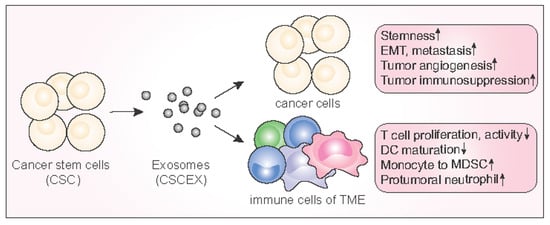
Exosomes and Cancer Stem Cells in Cancer Immunity: Current Reports and Future Directions
Abstract
1. Introduction
2. Exosomes Derived from Cancer Stem Cells (CSCs)
2.1. CSC-Derived Exosomes (CSCEXs)
2.1.1. Expression of CSC Biomarkers in Different Cancer Types
2.1.2. CSCEXs and Their Cargo
2.1.3. The Current Efforts to Define Differences between CSCEXs and TEXs
3. Biological and Physiological Roles of CSCEXs
3.1. Role of Exosomes in the Maintenance of Homeostasis between CSCs and Non-Stem Cancer Cells
3.2. EMT, Exosomes, and CSCs
3.3. Transport of Reprogramming Transcription Factor
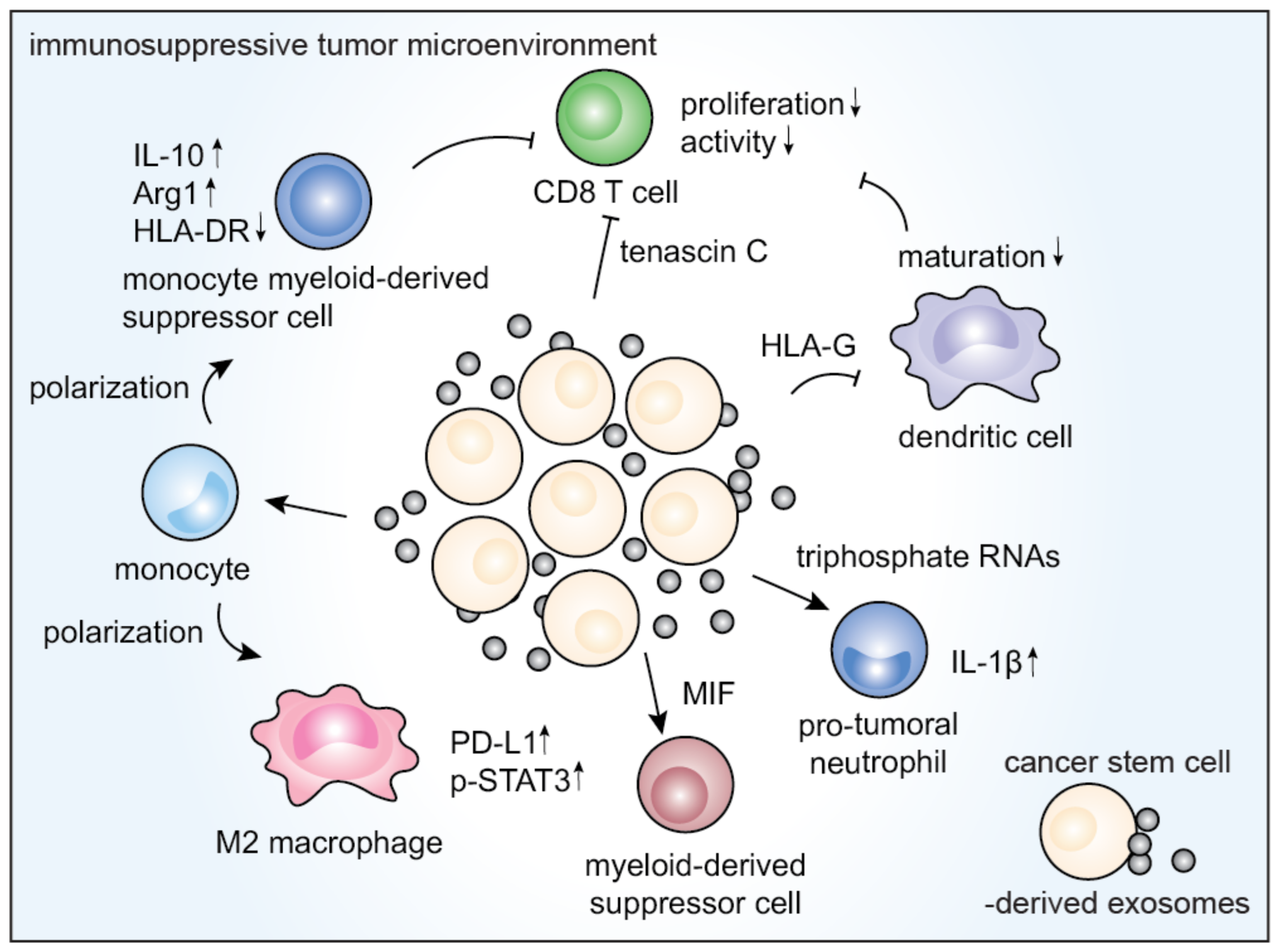
3.4. Immunological Effects of CSCEXs
4. Potential for Exploiting CSCEXs
4.1. Effects of Exosomes on Cancer Stem Cells
4.2. Targeting Cancer Stem Cells through Engineered Exosomes
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, K.D.; Visvader, J.E. Cellular Mechanisms Underlying Intertumoral Heterogeneity. Trends Cancer 2015, 1, 15–23. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Scheele, C.L.; Maynard, C.; van Rheenen, J. Intravital Insights into Heterogeneity, Metastasis, and Therapy Responses. Trends Cancer 2016, 2, 205–216. [Google Scholar] [CrossRef]
- Schepers, A.G.; Snippert, H.J.; Stange, D.E.; van den Born, M.; van Es, J.H.; van de Wetering, M.; Clevers, H. Lineage Tracing Reveals Lgr5+ Stem Cell Activity in Mouse Intestinal Adenomas. Science 2012, 337, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Yu, Z.; Pestell, T.G.; Lisanti, M.P.; Pestell, R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef]
- Mimeault, M.; Hauke, R.; Mehta, P.P.; Batra, S.K. Recent advances in cancer stem/progenitor cell research: Therapeutic implications for overcoming resistance to the most aggressive cancers. J. Cell. Mol. Med. 2007, 11, 981–1011. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.-C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.-M.; Nephew, K.P. Identification and Characterization of Ovarian Cancer-Initiating Cells from Primary Human Tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef]
- Matsui, W.; Huff, C.A.; Wang, Q.; Malehorn, M.T.; Barber, J.; Tanhehco, Y.; Smith, B.D.; Civin, C.I.; Jones, R.J. Characterization of clonogenic multiple myeloma cells. Blood 2004, 103, 2332–2336. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Sharma, P.; Dogra, N.; Singh, S. Eradicating Cancer Stem Cells: Concepts, Issues, and Challenges. Curr. Treat. Options Oncol. 2018, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells—What challenges do they pose? Nat. Rev. Drug Discov. 2014, 13, 497–512. [Google Scholar] [CrossRef]
- Sreepadmanabh, M.; Toley, B.J. Investigations into the cancer stem cell niche using in-vitro 3-D tumor models and microfluidics. Biotechnol. Adv. 2018, 36, 1094–1110. [Google Scholar] [CrossRef]
- Wang, K.-X.; Xu, L.-L.; Rui, Y.-F.; Huang, S.; Lin, S.-E.; Xiong, J.-H.; Li, Y.-H.; Lee, W.Y.-W.; Li, G. The Effects of Secretion Factors from Umbilical Cord Derived Mesenchymal Stem Cells on Osteogenic Differentiation of Mesenchymal Stem Cells. PLoS ONE 2015, 10, e0120593. [Google Scholar] [CrossRef] [PubMed]
