Protective Vaccination Reshapes Hepatic Response to Blood-Stage Malaria of Genes Preferentially Expressed by NK Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Protective Vaccination
2.3. Blood-Stage Malaria of P. chabaudi
2.4. Liver Sampling During Primary Blood-Stage Infections
2.5. Hybridization of Mouse Whole Genome Oligo Microarrays
2.6. Analyses of Microarrays
2.7. Selection of DEGs
2.8. Chromosomal Landscape Analysis of DEGs
2.9. Quantitative real-time PCR
3. Results
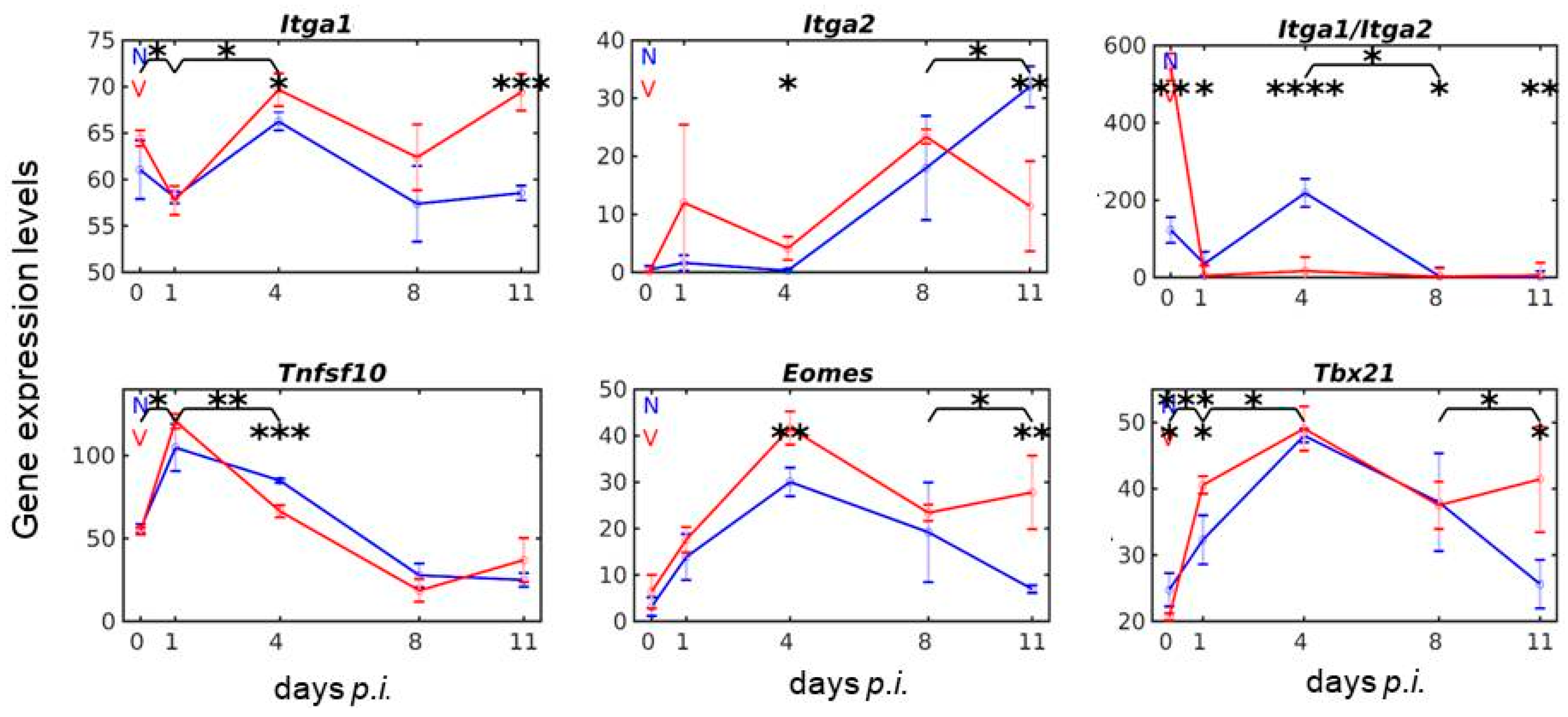
3.1. Vaccination Reshapes the Response to Blood-Stage Malaria of Genes Expressed by lrNK and cNK Cells in the Liver
3.2. Vaccination Reshapes Malaria-Induced Expression of Ncr1, Prf1 and Gzm Genes in the Liver
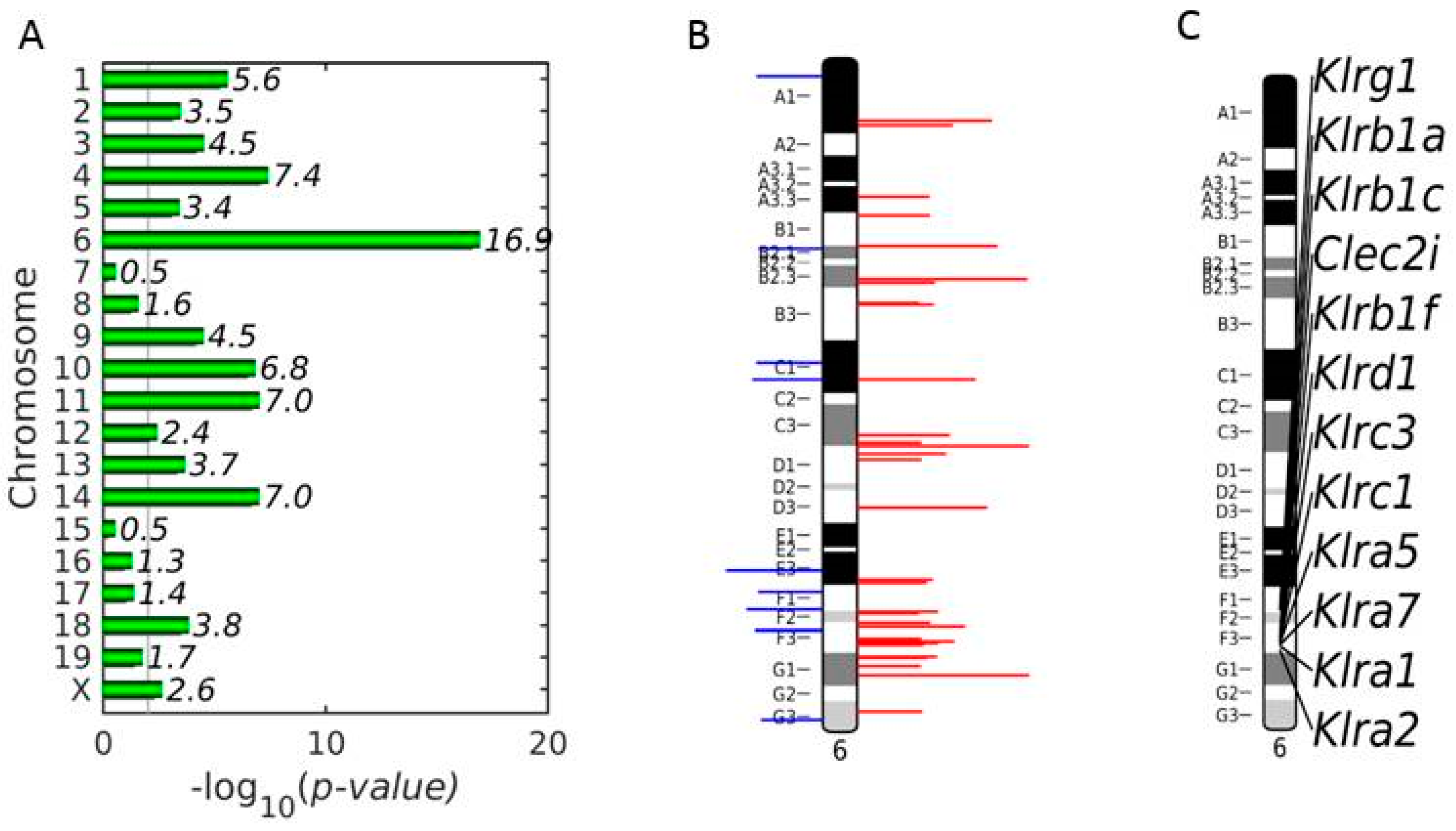
3.3. Enrichment of NKC with Up-Regulated DEGs on Chromosome 6 in Vaccinated Mice on Day 4 p.i.
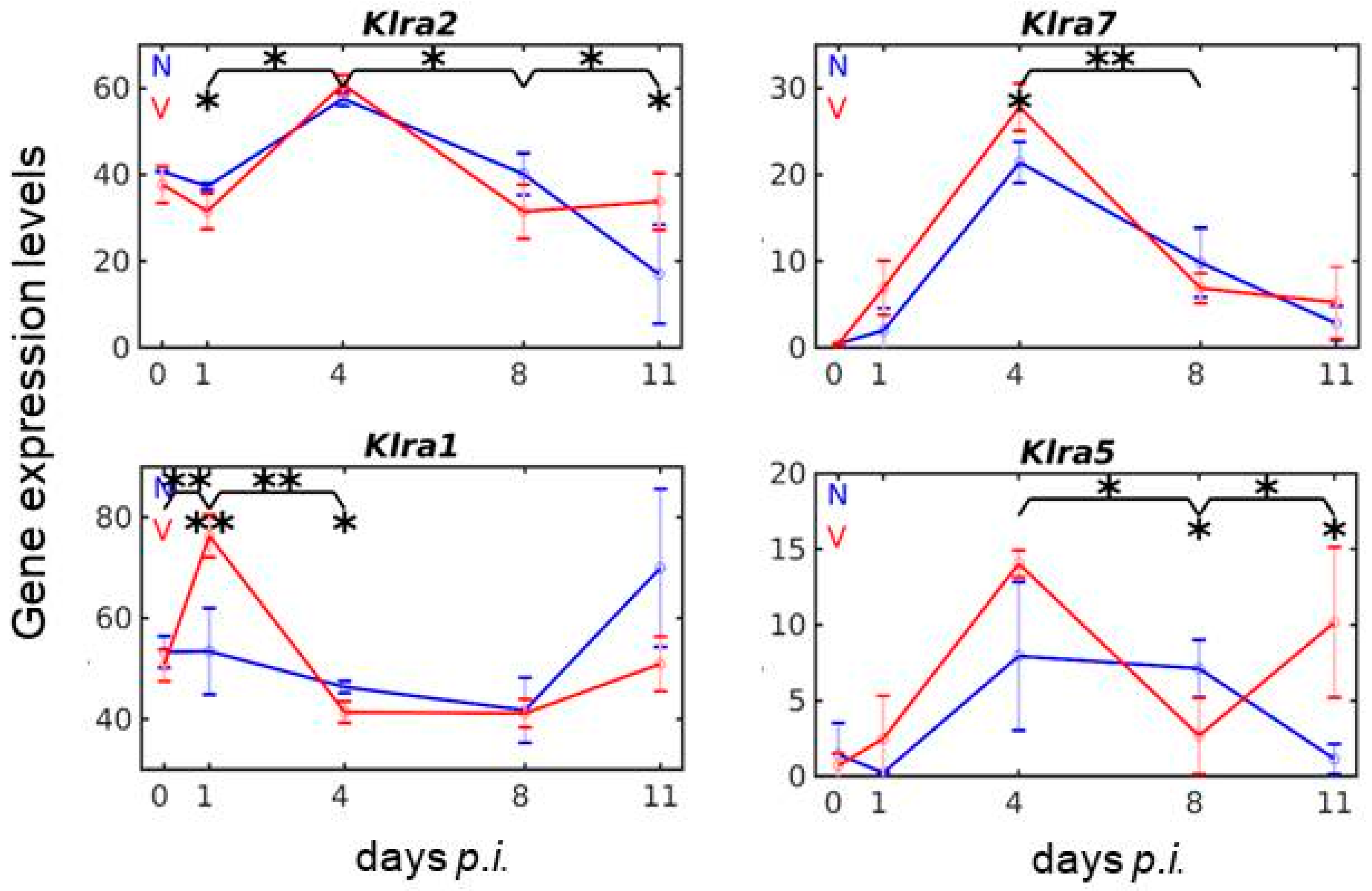
3.4. Vaccination Reshapes Malaria-Responsive Expression of Klra Genes in the Liver
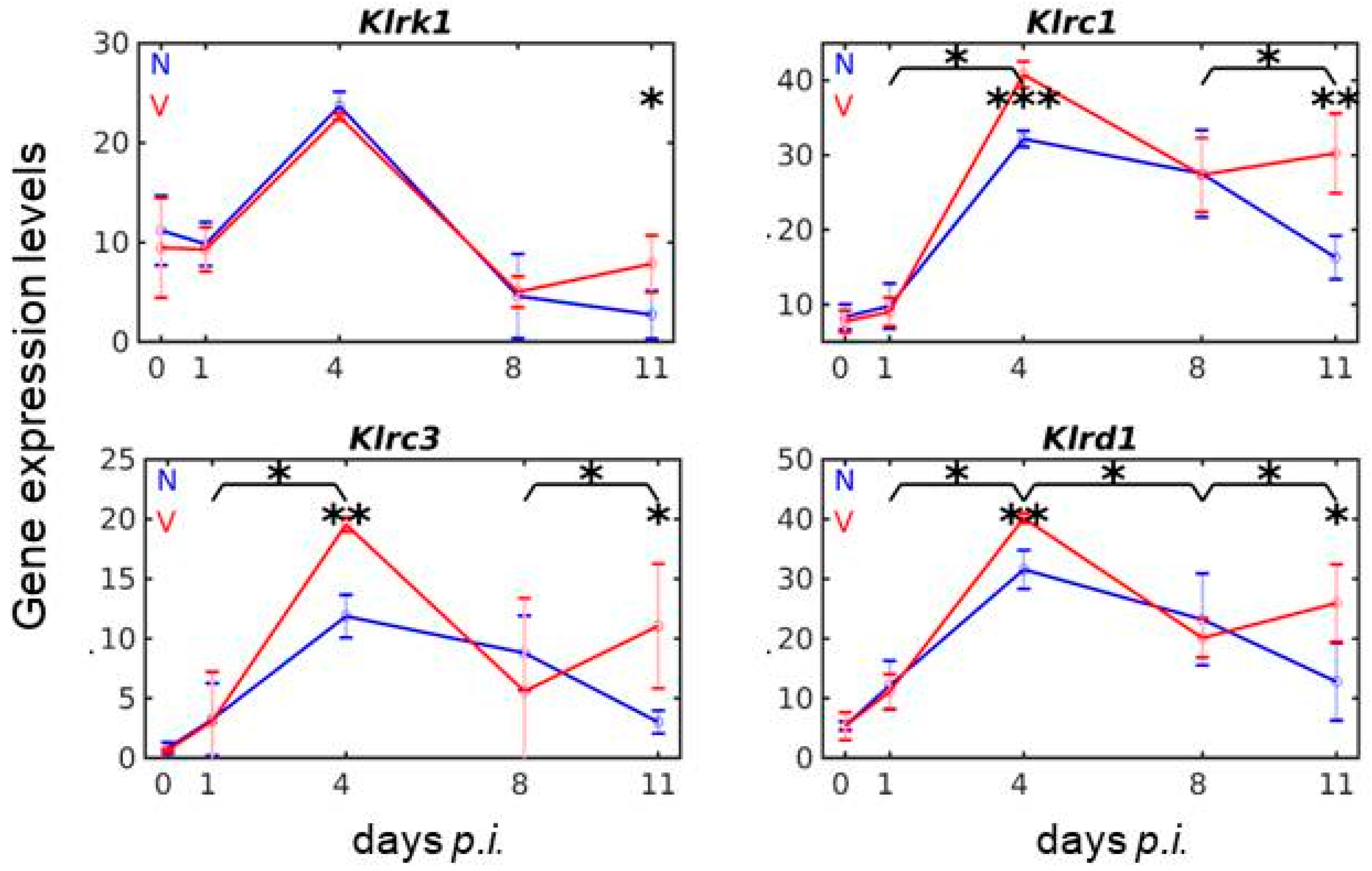
3.5. Vaccination Reshapes Malaria-Induced Expression of Cd94/Nkg2 Genes
3.6. Vaccination Reshapes Responsiveness of Klrb1/Nkrp1 Gene Expression to Malaria
3.7. Vaccination reshapes malaria-induced expression of Clec2 genes in the liver
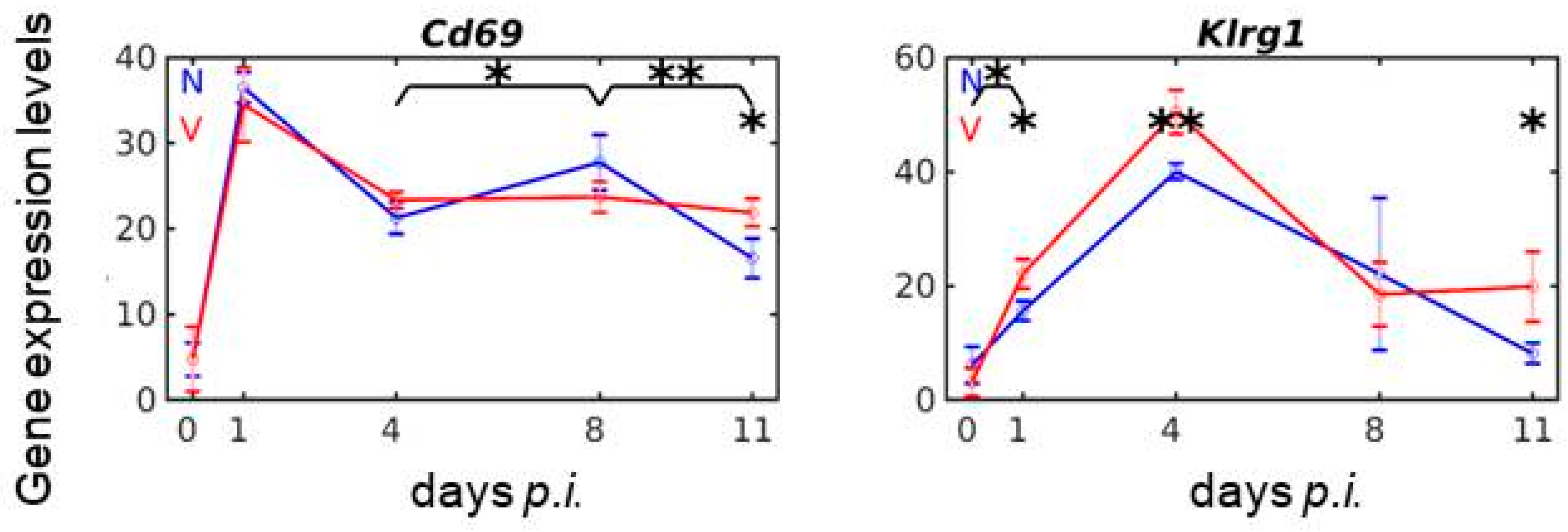
3.8. Vaccination Reshapes Malaria-Responsive Expression of Cd69 and Klrg1 in the Liver
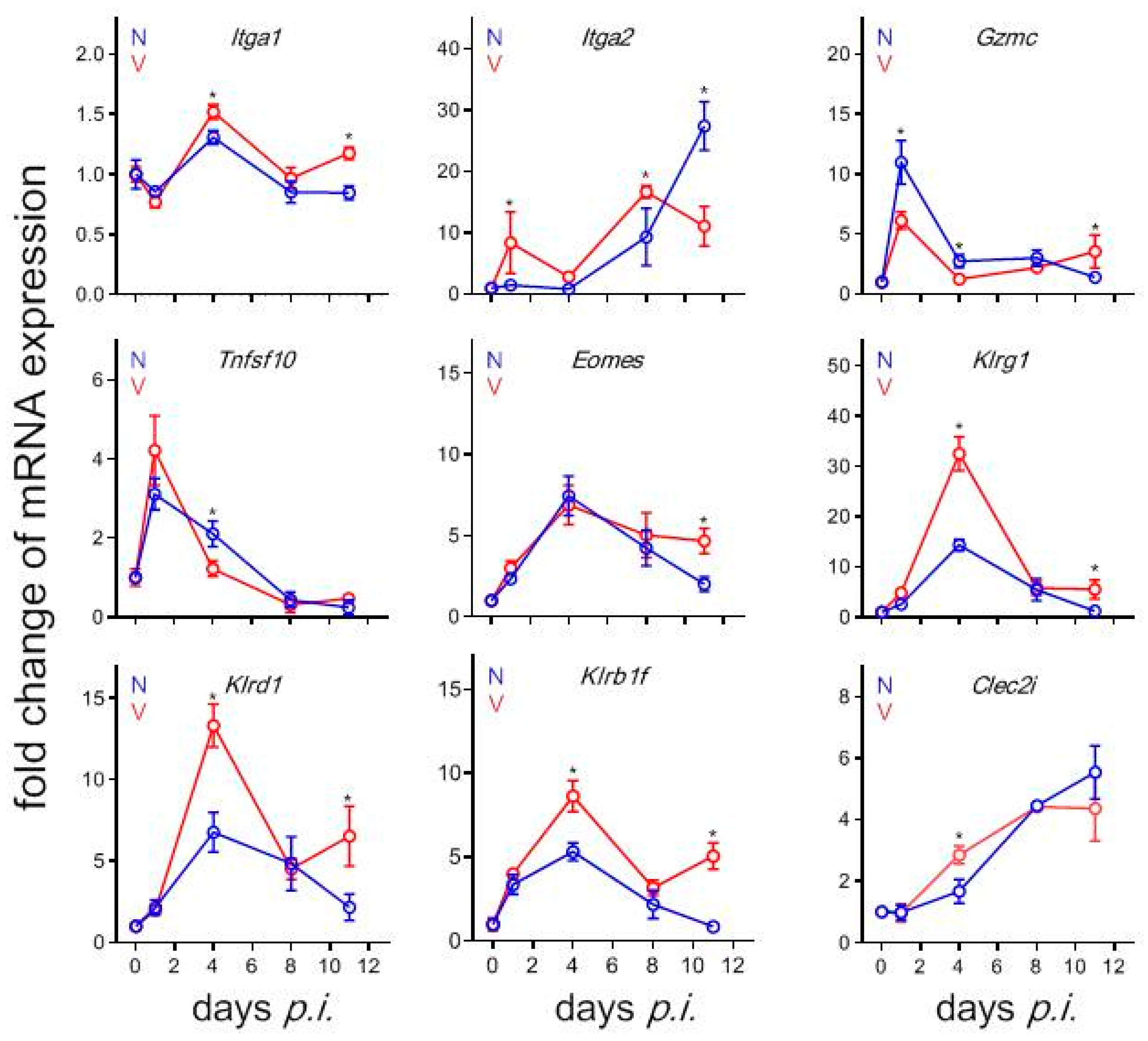
3.9. Validation by Quantitative qRT-PCR of Microarrays Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Statement of Ethics
References
- WHO. World Malaria Report 2019; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- De Vrieze, J.; Chikondi, T. A shot of hope. Science 2019, 366, 1062–1065. [Google Scholar] [CrossRef]
- Draper, S.J.; Sack, B.K.; Richter King, C.; Nielsen, C.N.; Rayner, J.C.; Higgins, M.K.; Long, C.A.; Seder, R.A. Malaria vaccines: Recent advances and new horizons. Cell Host Microbe 2018, 24, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Del Portillo, H.A.; Ferrer, M.; Brugat, T.; Martin-Jaular, L.; Langhorne, J.; Lacerda, M.V.G. The role of the spleen in malaria. Cell. Microbiol. 2012, 14, 343–355. [Google Scholar] [CrossRef]
- White, N.J. Malaria parasite clearence. Malar. J. 2017, 16, 88. [Google Scholar] [CrossRef] [Green Version]
- Wunderlich, F.; Al-Quraishy, S.; Dkhil, M. Liver-inherent immune system: Its role in blood-stage malaria. Front. Microbiol. 2014, 5, 559. [Google Scholar] [CrossRef]
- Terpstra, V.; Berkel, T.J. Scavenger receptors on liver Kupffer cells mediate the in vitro uptake of oxidatively damaged red blood cells in mice. Blood 2000, 15, 2157–2163. [Google Scholar] [CrossRef]
