Selection and Evaluation of Porcine circovirus (PCV) 2d Vaccine Strains to Protect against Currently Prevalent PCV2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses and Cells
2.2. Production of Recombinant VLP
2.3. Guinea Pig Immunization
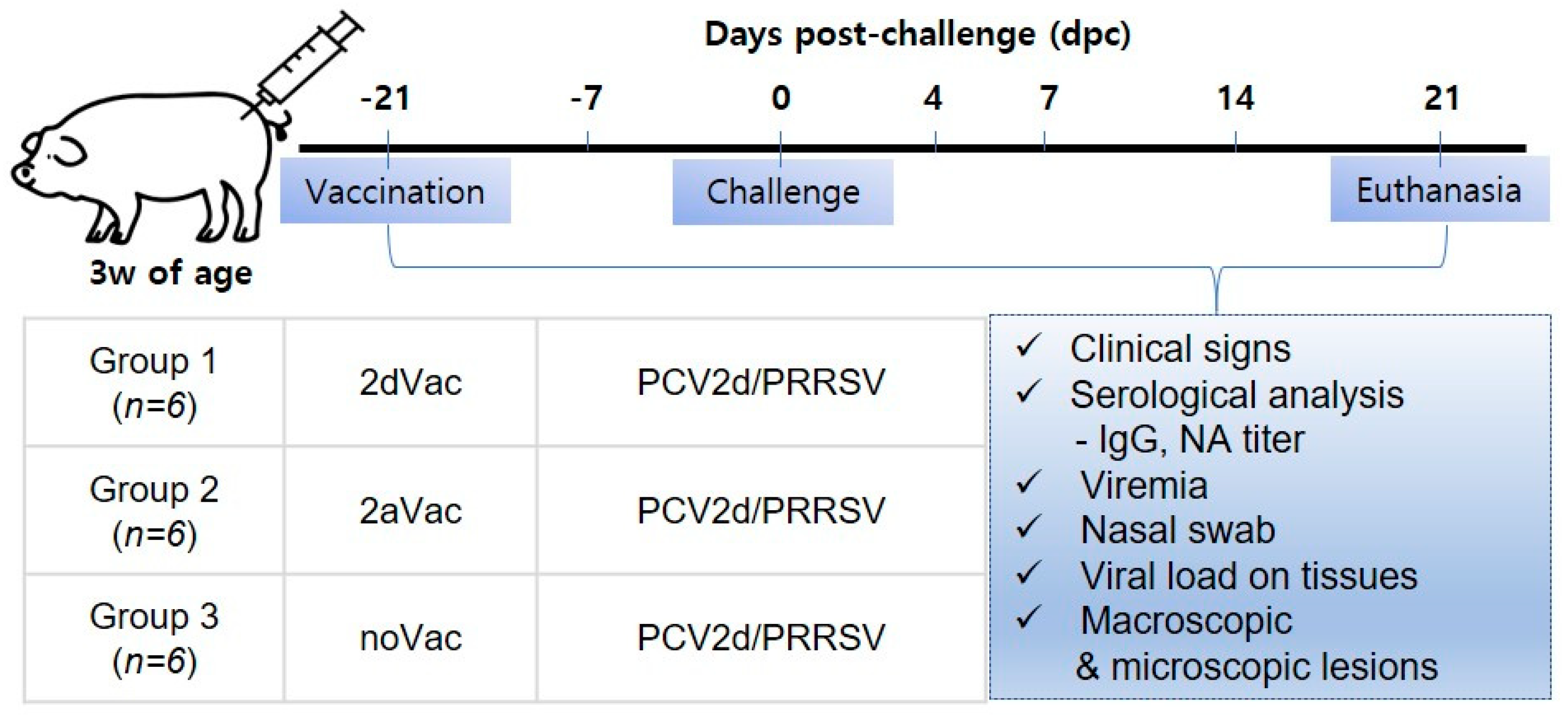
2.4. Pig Studies
2.5. Clinical Signs and Body Weight
2.6. Quantification of the Virus in Serum and Tissues
2.7. Viral Neutralization Assay
2.8. Gross Pathology and Histopathology
2.9. Statistical Analyses
3. Results
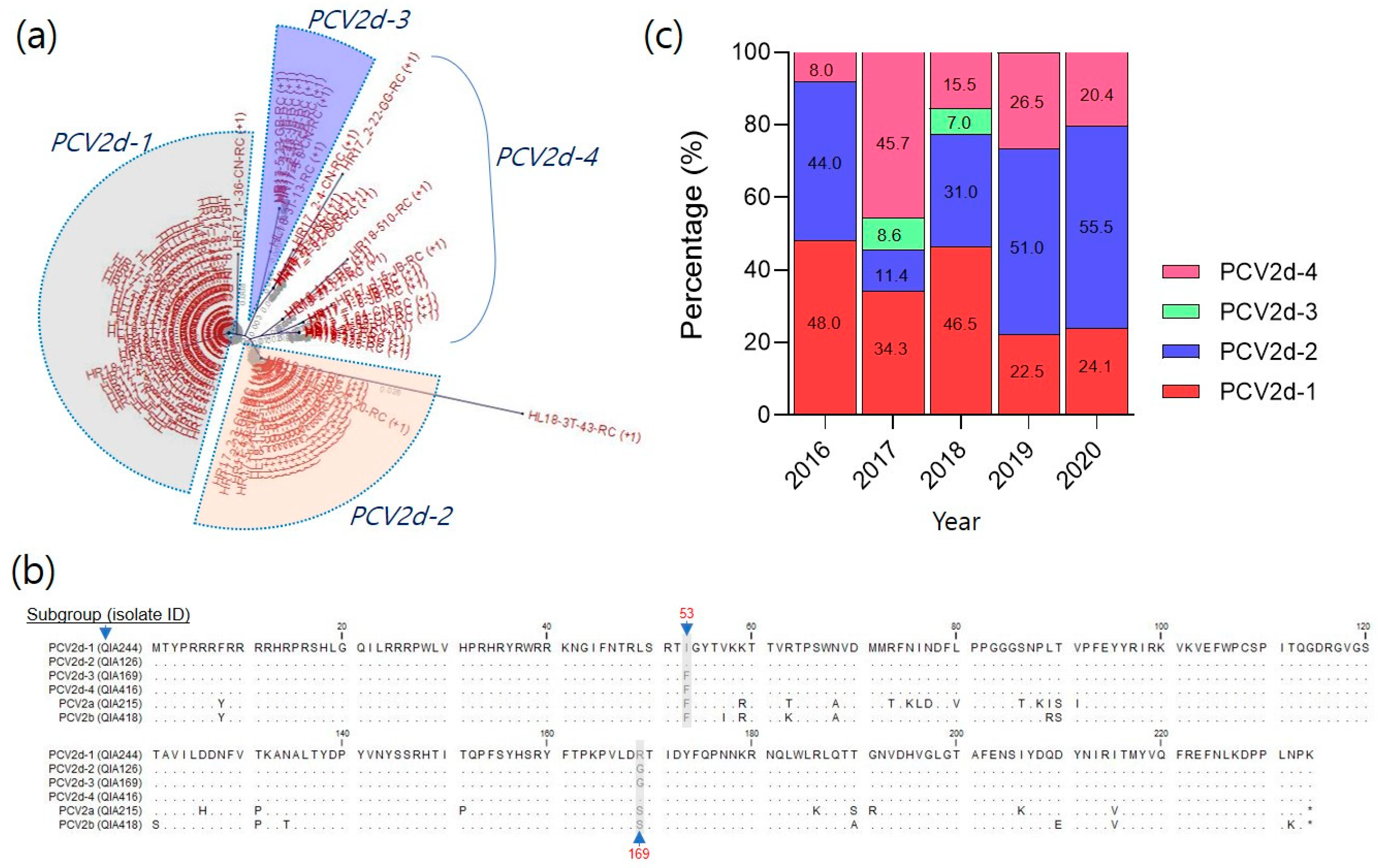
3.1. Antigenicity of PCV2d Field Isolates in South Korea
3.1.1. Analysis of Cap Proteins in the Recently Prevalent PCV2d
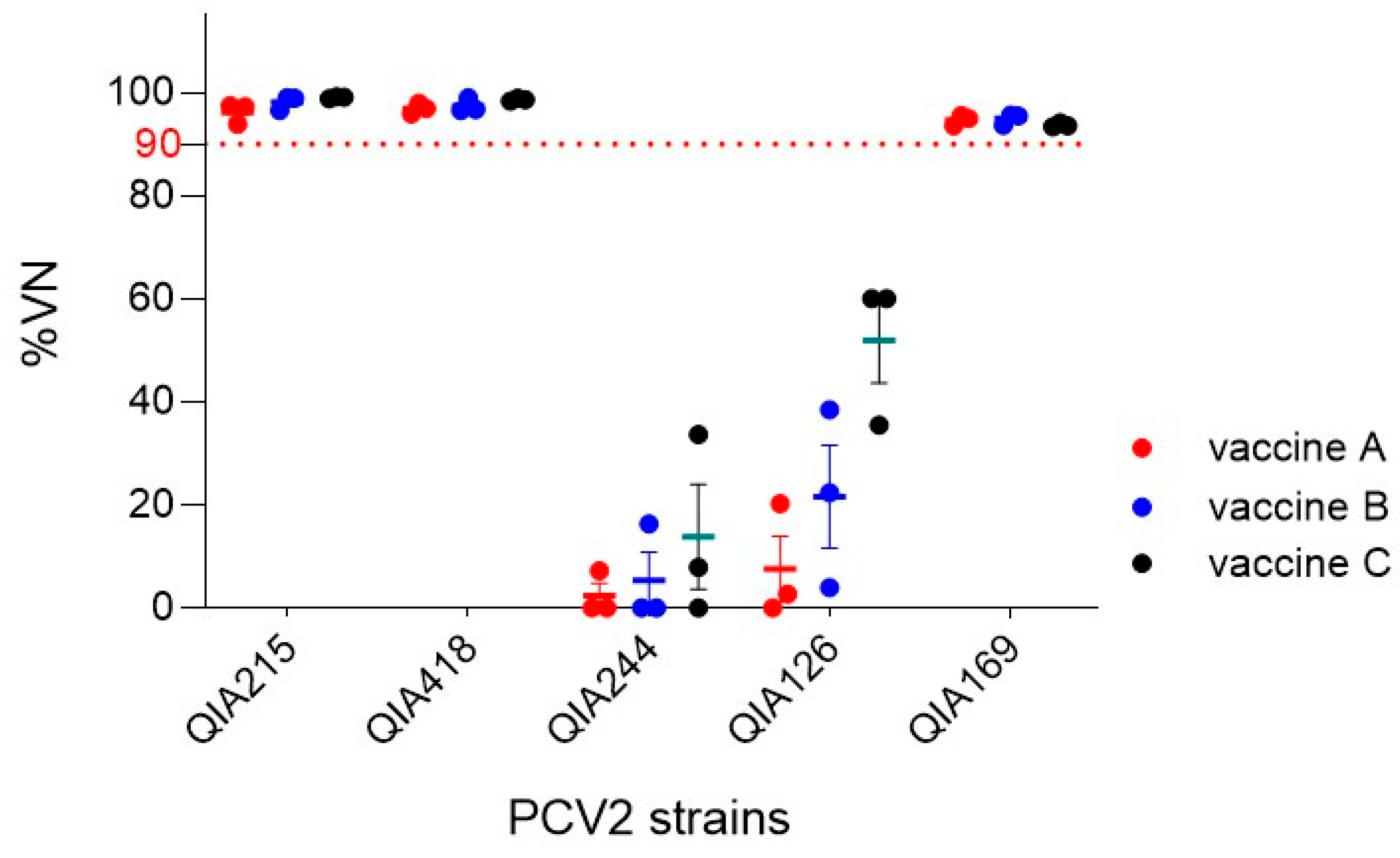
3.1.2. Neutralization of Commercial PCV2a-Based Vaccines against PCV2 Strains
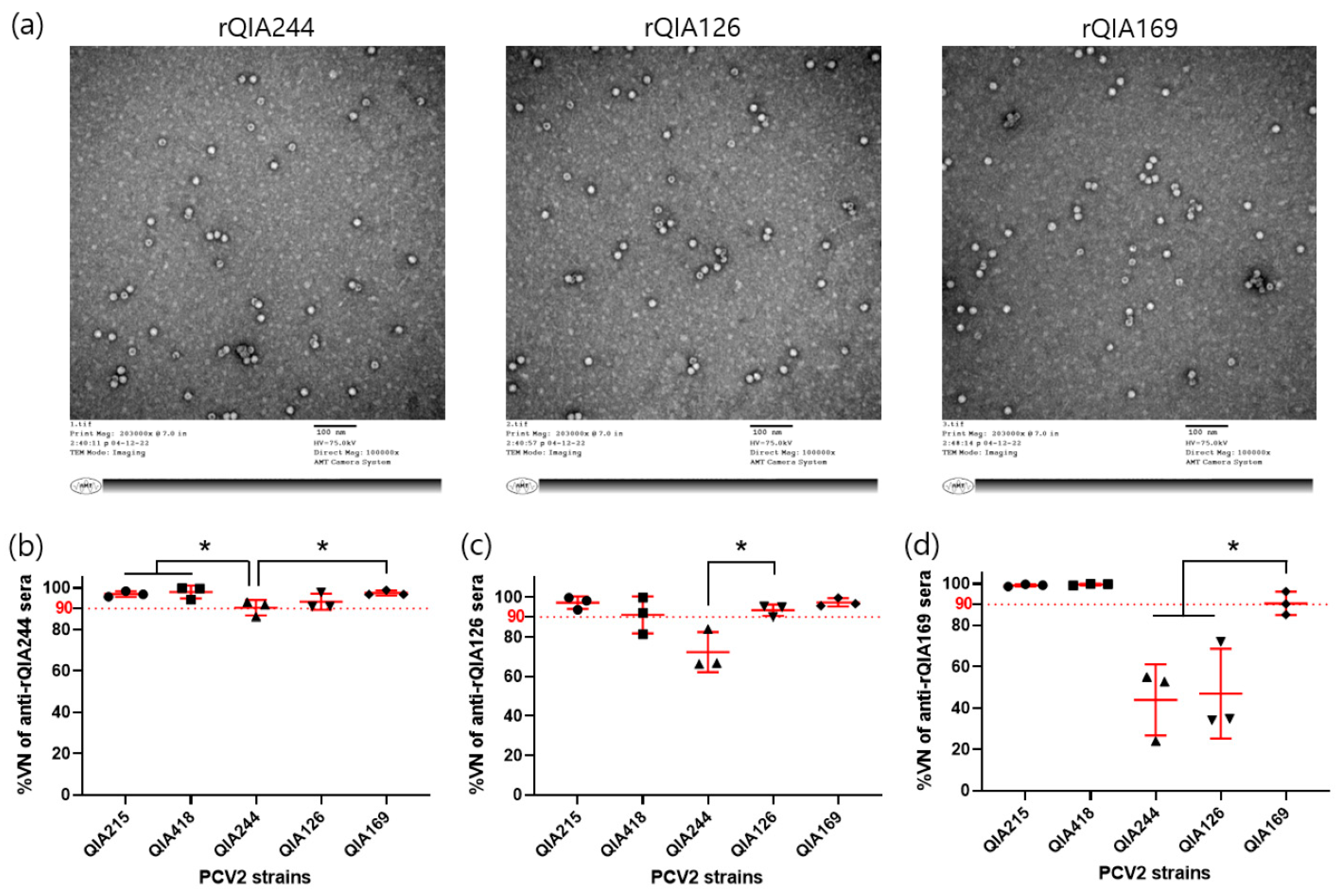
3.1.3. Cross-Neutralization of PCV2d-VLPs against PCV2 Strains
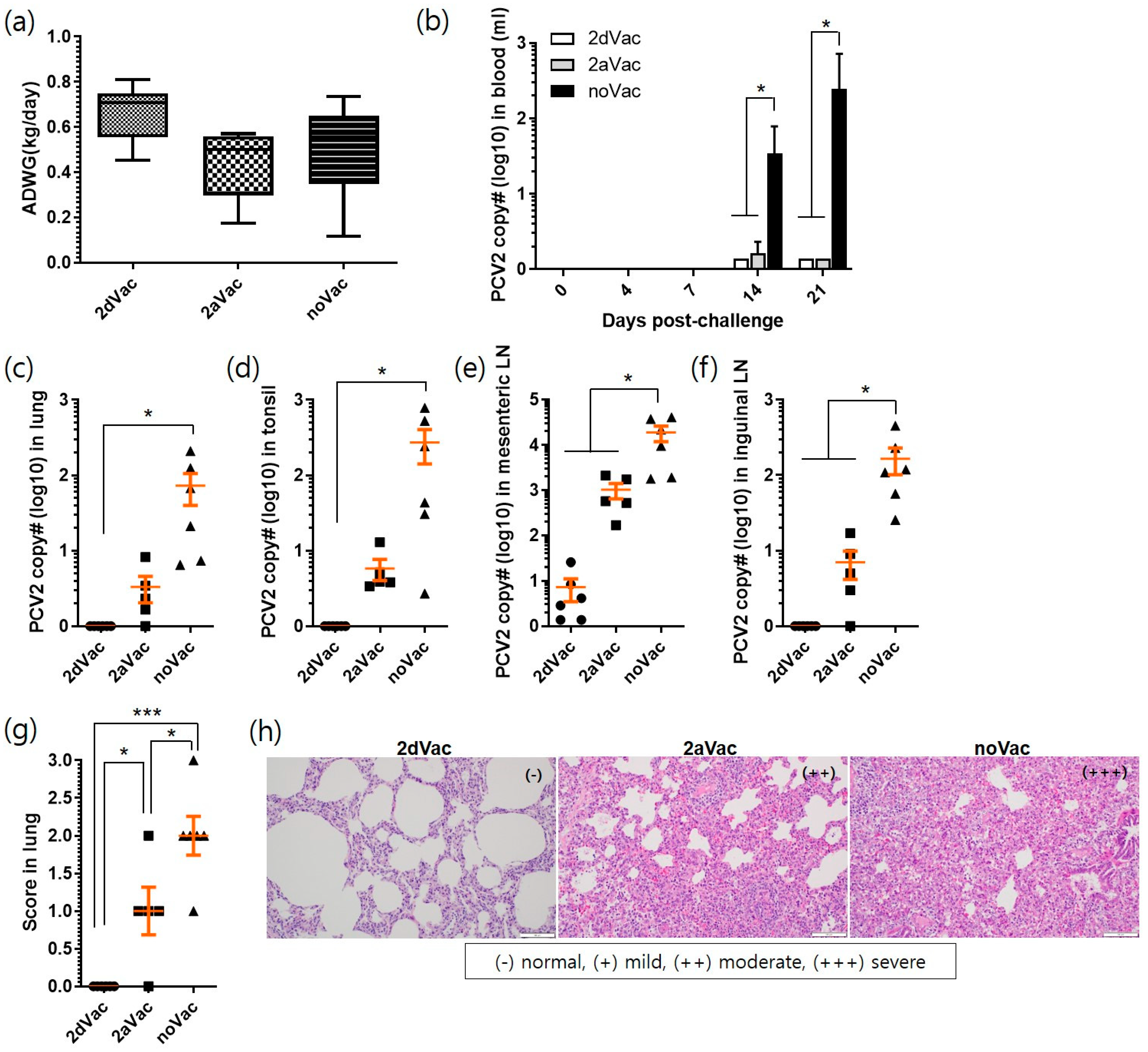
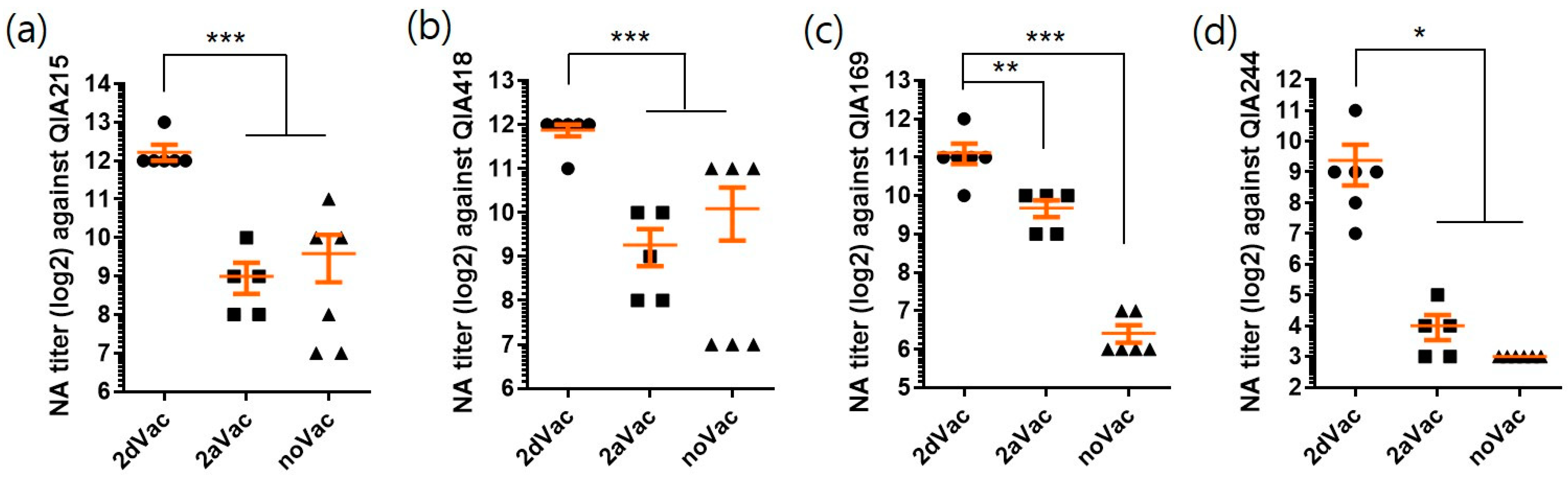
3.2. In Vivo Protection of 2dVac and 2aVac
3.2.1. Evaluation of Vaccine Efficacy against PCV2d Challenge