- Van Der Goot, F.G.; Gruenberg, J. Intra-endosomal membrane traffic. Trends Cell Biol. 2006, 16, 514–521. [Google Scholar] [CrossRef]
- Li, W.; Zhang, L.; Guo, B.; Deng, J.; Wu, S.; Li, F.; Wang, Y.; Lu, J.; Zhou, Y. Exosomal FMR1-AS1 facilitates maintaining cancer stem-like cell dynamic equilibrium via TLR7/NFκB/c-Myc signaling in female esophageal carcinoma. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. Exosomes/microvesicles: Mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol. 2011, 33, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Seo, N.; Akiyoshi, K.; Shiku, H. Exosome-mediated regulation of tumor immunology. Cancer Sci. 2018, 109, 2998–3004. [Google Scholar] [CrossRef]
- Lugini, L.; Cecchetti, S.; Huber, V.; Luciani, F.; Macchia, G.; Spadaro, F.; Paris, L.; Abalsamo, L.; Colone, M.; Molinari, A.; et al. Immune surveillance properties of human NK cell-derived exosomes. J. Immunol. 2012, 189, 2833–2842. [Google Scholar] [CrossRef]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.-P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. OncoImmunology 2016, 5, e1071008. [Google Scholar] [CrossRef]
- Dai, S.; Wei, D.; Wu, Z.; Zhou, X.; Wei, X.; Huang, H.; Li, G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther. 2008, 16, 782–790. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, Y.; Zhao, M. Exosome-Based Cancer Therapy: Implication for Targeting Cancer Stem Cells. Front. Pharmacol. 2017, 7, 533. [Google Scholar] [CrossRef] [PubMed]
- Al-Sowayan, B.S.; Al-Shareeda, A.T.; Alrfaei, B.M. Cancer Stem Cell-Exosomes, Unexposed Player in Tumorigenicity. Front. Pharmacol. 2020, 11, 384. [Google Scholar] [CrossRef]
- Gabrusiewicz, K.; Li, X.; Wei, J.; Hashimoto, Y.; Marisetty, A.L.; Ott, M.; Wang, F.; Hawke, D.; Yu, J.; Healy, L.M.; et al. Glioblastoma stem cell-derived exosomes induce M2 macrophages and PD-L1 expression on human monocytes. OncoImmunology 2018, 7, e1412909. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, G.; Zhao, D.; Wang, J.; Bai, Y.; Peng, Q.; Wang, H.; Fang, R.; Chen, G.; Wang, Z.; et al. CD103-positive CSC exosome promotes EMT of clear cell renal cell carcinoma: Role of remote MiR-19b-3p. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef]
- Cheng, Z.; Lei, Z.; Yang, P.; Si, A.; Xiang, D.; Tang, X.; Guo, G.; Zhou, J.; Hüser, N. Exosome-transmitted p120-catenin suppresses hepatocellular carcinoma progression via STAT3 pathways. Mol. Carcinog. 2019, 58, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhao, N.; Cui, J.; Wu, H.; Xiong, J.; Peng, T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell. Oncol. 2020, 43, 123–136. [Google Scholar] [CrossRef]
- Boelens, M.C.; Wu, T.J.; Nabet, B.Y.; Xu, B.; Qiu, Y.; Yoon, T.; Azzam, D.J.; Victor, C.T.-S.; Wiemann, B.Z.; Ishwaran, H.; et al. Exosome Transfer from Stromal to Breast Cancer Cells Regulates Therapy Resistance Pathways. Cell 2014, 159, 499–513. [Google Scholar] [CrossRef]
- Lee, C.J.; Dosch, J.; Simeone, D.M. Pancreatic Cancer Stem Cells. J. Clin. Oncol. 2008, 26, 2806–2812. [Google Scholar] [CrossRef]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef]
- Yang, Z.F.; Ho, D.W.; Ng, M.N.; Lau, C.K.; Yu, W.C.; Ngai, P.; Chu, P.W.; Lam, C.T.; Poon, R.T.; Fan, S.T. Significance of CD90+ Cancer Stem Cells in Human Liver Cancer. Cancer Cell 2008, 13, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- Dalerba, P.; Dylla, S.J.; Park, I.-K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef]