- Delic, D.; Warskulat, U.; Borsch, E.; Al-Qahtani, S.; Al-Guraishy, S.; Häussinger, D.; Wunderlich, F. Loss of ability to self-heal malaria upon taurine transporter deletion. Infect. Immun. 2010, 78, 1642–1649. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Park, S.Y.; Jung, M.Y.; Bae, S.M.; Kim, I.S. Mechanism for phosphatidylserine-dependent erythrophagocytosis in mouse liver. Blood 2011, 117, 5215–5223. [Google Scholar] [CrossRef] [Green Version]
- Theurl, I.; Hilgendorf, I.; Nairz, N.; Tymoszuk, P.; Haschka, D.; Asshoff, M.; He, S.; Gerhardt, L.M.S.; Holderried, T.A.W.; Seifertet, M.; et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat. Med. 2016, 22, 945–951. [Google Scholar] [CrossRef]
- Wolf, A.S.; Sherrat, S.; Riley, E.M. NK cells: Uncertain allies against malaria. Front. Immunol. 2017, 8, 212. [Google Scholar] [CrossRef] [Green Version]
- Ng, S.S.; Engwerda, C.R. Innate lymphcytes and malaria—Players or spectators? Trends Parasitol. 2019, 35, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Burrack, K.S.; Hart, G.T.; Hamilton, S.E. Contributions of natural killer cells to the immune response against Plasmodium. Malar. J. 2019, 18, 321. [Google Scholar] [CrossRef] [PubMed]
- Goodier, M.R.; Wolf, A.S.; Riley, E.M. Differentiation and adaptation of natural killer cells for antiviral immunity. Immunol. Rev. 2020, 293, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Elliott, J.M.; Keyel, P.A.; Yang, L.; Carrero, J.A.; Yokoyama, W.M. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA 2009, 106, 1915–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.C.; Beilke, J.N.; Lanier, L.L. Adaptive immune features of natural killer cells. Nature 2009, 457, 557–561. [Google Scholar] [CrossRef]
- Wight, A.; Mahmoud, A.B.; Scur, M.; Tu, M.M.; Rahim, M.M.A.; Sad, S.; Makrigiannis, A.P. Critical role for the Ly49 family of class I MHC receptors in adaptive natural killer cell responses. Proc. Natl. Acad. Sci. USA 2018, 115, 11579–11584. [Google Scholar] [CrossRef] [Green Version]
- Barrow, A.D.; Martin, C.J.; Colonna, M. The natural cytotoxicity receptors in health and disease. Front. Immunol. 2019, 10, 909. [Google Scholar] [CrossRef] [Green Version]
- Ewen, C.L.; Kane, K.P.; Bleackley, R.C. A quarter century of granzymes. Cell Death Differ. 2012, 19, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Prager, I.; Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef]
- Shifrin, N.; Raulet, D.H.; Ardolino, M. NK cell self tolerance, responsiveness and missing self recognition. Semin. Immunol. 2014, 26, 138–144. [Google Scholar] [CrossRef] [Green Version]