3.2.2. NA Titers of 2dVac and 2aVac against PCV2 Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tischer, I.; Gelderblom, H.; Vettermann, W.; Koch, M.A. A very small porcine virus with circular single-stranded DNA. Nature 1982, 295, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl. Bakteriol. Orig. A 1974, 226, 153–167. [Google Scholar]
- Kekarainen, T.; Segales, J. Porcine circovirus 2 immunology and viral evolution. Porcine Health Manag. 2015, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Segales, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.M.; Nguyen, B.H.; Nguyen, V.G.; Dang, H.A.; Mai, T.N.; Tran, T.H.; Ngo, M.H.; Le, V.T.; Vu, T.N.; Ta, T.K.; et al. Phylogenetic and phylogeographic analyses of porcine circovirus type 2 among pig farms in Vietnam. Transbound. Emerg. Dis. 2014, 61, e25–e34. [Google Scholar] [CrossRef]
- Bao, F.; Mi, S.; Luo, Q.; Guo, H.; Tu, C.; Zhu, G.; Gong, W. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound. Emerg. Dis. 2018, 65, 432–440. [Google Scholar] [CrossRef]
- Franzo, G.; Cortey, M.; Segales, J.; Hughes, J.; Drigo, M. Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol. Phylogenet. Evol. 2016, 100, 269–280. [Google Scholar] [CrossRef]
- Karuppannan, A.K.; Opriessnig, T. Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology. Viruses 2017, 9, 99. [Google Scholar] [CrossRef]
- Kwon, T.; Lee, D.U.; Yoo, S.J.; Je, S.H.; Shin, J.Y.; Lyoo, Y.S. Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res. 2017, 228, 24–29. [Google Scholar] [CrossRef]
- Kim, S.C.; Nazki, S.; Kwon, S.; Juhng, J.H.; Mun, K.H.; Jeon, D.Y.; Jeong, C.G.; Khatun, A.; Kang, S.J.; Kim, W.I. The prevalence and genetic characteristics of porcine circovirus type 2 and 3 in Korea. BMC Vet. Res. 2018, 14, 294. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.J.; Kang, H.; You, S.H.; Lee, H.J.; Lee, N.; Hyun, B.H.; Cha, S.H. Genetic diversity and different cross-neutralization capability of porcine circovirus type 2 isolates recently circulating in South Korea. BMC Vet. Res. 2020, 16, 334. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J. Gen. Virol. 