- Francipane, M.G.; Alea, M.P.; Lombardo, Y.; Todaro, M.; Medema, J.P.; Stassi, G. Crucial Role of Interleukin-4 in the Survival of Colon Cancer Stem Cells: Figure 1. Cancer Res. 2008, 68, 4022–4025. [Google Scholar] [CrossRef] [PubMed]
- Burger, P.E.; Xiong, X.; Coetzee, S.; Salm, S.N.; Moscatelli, D.; Goto, K.; Wilson, E.L. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc. Natl. Acad. Sci. USA 2005, 102, 7180–7185. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Salm, S.N.; Coetzee, S.; Xiong, X.; Burger, P.E.; Shapiro, E.; Lepor, H.; Moscatelli, D.; Wilson, E.L. Proximal Prostatic Stem Cells Are Programmed to Regenerate a Proximal-Distal Ductal Axis. STEM CELLS 2006, 24, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.-G.; Lee, S.-H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 1–16. [Google Scholar] [CrossRef]
- Eguchi, T.; Sogawa, C.; Okusha, Y.; Uchibe, K.; Iinuma, R.; Ono, K.; Nakano, K.; Murakami, J.; Itoh, M.; Arai, K.; et al. Organoids with cancer stem cell-like properties secrete exosomes and HSP90 in a 3D nanoenvironment. PLoS ONE 2018, 13, e0191109. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, F.A.; El-Magd, M.A.; Abdelfattah-Hassan, A.; Saleh, A.A.; Saadeldin, I.M.; El-Shetry, E.S.; Badawy, A.A.; Alkarim, S. Potential Effect of Exosomes Derived from Cancer Stem Cells and MSCs on Progression of DEN-Induced HCC in Rats. Stem Cells Int. 2018, 2018, 1–17. [Google Scholar] [CrossRef]
- Mirzaei, R.; Sarkar, S.; Dzikowski, L.; Rawji, K.S.; Khan, L.; Faissner, A.; Bose, P.; Yong, V.W. Brain tumor-initiating cells export tenascin-C associated with exosomes to suppress T cell activity. OncoImmunology 2018, 7, e1478647. [Google Scholar] [CrossRef]
- Sun, H.; Rana, S.; Wang, Z.; Zhao, K.; Schnölzer, M.; Provaznik, J.; Hackert, T.; Lv, Q.; Zöller, M. The Pancreatic Cancer-Initiating Cell Marker CD44v6 Affects Transcription, Translation, and Signaling: Consequences for Exosome Composition and Delivery. J. Oncol. 2019, 2019, 3516973-24. [Google Scholar] [CrossRef]
- Sun, W.; Ren, Y.; Lu, Z.; Zhao, X. The potential roles of exosomes in pancreatic cancer initiation and metastasis. Mol. Cancer 2020, 19, 1–18. [Google Scholar] [CrossRef]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles Released from Human Renal Cancer Stem Cells Stimulate Angiogenesis and Formation of Lung Premetastatic Niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef]
- Zhang, D.; Li, D.; Shen, L.; Hu, D.; Tang, B.; Guo, W.; Wang, Z.; Zhang, Z.; Wei, G.; He, D. Exosomes derived from Piwil2-induced cancer stem cells transform fibroblasts into cancer-associated fibroblasts. Oncol. Rep. 2020, 43, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Zhang, J.; Yarden, Y.; Fu, L. The key roles of cancer stem cell-derived extracellular vesicles. Signal Transduct. Target. Ther. 2021, 6, 1–15. [Google Scholar] [CrossRef]
- Turdo, A.; Porcelli, G.; D’Accardo, C.; Di Franco, S.; Verona, F.; Forte, S.; Giuffrida, D.; Memeo, L.; Todaro, M.; Stassi, G. Metabolic Escape Routes of Cancer Stem Cells and Therapeutic Opportunities. Cancers 2020, 12, 1436. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Investig. 2016, 126, 1216–1223. [Google Scholar] [CrossRef]