- Rahim, M.M.A.; Tu, M.M.; Mahmoud, A.B.; Wight, A.; Abou-Samra, E.; Lima, P.D.A.; Makrigiannis, A.P. Ly49 receptors: Innate and adaptive immune paradigms. Front. Immunol. 2014, 5, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, W.M.; Seaman, W.E. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: The NK gene complex. Annu. Rev. Immunol. 1993, 11, 613–635. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Klein, J.; Nei, M. Heterogenous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc. Natl. Acad. Sci. USA 2006, 103, 3192–3197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkham, C.L.; Carlyle, J.R. Complexity and diversity of the NKR-P1:Clr (Klrb1:Clec2) recognition systems. Front. Immunol. 2014, 5, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parham, P.; Normn, P.J.; Abi-Rached, L.; Guethlein, L.A. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos. Trans. R. Soc. B 2012, 367, 800–811. [Google Scholar] [CrossRef]
- Vance, R.E.; Jamieson, A.M.; Raulet, D.H. Recognition of the class Ib molecule Qa-1b by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J. Exp. Med. 1999, 190, 1801–1812. [Google Scholar] [CrossRef]
- Yokoyama, W.M.; Plougastel, B.F. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 2003, 3, 304–316. [Google Scholar] [CrossRef]
- Zhang, Q.; Rahim, M.M.A.; Allan, D.S.J.; Tu, M.M.; Belanger, S.; Abou-Samra, E.; Ma, J.; Sekhon, H.S.; Fairhead, T.; Zein, H.S.; et al. Mouse Nkrp1-Clr gene cluster sequence and expression analyses reveal conservation of tissue-specific MHC-independent immunosurveillance. PLoS ONE 2012, 7, e50561. [Google Scholar] [CrossRef]
- Sojka, D.K.; Plougastel, B.; Yang, L.; Pak-Wittel, M.A.; Artyomov, M.N.; Ivanova, Y.; Zhong, C.; Chase, J.M.; Rothman, P.B. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife 2014, 3, e01659. [Google Scholar] [CrossRef]
- Sojka, D.K.; Tian, Z.; Yokoyama, W.M. Tissue resident natural killer cells and their potential diversity. Semin. Immunol. 2014, 26, 127–131. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Wisse, E.; Tian, Z. Liver natural killer cells: Subsets and roles in liver immunity. Cell. Mol. Immunol. 2016, 13, 328–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Sun, R. Liver-resident NK cells and their potential functions. Cell. Mol. Immunol. 2017, 14, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Longley, R.; Smith, C.; Fortin, A.; Berghout, J.; McMorran, B.; Burgio, G.; Foote, S.; Gros, P. Host resistance to malaria: Using mouse models to explore the host response. Mamm. Genome 2011, 22, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Stephens, R.; Culleton, R.L.; Lamb, T.J. The contribution of Plasmodium chabaudi to our understanding of malaria. Trends Parasitol. 2012, 28, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Wunderlich, F.; Brenner, H.; Helwig, M. Plasmodium chabaudi malaria: Protective immunization with surface membranes of erythrocytes infected with Plasmodium chabaudi. Infect. Immun. 1988, 56, 3326–3328. [Google Scholar] [CrossRef] [Green Version]