2015, 96, 1830–1841. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Jin, M.; Yoon, I.; Yoo, H.S. Efficacy of bivalent vaccines of porcine circovirus type 2 and Mycoplasma hyopneumoniae in specific pathogen-free pigs challenged with porcine circovirus type 2d. J. Vet. Sci. 2022, 23, e49. [Google Scholar] [CrossRef] [PubMed]
- Dhindwal, S.; Avila, B.; Feng, S.; Khayat, R. Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites to Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid. J Virol. 2019, 93, e02222-18. [Google Scholar] [CrossRef]
- Mao, Q.; Zhang, W.; Ma, S.; Qiu, Z.; Li, B.; Xu, C.; He, H.; Fan, S.; Wu, K.; Chen, J.; et al. Fusion Expression and Immune Effect of PCV2 Cap Protein Tandem Multiantigen Epitopes with CD154/GM-CSF. Vet. Sci. 2021, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Chae, C. Commercial porcine circovirus type 2 vaccines: Efficacy and clinical application. Vet. J. 2012, 194, 151–157. [Google Scholar] [CrossRef]
- Park, K.H.; Oh, T.; Yang, S.; Cho, H.; Kang, I.; Chae, C. Evaluation of a porcine circovirus type 2a (PCV2a) vaccine efficacy against experimental PCV2a, PCV2b, and PCV2d challenge. Vet. Microbiol. 2019, 231, 87–92. [Google Scholar] [CrossRef]
- Opriessnig, T.; O’Neill, K.; Gerber, P.F.; de Castro, A.M.; Gimenez-Lirola, L.G.; Beach, N.M.; Zhou, L.; Meng, X.J.; Wang, C.; Halbur, P.G. A PCV2 vaccine based on genotype 2b is more effective than a 2a-based vaccine to protect against PCV2b or combined PCV2a/2b viremia in pigs with concurrent PCV2, PRRSV and PPV infection. Vaccine 2013, 31, 487–494. [Google Scholar] [CrossRef]
- Kang, S.J.; Bae, S.M.; Lee, H.J.; Jeong, Y.J.; Lee, M.A.; You, S.H.; Lee, H.S.; Hyun, B.H.; Lee, N.; Cha, S.H. Porcine Circovirus (PCV) Genotype 2d-Based Virus-like Particles (VLPs) Induced Broad Cross-Neutralizing Antibodies against Diverse Genotypes and Provided Protection in Dual-Challenge Infection of a PCV2d Virus and a Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV). Pathogens 2021, 10, 1145. [Google Scholar] [CrossRef]