- Lecerf, C.; Peperstraete, E.; Le Bourhis, X.; Adriaenssens, E. Propagation and Maintenance of Cancer Stem Cells: A Major Influence of the Long Non-Coding RNA H19. Cells 2020, 9, 2613. [Google Scholar] [CrossRef]
- Rodríguez, M.; Silva, J.; Herrera, A.; Herrera, M.; Peña, C.; Martín, P.; Gil-Calderón, B.; Larriba, M.J.; Coronado, M.J.; Soldevilla, B.; et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget 2015, 6, 40575–40587. [Google Scholar] [CrossRef]
- Santos, P.; Almeida, F. Role of Exosomal miRNAs and the Tumor Microenvironment in Drug Resistance. Cells 2020, 9, 1450. [Google Scholar] [CrossRef]
- Du, Q.; Ye, X.; Lu, S.-R.; Li, H.; Liu, H.-Y.; Zhai, Q.; Yu, B. Exosomal miR-30a and miR-222 derived from colon cancer mesenchymal stem cells promote the tumorigenicity of colon cancer through targeting MIA3. J. Gastrointest. Oncol. 2021, 12, 52–68. [Google Scholar] [CrossRef]
- Sun, Z.-P.; Li, A.-Q.; Jia, W.-H.; Ye, S.; Van Eps, G.; Yu, J.-M.; Yang, W.-J. MicroRNA expression profiling in exosomes derived from gastric cancer stem-like cells. Oncotarget 2017, 8, 93839–93855. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ma, X.; Wang, J.; Zhao, Y.; Wang, Y.; Bihl, J.C.; Chen, Y.; Jiang, C. Glioma stem cells-derived exosomes promote the angiogenic ability of endothelial cells through miR-21/VEGF signal. Oncotarget 2017, 8, 36137–36148. [Google Scholar] [CrossRef]
- Li, J.; Liao, T.; Liu, H.; Yuan, H.; Ouyang, T.; Wang, J.; Chai, S.; Li, J.; Chen, J.; Li, X.; et al. Hypoxic Glioma Stem Cell-Derived Exosomes Containing Linc01060 Promote Progression of Glioma by Regulating the MZF1/c-Myc/HIF1alpha Axis. Cancer Res. 2021, 81, 114–128. [Google Scholar] [PubMed]
- Wang, L.; He, J.; Hu, H.; Tu, L.; Sun, Z.; Liu, Y.; Luo, F. Lung CSC-derived exosomal miR-210-3p contributes to a pro-metastatic phenotype in lung cancer by targeting FGFRL1. J. Cell Mol. Med. 2020, 24, 6324–6339. [Google Scholar] [CrossRef]
- Shoff, M.; Booker, T.; Leavitt, B.; Harmon, D.; Kingsley, K.; Howard, K.M. Differential exosome miRNA expression in oral cancer stem cells. ExRNA 2020, 2, 1–9. [Google Scholar] [CrossRef]
- Sánchez, C.A.; Andahur, E.I.; Valenzuela, R.; Castellón, E.A.; Fullá, J.A.; Ramos, C.G.; Triviño, J.C. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget 2016, 7, 3993–4008. [Google Scholar] [CrossRef] [PubMed]
- Hardin, H.; Helein, H.; Meyer, K.; Robertson, S.; Zhang, R.; Zhong, W.; Lloyd, R.V. Thyroid cancer stem-like cell exosomes: Regulation of EMT via transfer of lncRNAs. Lab. Investig. 2018, 98, 1133–1142. [Google Scholar] [CrossRef]
- Wang, Z.; Zöller, M. Exosomes, metastases, and the miracle of cancer stem cell markers. Cancer Metastasis Rev. 2019, 38, 259–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, H.; Provaznik, J.; Hackert, T.; Zöller, M. Pancreatic cancer-initiating cell exosome message transfer into noncancer-initiating cells: The importance of CD44v6 in reprogramming. J. Exp. Clin. Cancer Res. 2019, 38, 1–20. [Google Scholar] [CrossRef]