- Krücken, J.; Delic, D.; Pauen, H.; Wojtalla, A.; El-Khadragy, M.; Dkhil, M.A.; Mossmann, H.; Wunderlich, F. Augmented particle trapping and attenuated inflammation in the liver by protective vaccination against Plasmodium chabaudi malaria. Malar. J. 2009, 8, 54–64. [Google Scholar] [CrossRef] [Green Version]
- Al-Quraishy, S.; Dkhil, M.A.; Alomar, S.L.; Abdel-Baki, A.A.; Delic, D.; Wunderlich, F.; Araúzo-Bravo, M.J. Blood-stage malaria of Plasmodium chabaudi induces differential Tlr expression in the liver of susceptible and vaccination-protected Balb/c mice. Parasitol. Res. 2016, 115, 1835–1843. [Google Scholar] [CrossRef]
- Al-Quraishy, S.A.; Dkhil, M.A.; Abdel-Baki, A.A.; Delic, D.; Wunderlich, F. Protective vaccination against blood-stage malaria of Plasmodium chabaudi: Differential gene expression in the liver of Balb/c mice toward the end of crisis phase. Front. Microbiol. 2016, 7, 1087. [Google Scholar] [CrossRef] [Green Version]
- Al-Quraishy, S.; Dkhil, M.A.; Abdel-Baki, A.S.; Ghanjati, F.; Erichsen, L.; Santourlidis, S.; Wunderlich, F.; Araúzo-Bravo, M.J. Protective vaccination and blood-stage malaria modify DNA methylation of gene promoters in the liver of Blab/c mice. Parasitol. Res. 2017, 116, 1463–1477. [Google Scholar] [CrossRef]
- Dkhil, M.A.; Al-Quraishy, S.A.; Abdel-Baki, A.S.; Delic, D.; Wunderlich, F. Differential miRNA expression in the liver of Balb/c protected by vaccination during crisis of Plasmodium chabaudi blood-stage malaria. Front. Microbiol. 2017, 7, 2155. [Google Scholar] [CrossRef] [Green Version]
- Al-Quraishy, S.; Dkhil, M.A.; Al-Shaebi, E.M.; Abdel-Baki, A.A.S.; Araúzo-Bravo, M.J.; Delic, D.; Wunderlich, F. Gene expression of the liver of vaccination-protected mice in response to early patent infections of Plasmodium chabaudi blood-stage malaria. Malar. J. 2018, 17, 215. [Google Scholar] [CrossRef] [PubMed]
- Al-Quraishy, S.; Dkhil, M.A.; Abdel-Baki, A.A.S.; Delic, D.; Wunderlich, F. Protective vaccination alters gene expression of the liver of Balb/c mice in response to early prepatent blood-stage malaria of Plasmodium chabaudi. Parasitol. Res. 2018, 117, 1115–1129. [Google Scholar] [CrossRef] [PubMed]