- Meerts, P.; Van Gucht, S.; Cox, E.; Vandebosch, A.; Nauwynck, H.J. Correlation between type of adaptive immune response against porcine circovirus type 2 and level of virus replication. Viral Immunol. 2005, 18, 333–341. [Google Scholar] [CrossRef]
- Xiao, C.T.; Harmon, K.M.; Halbur, P.G.; Opriessnig, T. PCV2d-2 is the predominant type of PCV2 DNA in pig samples collected in the U.S. during 2014–2016. Vet. Microbiol. 2016, 197, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.W.; Park, C.; Kang, I.; Choi, K.; Jeong, J.; Park, S.J.; Chae, C. Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Arch. Virol. 2014, 159, 3107–3111. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhan, Y.; Wang, A.; Zhang, L.; Khayat, R.; Yang, Y. In silico analysis of surface structure variation of PCV2 capsid resulting from loop mutations of its capsid protein (Cap). J. Gen. Virol. 2016, 97, 3331–3344. [Google Scholar] [CrossRef] [PubMed]
- Khayat, R.; Brunn, N.; Speir, J.A.; Hardham, J.M.; Ankenbauer, R.G.; Schneemann, A.; Johnson, J.E. The 2.3-angstrom structure of porcine circovirus 2. J. Virol. 2011, 85, 7856–7862. [Google Scholar] [CrossRef]
