Genome-Based Multi-Antigenic Epitopes Vaccine Construct Designing against Staphylococcus hominis Using Reverse Vaccinology and Biophysical Approaches
Abstract
:1. Introduction
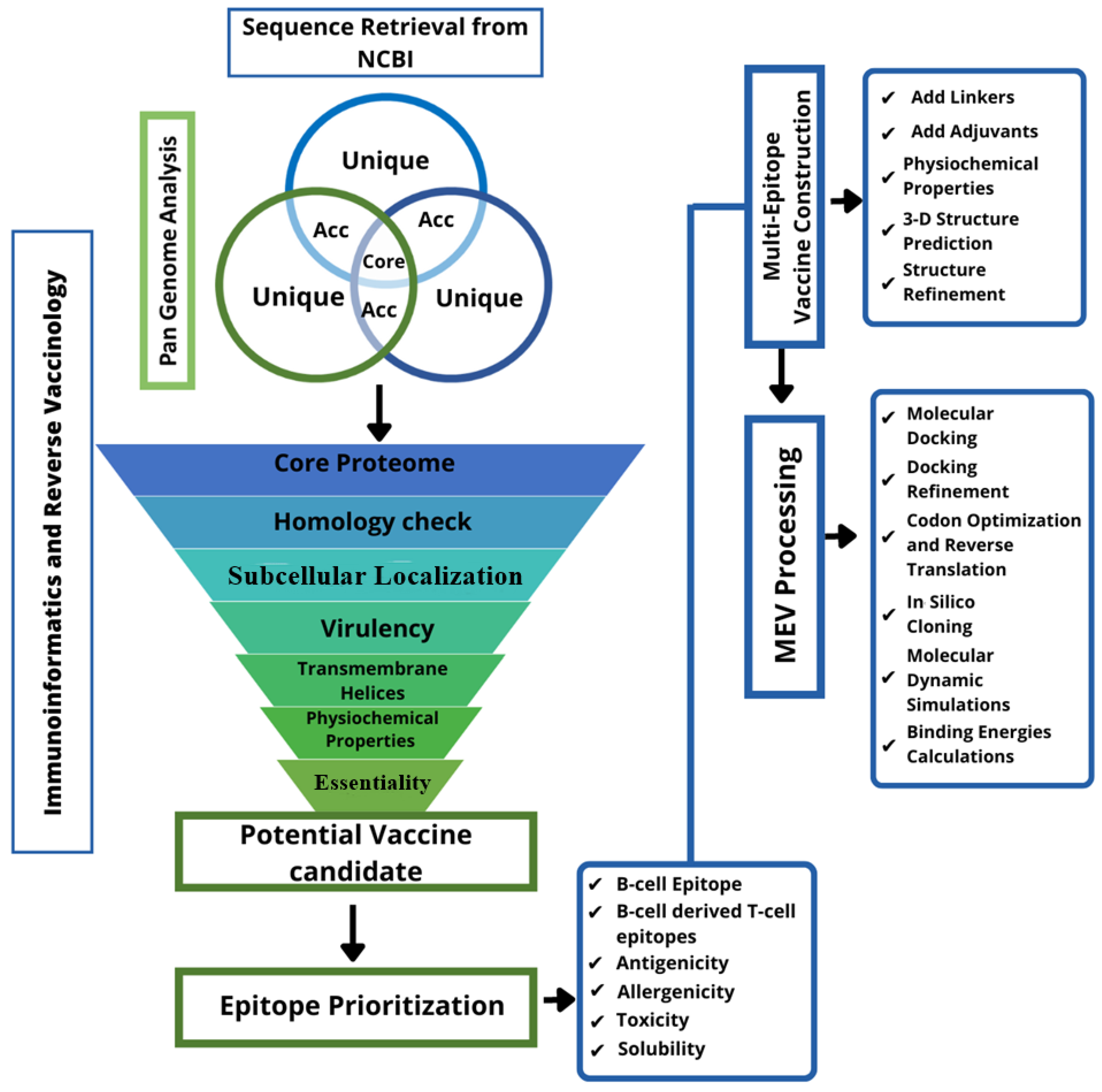
2. Materials and Methods