- Delic, D.; Wunderlich, F.; Al-Quraishy, A.; Abdel-Baki, A.; Dkhil, M.; Araúzo-Bravo, M.J. Vaccination accelerates hepatic erythroblastosis induced by blood-stage malaria. Malar. J. 2020, 19, 49. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, F.; Helwig, M.; Schillinger, G.; Vial, H.; Philippot, J.; Speth, V. Isolation and characterization of parasites and host cell ghosts from erythrocytes infected with Plasmodium chabaudi. Mol. Biochem. Parasitol. 1989, 23, 103–115. [Google Scholar] [CrossRef]
- Wunderlich, F.; Helwig, M.; Schillinger, G.; Speth, V. Cryptic disposition of antigenic parasite proteins in plasma membranes of erythrocytes infected with Plasmodium chabaudi. Mol. Biochem. Parasitol. 1988, 30, 55–65. [Google Scholar] [CrossRef]
- Wunderlich, F.; Helwig, M.; Schillinger, G.; Speth, V.; Wiser, M.F. Expression of the parasite protein Pc90 in plasma membranes of erythrocytes infected with Plasmodium chabaudi. Eur. J. Cell. Biol. 1988, 47, 157–164. [Google Scholar]
- Wunderlich, F.; Maurien, W.; Benten, W.P.M.; Schmitt-Wrede, H.P. Testosterone impairs efficacy of protective vaccination against P. chabaudi malaria. Vaccine 1993, 11, 1097–1099. [Google Scholar] [CrossRef]
- Wunderlich, F.; Mossmann, H.; Helwig, M.; Schillinger, G. Resistance to Plasmodium chabaudi in B10 mice: Influence of the H-2 complex and testosterone. Infect. Immun. 1988, 56, 2400–2406. [Google Scholar] [CrossRef] [Green Version]
- Wunderlich, F.; Stuebig, H.; Koenigk, E. Development of Plasmodium chabaudi in mouse red blood cells: Structural properties of the host and parasite membranes. J. Protozool. 1982, 29, 60–66. [Google Scholar] [CrossRef]
- Wunderlich, F.; Dkhil, M.; Mehnert, L.; Braun, J.; El-Khadragy, M.; Borsch, E.; Hermsen, D.; Benten, W.P.M.; Pfeffer, K.; Mossmaan, H. Testosterone responsiveness of spleen and liver in female lymphotoxin beta receptor-deficient mice resistant to blood-stage malaria. Microbes Infect. 2005, 7, 399–409. [Google Scholar] [CrossRef]
- Gerovska, D.; Araúzo-Bravo, M.J. Computational analysis of single cell transcriptomics data elucidates the stabilization of Oct4 expression in the E3.25 mouse preimplantation embryo. Sci. Rep. 2019, 9, 8930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.K.; Dewar, K.; Goulet, M.L.; Leveque, G.; Makrigiannis, A.P. Complete elucidation of a minimal class I MHC natural killer cell receptor haplotype. Genes Immun. 2005, 6, 481–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenkel, A.R.; Kingry, L.C.; Slayden, R.A. The ly49 gene family. A brief guide to the nomenclature, genetics, and role in intracellular infection. Front. Immunol. 2013, 4, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jelenčić, V.; Šestan, M.; Kavazović, I.; Lenartić, M.; Marinović, S.; Holmes, T.D.; Prchal-Murphy, M.; Lisnic, B.; Sexl, V.; Bryceson, Y.T. NK cell receptor NKG2D sets activation threshold for the NCR1 receptor early in NK cell development. Nat. Immunol. 2018, 19, 1083–1092. [Google Scholar] [CrossRef]