- Mahe, D.; Blanchard, P.; Truong, C.; Arnauld, C.; Le Cann, P.; Cariolet, R.; Madec, F.; Albina, E.; Jestin, A. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 2000, 81, 1815–1824. [Google Scholar] [CrossRef]
- Guo, L.J.; Lu, Y.H.; Wei, Y.W.; Huang, L.P.; Liu, C.M. Porcine circovirus type 2 (PCV2): Genetic variation and newly emerging genotypes in China. Virol. J. 2010, 7, 273. [Google Scholar] [CrossRef]
- Huang, L.P.; Lu, Y.H.; Wei, Y.W.; Guo, L.J.; Liu, C.M. Identification of one critical amino acid that determines a conformational neutralizing epitope in the capsid protein of porcine circovirus type 2. BMC Microbiol. 2011, 11, 188. [Google Scholar] [CrossRef]
- Lekcharoensuk, P.; Morozov, I.; Paul, P.S.; Thangthumniyom, N.; Wajjawalku, W.; Meng, X.J. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 2004, 78, 8135–8145. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, L.; You, S.-H.; Lee, M.-A.; Jayaramaiah, U.; Jeong, Y.-J.; Lee, H.-S.; Hyun, B.-H.; Lee, N.; Kang, S.-J. Selection and Evaluation of Porcine circovirus (PCV) 2d Vaccine Strains to Protect against Currently Prevalent PCV2. Vaccines 2023, 11, 1447. https://doi.org/10.3390/vaccines11091447