2.1. Pre-Screening
2.2. Vaccine Candidate Prioritization
2.3. Prediction of Immune Cells Epitopes
2.4. Epitopes Prioritization
2.5. Multi-Epitope Peptide Designing
2.6. Molecular Docking
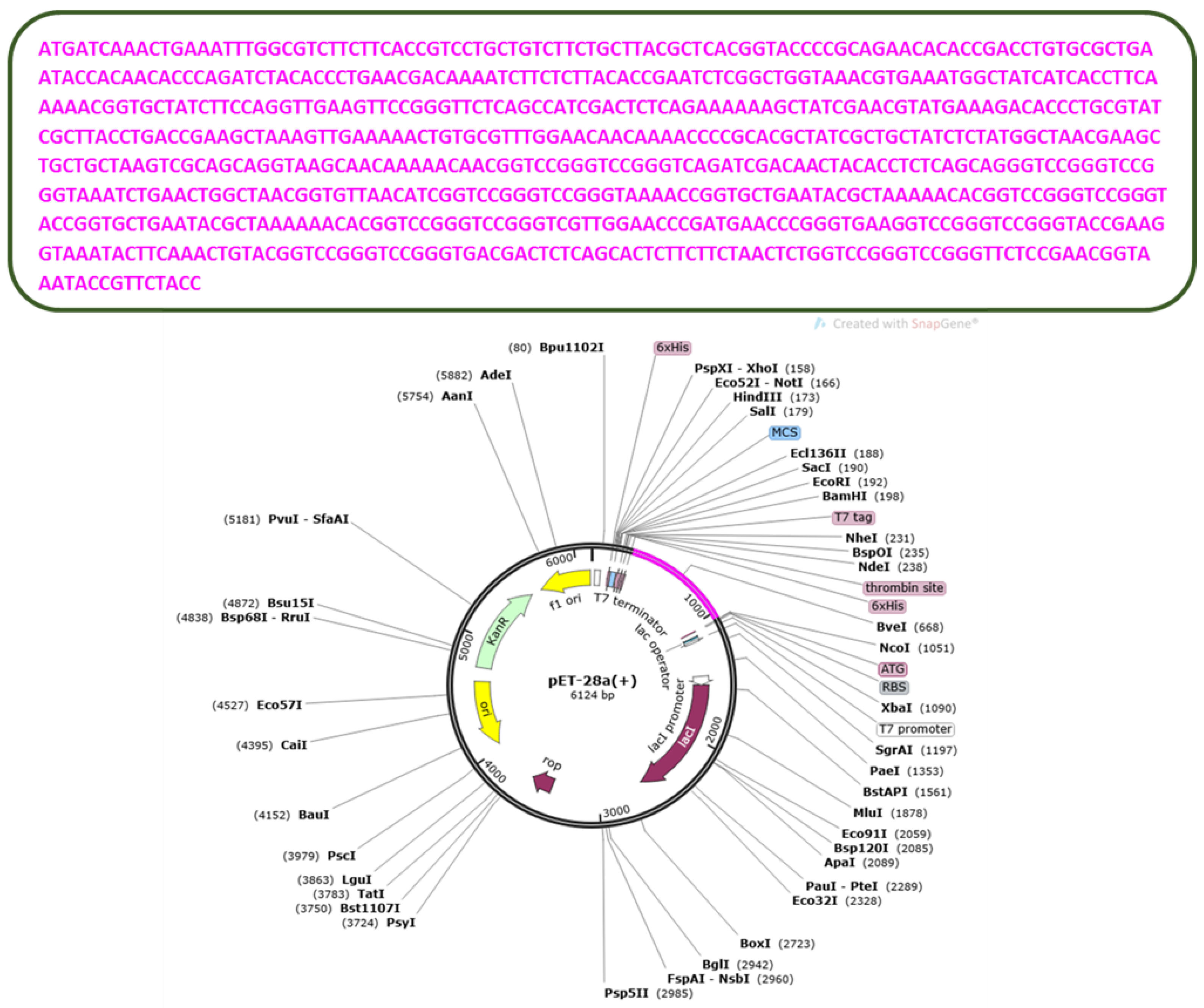