- Wensveen, F.M.; Jelencic, V.; Polic, B. NKG2D: A master regulator of immune cell responsiveness. Front. Immunol. 2018, 9, 441. [Google Scholar] [CrossRef]
- Bartel, Y.; Bauer, B.; Steinle, A. Modulation of NK cell function by genetically coupled C-type lectin-like receptor/ligand pairs encoded in the human natural killer gene complex. Front. Immunol. 2013, 4, 362. [Google Scholar] [CrossRef] [Green Version]
- Aguilar, O.A.; Berry, R.; Rahim, M.M.A.; Reichel, J.J.; Popovi, B.; Tanaka, M.; Fu, Z.; Balaji, G.R.; Lau, T.N.H.; Tu, M.M.; et al. A viral immunoevasion controls innate immunity by targeting the prototypical natural killer cell receptor family. Cell 2017, 169, 58–71. [Google Scholar] [CrossRef] [Green Version]
- Vogler, I.; Steinle, A. Vis-à-Vis in the NKC: Genetically Linked Natural Killer Cell Receptor/Ligand Pairs in the Natural Killer Gene Complex (NKC). J. Innate Immun. 2011, 3, 227–235. [Google Scholar] [CrossRef]
- Borrego, F.; Robertson, M.J.; Ritz, J.; Pena, J.; Soilana, R. CD69 is a stimulatory receptor for natural killer cell and its cytotoxic effect is blocked by CD94 inhibitory receptor. Immunology 1999, 97, 159–165. [Google Scholar] [CrossRef]
- Marden, C.M.; North, J.; Anderson, R.; Baksch, I.A.; Addison, E.; Pittmann, H. CD69 is required for activated NK cell-mediated killing of resistant targets. Blood 2005, 106, 3322. [Google Scholar] [CrossRef]
- Marzio, R.; Mauel, J.; Betz-Corradin, S. CD69 and regulation of the immune function. Immunopharmacol. Immunotoxicol. 1999, 21, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Robbins, S.H.; Nguyen, K.B.; Takahashi, N.; Mikayama, T.; Biron, C.A.; Brossay, L. Inhibitory functions of the killer cell liectin-like receptor G1 molecule during the activation of mouse NK cells. J. Immunol. 2002, 168, 2585–2589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krueger, P.D.; Lassen, M.G.; Qiao, H.; Hahn, Y.S. Regulation of NK cell repertoire and function in the liver. Crit. Rev. Immunol. 2011, 31, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.C.; Parikh, S.; Sun, J.C.; Myrick, A.; Lanier, L.L.; Rosenthal, P.J.; De Risi, J.L. Experimental malaria infection triggers early expansion of natural killer cells. Infect. Immun. 2008, 76, 5873–5882. [Google Scholar] [CrossRef] [Green Version]