- Hwang, W.-L.; Lan, H.-Y.; Cheng, W.-C.; Huang, S.-C.; Yang, M.-H. Tumor stem-like cell-derived exosomal RNAs prime neutrophils for facilitating tumorigenesis of colon cancer. J. Hematol. Oncol. 2019, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Clayton, S.M.; Archard, J.A.; Wagner, J.; Farwell, D.G.; Bewley, A.F.; Beliveau, A.; Birkeland, A.; Rao, S.; Abouyared, M.; Belafsky, P.C.; et al. Immunoregulatory Potential of Exosomes Derived from Cancer Stem Cells. Stem Cells Dev. 2020, 29, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, S.; Fu, Q. Exosomes from CD133 + cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J. Cell. Biochem. 2020, 121, 3286–3297. [Google Scholar] [CrossRef]
- Kyuno, D.; Zhao, K.; Schnölzer, M.; Provaznik, J.; Hackert, T.; Zöller, M. Claudin7-dependent exosome-promoted reprogramming of nonmetastasizing tumor cells. Int. J. Cancer 2019, 145, 2182–2200. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, L.; Zhou, Y.; Dong, L.; Ma, W.; Lv, L.; Zhang, J.; Wang, X. Glioblastoma Stem Cell-Derived Exosomes Enhance Stemness and Tumorigenicity of Glioma Cells by Transferring Notch1 Protein. Cell. Mol. Neurobiol. 2019, 40, 767–784. [Google Scholar] [CrossRef]
- Hu, Y.; Yan, C.; Mu, L.; Huang, K.; Li, X.; Tao, D.; Wu, Y.; Qin, J. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS ONE 2015, 10, e0125625. [Google Scholar] [CrossRef]
- Lin, R.; Wang, S.; Zhao, R.C. Exosomes from human adipose-derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Mol. Cell. Biochem. 2013, 383, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Baran, J.; Baj-Krzyworzeka, M.; Weglarczyk, K.; Szatanek, R.; Zembala, M.; Barbasz, J.; Czupryna, A.; Szczepanik, A.; Zembala, M. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol. Immunother. 2010, 59, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Raimondo, S.; Saieva, L.; Corrado, C.; Fontana, S.; Flugy, A.; Rizzo, A.; De Leo, G.; Alessandro, R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun. Signal. 2015, 13, 1–12. [Google Scholar] [CrossRef]
- Philip, R.; Heiler, S.; Mu, W.; Büchler, M.W.; Zöller, M.; Thuma, F. Claudin-7 promotes the epithelial—Mesenchymal transition in human colorectal cancer. Oncotarget 2014, 6, 2046–2063. [Google Scholar] [CrossRef]
- Wang, L.; Yang, G.; Zhao, D.; Wang, J.; Bai, Y.; Peng, Q.; Wang, H.; Fang, R.; Chen, G.; Wang, Z.; et al. Correction to: CD103-positive CSC exosome promotes EMT of clear cell renal cell carcinoma: Role of remote MiR-19b-3p. Mol. Cancer 2020, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M. Extracellular vesicle-mediated transport of non-coding RNAs between stem cells and cancer cells: Implications in tumor progression and therapeutic resistance. Stem Cell Investig. 2017, 4, 83. [Google Scholar] [CrossRef]
- Gernapudi, R.; Yao, Y.; Zhang, Y.; Wolfson, B.; Roy, S.; Duru, N.; Eades, G.; Yang, P.; Zhou, Q. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res. Treat. 2015, 150, 685–695. [Google Scholar] [CrossRef]
- Felicetti, F.; De Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Carè, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 1–15. [Google Scholar] [CrossRef]
- Liao, J.; Liu, R.; Shi, Y.-J.; Yin, L.-H.; Pu, Y.-P. Exosome-shuttling microRNA-21 promotes cell migration and invasion-targeting PDCD4 in esophageal cancer. Int. J. Oncol. 2016, 48, 2567–2579. [Google Scholar] [CrossRef] [PubMed]