Ju L, You S-H, Lee M-A, Jayaramaiah U, Jeong Y-J, Lee H-S, Hyun B-H, Lee N, Kang S-J. Selection and Evaluation of Porcine circovirus (PCV) 2d Vaccine Strains to Protect against Currently Prevalent PCV2. Vaccines. 2023; 11(9):1447. https://doi.org/10.3390/vaccines11091447
Chicago/Turabian StyleJu, Lanjeong, Su-Hwa You, Min-A Lee, Usharani Jayaramaiah, Young-Ju Jeong, Hyang-Sim Lee, Bang-Hun Hyun, Nakhyung Lee, and Seok-Jin Kang. 2023. "Selection and Evaluation of Porcine circovirus (PCV) 2d Vaccine Strains to Protect against Currently Prevalent PCV2" Vaccines 11, no. 9: 1447. https://doi.org/10.3390/vaccines11091447
APA StyleJu, L., You, S.-H., Lee, M.-A., Jayaramaiah, U., Jeong, Y.-J., Lee, H.-S., Hyun, B.-H., Lee, N., & Kang, S.-J. (2023). Selection and Evaluation of Porcine circovirus (PCV) 2d Vaccine Strains to Protect against Currently Prevalent PCV2. Vaccines, 11(9), 1447. https://doi.org/10.3390/vaccines11091447