- Kazmin, D.; Nakaya, H.I.; Lee, E.K.; Johnson, M.J.; van der Most, R.; van den Berg, R.A.; Ballou, W.R.; Jongert, E.; Wille-Reece, U.; Ockenhouse, C.; et al. Systems analysis of protective immune responses to RTS, S malaria vaccination in humans. Proc. Natl. Acad. Sci. USA 2017, 114, 2425–2430. [Google Scholar] [CrossRef] [Green Version]
- Wagstaffe, H.R.; Mooney, J.P.; Riley, E.M.; Goodier, M.R. Vaccinating for natural killer cell effector functions. Clin. Transl. Immunol. 2018, 7, e1010. [Google Scholar] [CrossRef]
- De Poot, S.A.H.; Bovenschen, N. Granzyme M: Behind enemy lines. Cell Death Differ. 2014, 21, 359–388. [Google Scholar] [CrossRef] [Green Version]
- Sheppard, S.; Schuster, I.S.; Andonou, C.E.; Cocita, C.; Adejumo, T.; Kung, S.K.P.; Sun, J.C.; Degli-Esposti, M.A.; Guerra, N. The murine cytotoxic receptor NKp46/NCR1 controls TRAIL protein expression in NK cells and ILC1s. Cell Rep. 2018, 22, 3385–3392. [Google Scholar] [CrossRef] [Green Version]
- Dunn, C.; Brunetto, M.; Reynolds, G.; Christophidis, T.; Kennedy, P.T.; Lampertico, P.; Das, A.; Lopes, A.R.; Borrow, P.; Williams, K.; et al. Cytokines induced during chronic hepatitis B virus infection promote a pathway for NK cell-mediated liver damage. J. Exp. Med. 2007, 204, 667–680. [Google Scholar] [CrossRef] [Green Version]
- Goodall, K.J.; Nguyen, A.; Sullivan, L.C.; Anrews, D.M. The expanding role of murine class Ib MHC in the development and activation of natural killer cells. Mol. Immunol. 2019, 115, 31–98. [Google Scholar] [CrossRef] [PubMed]
- Orr, M.T.; Wu, J.; Fang, M.; Sigal, L.J.; Spee, P.; Egebjerg, T.; Dissen, E.; Fossum, S.; Phillips, J.H.; Lanier, L.L. Development and function of CD94-deficient natural killer cells. PLoS ONE 2010, 5, e15184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Reyes-Vargas, E.; Dai, H.; Escobar, H.; Rudd, B.; Fairbanks, J.; Ho, A.; Cusick, M.F.; Kumanovics, A.; Delgado, J.; et al. Expression of the mouse MHC calss Ib H2-T11 gene product, a paralog of H2-T23 (Qa-1) with shead peptide-binding specificity. J. Immunol. 2014, 193, 1427–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, C.E.; Key, P.N.; Zhu, T.; Shabsovich, M.; Ni, A.; Tripathy, S.K. Expression of the inhibitory receptor NKG2A correlates with increased liver and splenic NK cell response to activating receptor engagement. Immun. Inflamm. Dis. 2017, 5, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sovova, Z.; Kopecky, V., Jr.; Pazderka, T.; Hofbauerova, K.; Rozbesky, D.; Vanek, O.; Bezouska, K.; Ettrich, E. Structural analysis of natural killer cell recptor protein (NKR-P1) extracellular domains suggests a conserved long loop region involved in ligand specificity. J. Mol. Model. 