- Falcone, G.; Felsani, A.; D’Agnano, I. Signaling by exosomal microRNAs in cancer. J. Exp. Clin. Cancer Res. 2015, 34, 1–10. [Google Scholar] [CrossRef]
- Pastò, A.; Consonni, F.M.; Sica, A. Influence of Innate Immunity on Cancer Cell Stemness. Int. J. Mol. Sci. 2020, 21, 3352. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Hsu, W.-H.; Han, J.; Xia, Y.; DePinho, R.A. Cancer Stemness Meets Immunity: From Mechanism to Therapy. Cell Rep. 2021, 34, 108597. [Google Scholar] [CrossRef]
- Maccalli, C.; Rasul, K.I.; Elawad, M.; Ferrone, S. The role of cancer stem cells in the modulation of anti-tumor immune responses. Semin. Cancer Biol. 2018, 53, 189–200. [Google Scholar] [CrossRef]
- Naseri, M.; Zöller, M.; Hadjati, J.; Ghods, R.; Pirmardan, E.R.; Kiani, J.; Eini, L.; Bozorgmehr, M.; Madjd, Z. Dendritic cells loaded with exosomes derived from cancer stem cell-enriched spheroids as a potential immunotherapeutic option. J. Cell. Mol. Med. 2021, 25, 3312–3326. [Google Scholar] [CrossRef]
- Domenis, R.; Cesselli, D.; Toffoletto, B.; Bourkoula, E.; Caponnetto, F.; Manini, I.; Beltrami, A.P.; Ius, T.; Skrap, M.; Di Loreto, C.; et al. Systemic T Cells Immunosuppression of Glioma Stem Cell-Derived Exosomes Is Mediated by Monocytic Myeloid-Derived Suppressor Cells. PLoS ONE 2017, 12, e0169932. [Google Scholar] [CrossRef]
- Otvos, B.; Silver, D.J.; Mulkearns-Hubert, E.; Alvarado, A.G.; Turaga, S.M.; Sorensen, M.D.; Rayman, P.; Flavahan, W.A.; Hale, J.S.; Stoltz, K.; et al. Cancer Stem Cell-Secreted Macrophage Migration Inhibitory Factor Stimulates Myeloid Derived Suppressor Cell Function and Facilitates Glioblastoma Immune Evasion. STEM CELLS 2016, 34, 2026–2039. [Google Scholar] [CrossRef]
- Grange, C.; Tapparo, M.; Tritta, S.; Deregibus, M.C.; Battaglia, A.; Gontero, P.; Frea, B.; Camussi, G. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Cancer 2015, 15, 1–11. [Google Scholar] [CrossRef]
- Cheng, W.; Liao, T.; Lin, C.; Yuan, L.E.; Lan, H.; Lin, H.; Teng, H.; Chang, H.; Lin, C.; Yang, C.; et al. RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int. J. Cancer 2019, 145, 2209–2224. [Google Scholar] [CrossRef]
- Müller, L.; Tunger, A.; Plesca, I.; Wehner, R.; Temme, A.; Westphal, D.; Meier, F.; Bachmann, M.; Schmitz, M. Bidirectional Crosstalk Between Cancer Stem Cells and Immune Cell Subsets. Front. Immunol. 2020, 11, 140. [Google Scholar] [CrossRef]
- Zhou, X.; Xie, F.; Wang, L.; Zhang, L.; Zhang, S.; Fang, M.; Zhou, F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell. Mol. Immunol. 2020, 17, 323–334. [Google Scholar] [CrossRef]
- Wolfson, B.; Eades, G.; Zhou, Q. Roles of microRNA-140 in stem cell-associated early stage breast cancer. World J. Stem Cells 2014, 6, 591–597. [Google Scholar] [CrossRef]
- Li, Q.; Eades, G.; Yao, Y.; Zhang, Y.; Zhou, Q. Characterization of a Stem-like Subpopulation in Basal-like Ductal Carcinoma in Situ (DCIS) Lesions. J. Biol. Chem. 2014, 289, 1303–1312. [Google Scholar] [CrossRef]
