.; Imai, K.; Miyoshi, K.; Kishigami, S.; Ueno, T.; Iwatani, Y.; et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 2021, 12, 848. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; Graham, B.S.; et al. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488. [Google Scholar] [CrossRef]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Hsieh, C.L.; Goldsmith, J.A.; Schaub, J.M.; DiVenere, A.M.; Kuo, H.C.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Soullié, T.; van Nimwegen, M.; Willart, M.A.; Muskens, F.; Jung, S.; Hoogsteden, H.C.; Hammad, H.; Lambrecht, B.N. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J. Exp. Med. 2008, 205, 869–882. [Google Scholar] [CrossRef]
- Kool, M.; Pétrilli, V.; De Smedt, T.; Rolaz, A.; Hammad, H.; van Nimwegen, M.; Bergen, I.M.; Castillo, R.; Lambrecht, B.N.; Tschopp, J. Cutting edge: Alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J. Immunol. 2008, 181, 3755–3759. [Google Scholar] [CrossRef] [PubMed]
- HogenEsch, H.; O’Hagan, D.T.; Fox, C.B. Optimizing the utilization of aluminum adjuvants in vaccines: You might just get what you want. NPJ Vaccines 2018, 3, 51. [Google Scholar] [CrossRef]
- Ella, R.; Vadrevu, K.M.; Jogdand, H.; Prasad, S.; Reddy, S.; Sarangi, V.; Ganneru, B.; Sapkal, G.; Yadav, P.; Abraham, P.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: A double-blind, randomised, phase 1 trial. Lancet Infect. Dis. 2021, 21, 637–646. [Google Scholar] [CrossRef]
- Pace, D.; Khatami, A.; McKenna, J.; Campbell, D.; Attard-Montalto, S.; Birks, J.; Voysey, M.; White, C.; Finn, A.; Macloed, E.; et al. Immunogenicity of reduced dose priming schedules of serogroup C meningococcal conjugate vaccine followed by booster at 12 months in infants: Open label randomised controlled trial. BMJ 2015, 350, h1554. [Google Scholar] [CrossRef]
- Nitayaphan, S.; Pitisuttithum, P.; Karnasuta, C.; Eamsila, C.; de Souza, M.; Morgan, P.; Polonis, V.; Benenson, M.; VanCott, T.; Ratto-Kim, S.; et al. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J. Infect. Dis. 2004, 190, 702–706. [Google Scholar] [CrossRef]
- Sandström, E.; Nilsson, C.; Hejdeman, B.; Bråve, A.; Bratt, G.; Robb, M.; Cox, J.; Vancott, T.; Marovich, M.; Stout, R.; et al. Broad immunogenicity of a multigene, multiclade HIV-1 DNA vaccine boosted with heterologous HIV-1 recombinant modified vaccinia virus Ankara. J. Infect. Dis. 2008, 198, 1482–1490. [Google Scholar] [CrossRef]
- Kardani, K.; Bolhassani, A.; Shahbazi, S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine 2016, 34, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.J.; Mansouri, K.; Stalls, V.; Manne, K.; Watts, B.; Parks, R.; Janowska, K.; Gobeil, S.M.C.; Kopp, M.; Li, D.; et al. Cold sensitivity of the SARS-CoV-2 spike ectodomain. Nat. Struct. Mol. Biol. 2021, 28, 128–131. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seephetdee, C.; Buasri, N.; Bhukhai, K.; Srisanga, K.; Manopwisedjaroen, S.; Lertjintanakit, S.; Phueakphud, N.; Pakiranay, C.; Kangwanrangsan, N.; Srichatrapimuk, S.; et al. Mice Immunized with the Vaccine Candidate HexaPro Spike Produce Neutralizing Antibodies against SARS-CoV-2. Vaccines 2021, 9, 498. https://doi.org/10.3390/vaccines9050498
Seephetdee C, Buasri N, Bhukhai K, Srisanga K, Manopwisedjaroen S, Lertjintanakit S, Phueakphud N, Pakiranay C, Kangwanrangsan N, Srichatrapimuk S, et al. Mice Immunized with the Vaccine Candidate HexaPro Spike Produce Neutralizing Antibodies against SARS-CoV-2. Vaccines. 2021; 9(5):498. https://doi.org/10.3390/vaccines9050498
Chicago/Turabian StyleSeephetdee, Chotiwat, Nattawut Buasri, Kanit Bhukhai, Kitima Srisanga, Suwimon Manopwisedjaroen, Sarat Lertjintanakit, Nut Phueakphud, Chatbenja Pakiranay, Niwat Kangwanrangsan, Sirawat Srichatrapimuk, and et al. 2021. "Mice Immunized with the Vaccine Candidate HexaPro Spike Produce Neutralizing Antibodies against SARS-CoV-2" Vaccines 9, no. 5: 498. https://doi.org/10.3390/vaccines9050498
APA StyleSeephetdee, C., Buasri, N., Bhukhai, K., Srisanga, K., Manopwisedjaroen, S., Lertjintanakit, S., Phueakphud, N., Pakiranay, C., Kangwanrangsan, N., Srichatrapimuk, S., Kirdlarp, S., Sungkanuparph, S., Chutipongtanate, S., Thitithanyanont, A., Hongeng, S., & Wongtrakoongate, P. (2021). Mice Immunized with the Vaccine Candidate HexaPro Spike Produce Neutralizing Antibodies against SARS-CoV-2. Vaccines, 9(5), 498. https://doi.org/10.3390/vaccines9050498