2011, 17, 1353–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahim, M.M.A.; Chen, P.; Mottashed, A.N.; Mahmoud, A.B.; Thomas, M.J.; Zhu, Q.; Brooks, C.G.; Kartsogiannis, V.; Gillespie, M.T.; Carlyle, J.R.; et al. The mouse NKR-P1B:Clr-b recognition system is a negative regulator of innate immune responses. Blood J. Am. Soc. Hematol. 2015, 125, 2217–2227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkham, C.L.; Aguilar, O.A.; Yu, T.; Tanaka, M.; Mesci, A.; Chu, K.L.; Fine, J.H.; Mossman, K.L.; Brenner, R.; Allan, D.S.J.; et al. Interferon-dependent induction of Clr-b during mouse cytomegalovirus infection protects bystander cells from natural killer cells via NKR-P1B-mediated inhibition. J. Innate Immun. 2017, 9, 343–358. [Google Scholar] [CrossRef]
- Carlyle, J.R.; Jamieson, A.M.; Gasser, S.; Clingan, C.S.; Arase, H.; Raulet, D.H. Missing self recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 3527–3532. [Google Scholar] [CrossRef] [Green Version]
- Ott, K.J. Influence of reticulocytosis on the course of infection of Plasmodium chabaudi and P. berghei. J. Protozool. 1968, 15, 365–369. [Google Scholar] [CrossRef]
- Friede, M.E.; Leibelt, S.; Dudziak, D.; Steinle, A. Select Clr-g expression on activated dendritic cells facilitates cognate interaction with a minor subset of splenic NK cells expressing the inhibitory Nkrp1g receptor. J. Immunol. 2018, 200, 983–996. [Google Scholar] [CrossRef]
- Tian, W.; Nunez, R.; Cheng, S.; Ding, Y.; Tumang, J.; Lyddane, C.; Roman, C.; Liou, H.-C. C-type lectin OCILRP2/Clr-g and is ligand NKRP1f costimulate T cell proliferation and IL-2 production. Cell. Immunol. 2005, 234, 39–53. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araúzo-Bravo, M.J.; Delic, D.; Gerovska, D.; Wunderlich, F. Protective Vaccination Reshapes Hepatic Response to Blood-Stage Malaria of Genes Preferentially Expressed by NK Cells. Vaccines 2020, 8, 677. https://doi.org/10.3390/vaccines8040677
Araúzo-Bravo MJ, Delic D, Gerovska D, Wunderlich F. Protective Vaccination Reshapes Hepatic Response to Blood-Stage Malaria of Genes Preferentially Expressed by NK Cells. Vaccines. 2020; 8(4):677. https://doi.org/10.3390/vaccines8040677
Chicago/Turabian StyleAraúzo-Bravo, Marcos J., Denis Delic, Daniela Gerovska, and Frank Wunderlich. 2020. "Protective Vaccination Reshapes Hepatic Response to Blood-Stage Malaria of Genes Preferentially Expressed by NK Cells" Vaccines 8, no. 4: 677. https://doi.org/10.3390/vaccines8040677
APA StyleAraúzo-Bravo, M. J., Delic, D., Gerovska, D., & Wunderlich, F. (2020). Protective Vaccination Reshapes Hepatic Response to Blood-Stage Malaria of Genes Preferentially Expressed by NK Cells. Vaccines, 8(4), 677. https://doi.org/10.3390/vaccines8040677