- McNamara, M.A.; Nair, S.K.; Holl, E.K. RNA-Based Vaccines in Cancer Immunotherapy. J. Immunol. Res. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Feng, Z.; Meng, S.; Zhou, H.; Xu, Z.; Tang, Y.; Li, P.; Liu, C.; Huang, Y.; Wu, M. Functions and Potential Applications of Circular RNAs in Cancer Stem Cells. Front. Oncol. 2019, 9, 500. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Xu, J.; Liao, K.; Zhou, W. Exosomes Regulate the Transformation of Cancer Cells in Cancer Stem Cell Homeostasis. Stem Cells Int. 2018, 2018, 1–16. [Google Scholar] [CrossRef]
- Valcz, G.; Buzás, E.I.; Szállási, Z.; Kalmár, A.; Krenács, T.; Tulassay, Z.; Igaz, P.; Molnár, B. Perspective: Bidirectional exosomal transport between cancer stem cells and their fibroblast-rich microenvironment during metastasis formation. NPJ Breast Cancer 2018, 4, 1–7. [Google Scholar] [CrossRef]
- Wang, L.; He, M.; Fu, L.; Jin, Y. Exosomal release of microRNA-454 by breast cancer cells sustains biological properties of cancer stem cells via the PRRT2/Wnt axis in ovarian cancer. Life Sci. 2020, 257, 118024. [Google Scholar] [CrossRef]
- Lin, X.-J.; Fang, J.-H.; Yang, X.-J.; Zhang, C.; Yuan, Y.; Zheng, L.; Zhuang, S. Hepatocellular carcinoma cell-secreted exosomal microRNA-210 promotes angiogenesis in vitro and in vivo. Mol. Ther. Nucleic Acids 2018, 11, 243–252. [Google Scholar] [CrossRef]
- Garcia-Mayea, Y.; Mir, C.; Masson, F.; Paciucci, R.; Lleonart, M. Insights into new mechanisms and models of cancer stem cell multidrug resistance. Semin. Cancer Biol. 2020, 60, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Noren, H.; Jove, R.; Beljanski, V.; Grinnemo, K.-H. Differences and similarities between cancer and somatic stem cells: Therapeutic implications. Stem Cell Res. Ther. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudás, J.; Skvortsov, S.; Ganswindt, U.; Riechelmann, H.; Skvortsova, I.I. Therapy resistance mediated by exosomes. Mol. Cancer 2019, 18, 58. [Google Scholar] [CrossRef]
- Dandawate, P.R.; Subramaniam, D.; Jensen, R.A.; Anant, S. Targeting cancer stem cells and signaling pathways by phytochemicals: Novel approach for breast cancer therapy. Semin. Cancer Biol. 2016, 40–41, 192–208. [Google Scholar] [CrossRef]
- Huang, R.; Rofstad, E.K. Cancer stem cells (CSCs), cervical CSCs and targeted therapies. Oncotarget 2017, 8, 35351–35367. [Google Scholar] [CrossRef]
- Aramini, B.; Masciale, V.; Haider, K.H. Defining lung cancer stem cells exosomal payload of miRNAs in clinical perspective. World J. Stem Cells 2020, 12, 406–421. [Google Scholar] [CrossRef]
- Brocco, D.; Lanuti, P.; Simeone, P.; Bologna, G.; Pieragostino, D.; Cufaro, M.C.; Graziano, V.; Peri, M.; Marino, P.D.; Tursi, M.D.; et al. Circulating Cancer Stem Cell-Derived Extracellular Vesicles as a Novel Biomarker for Clinical Outcome Evaluation. J. Oncol. 2019, 2019, 5879616. [Google Scholar] [CrossRef]

| Source | Surface Marker/Cargo | Effect | Reference |
|---|---|---|---|
| Breast CSCs | miR-155 | Enhanced resistance to doxorubicin, paclitaxel | [58] |
| Colorectal CSCs | miR-30a, miR-222 | Promotion of tumorigenicity through targeting MIA-3 | [59] |
| Triphosphate RNAs | Promotion of tumor phenotype of neutrophils | [69] | |
| circRNA-ABCC1 | Promotion of cancer stemness | [71] | |
| Gastric CSCs | hsa-miR-1290, -1246, -21-5p, -100-5p, -20a-5p, -26a-5p, -24-3p, -182-5p, -378a-3p, -148a-3p, -17-5p, -23a-5p; has-let-7f-5p, -7a-5p, -7g-5p | Predictive biomarkers for metastasis | [60] |
| Claudin7 | Promotion of metastasis | [72] | |
| Glioblastoma CSCs | Tenascin C | Inhibition of T cell proliferation and activation | [47] |
| Linc01060 | Promotion of cancer progression by activation of pro-oncogenic signaling | [62] | |
| Notch1 | Promotion of cancer stemness | [73] | |
| Lung CSCs | miR-210-3p | Pro-metastatic phenotype in lung cancer cells by targeting the fibroblast growth factor receptor-like 1 (FGFRL1) | [63] |
| Oral squamous cell carcinoma CSCs | Downregulation of miR-34 | Cancer progression | [64] |
| Pancreatic CSCs | miR-210 | Gemcitabine-resistant phenotype transference to pancreatic cancer cells | [34] |
| CD44v6 | Promotion of cancer stemness and metastasis | [68] | |
| Prostate CSCs | Hsa-miR-1307-5p, -139-5p, -148a-3p, -183-5p | Contribution to the premetastatic niche | [65] |
| Renal cell CSCs | Proangiogenic mRNAs (VEGF, angiopoietin1, MMP-2, MMP-9) | Angiogenesis and promotion of premetastatic niche formation | [50] |
| miR-19b-3p | Promotion of EMT | [32] | |
| Papillary carcinoma CSCs | lncRNA DOCK9-AS2 | Promotion of proliferation, migration, and invasion by activation of Wnt/β-catenin pathway | [66] |
| Source | Surface Marker/Cargo | Immunological Effects | Reference |
|---|---|---|---|
| Colorectal CSCs | Triphosphate RNAs | Neutrophil transformation into pro-tumoral phenotypes, interleukin (IL)-1β secretion | [69] |
| CSCEX loaded onto dendritic cells (DCs) | Activation of CSC-specific T cell response | [90] | |
| miR-146a | Increase in tumor-infiltrating CD66+ neutrophils, decrease in tumor-infiltrating CD8+ T cells | [94] | |
| Glioblastoma CSCs | Tenascin-C | Inhibition of T cell activation, proliferation | [47] |
| Eukaryotic initiation factor 2, mTOR, and ephrin B signaling pathways | M1 to M2 transition of monocytes via the STAT-3 pathway, upregulation of PD-L1 in macrophages | [31] | |
| Increased IL-10 and Arg1, downregulated MHC class II cell surface receptor (HLA-DR) | Differentiation of peripheral blood monocytes into cells similar to M-MDSCs | [91] | |
| MIF | Activation of MDSCs | [92] | |
| Renal CSCs | HLA-G | Impairment of DC maturation and T cell-mediated immune response | [93] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, N.-K.; Kothandan, V.K.; Kothandan, S.; Byun, Y.; Hwang, S.-R. Exosomes and Cancer Stem Cells in Cancer Immunity: Current Reports and Future Directions. Vaccines 2021, 9, 441. https://doi.org/10.3390/vaccines9050441
Lee N-K, Kothandan VK, Kothandan S, Byun Y, Hwang S-R. Exosomes and Cancer Stem Cells in Cancer Immunity: Current Reports and Future Directions. Vaccines. 2021; 9(5):441. https://doi.org/10.3390/vaccines9050441
Chicago/Turabian StyleLee, Na-Kyeong, Vinoth Kumar Kothandan, Sangeetha Kothandan, Youngro Byun, and Seung-Rim Hwang. 2021. "Exosomes and Cancer Stem Cells in Cancer Immunity: Current Reports and Future Directions" Vaccines 9, no. 5: 441. https://doi.org/10.3390/vaccines9050441
APA StyleLee, N.-K., Kothandan, V. K., Kothandan, S., Byun, Y., & Hwang, S.-R. (2021). Exosomes and Cancer Stem Cells in Cancer Immunity: Current Reports and Future Directions. Vaccines, 9(5), 441. https://doi.org/10.3390/vaccines9050441