2.7. In Silico Cloning and Codon Optimization
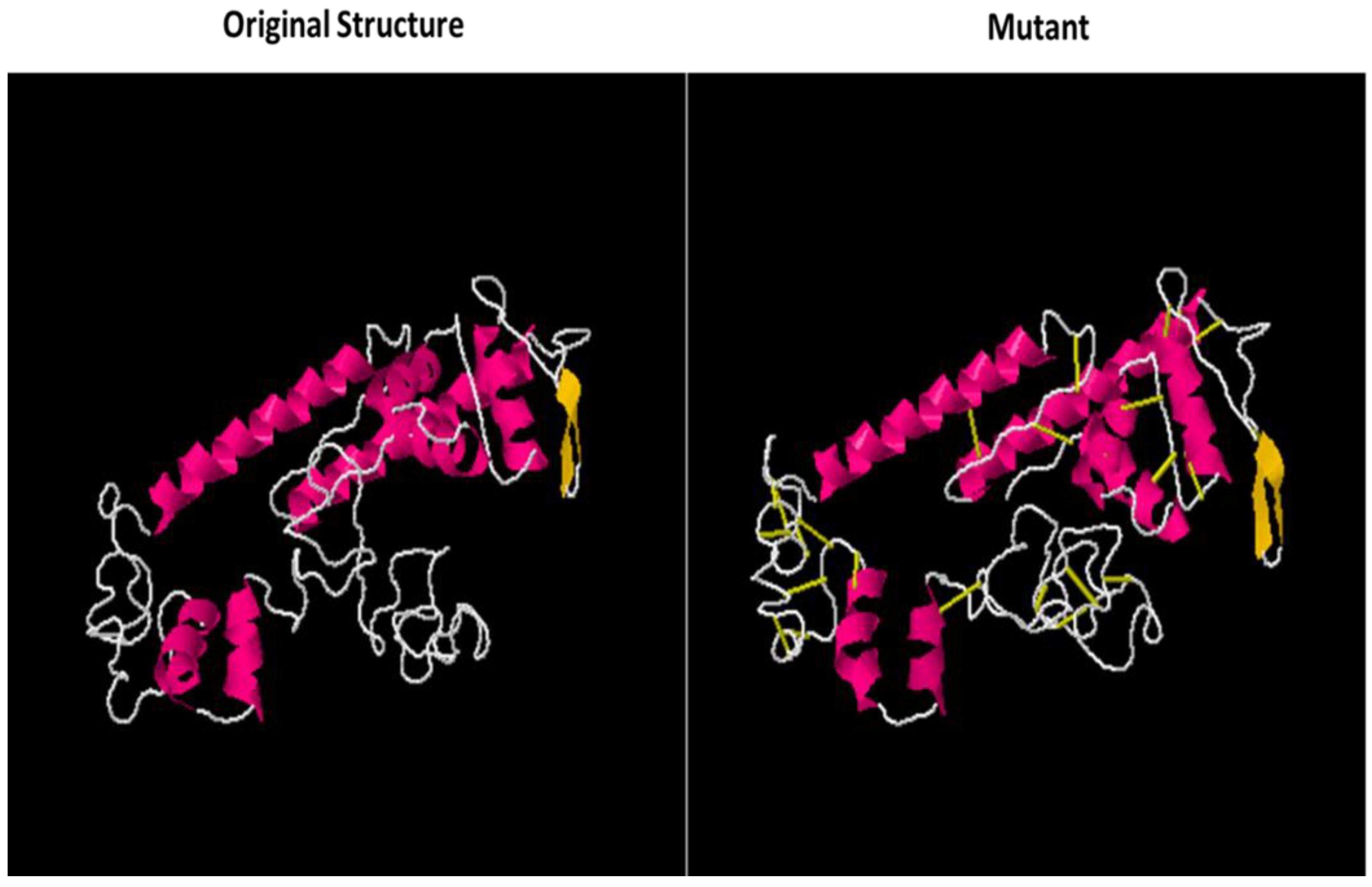
2.8. Disulfide Engineering
2.9. Molecular Dynamic (MD) Simulations
2.10. Binding Free Energies Calculation
3. Results
3.1. Proteins Sequence Retrieval
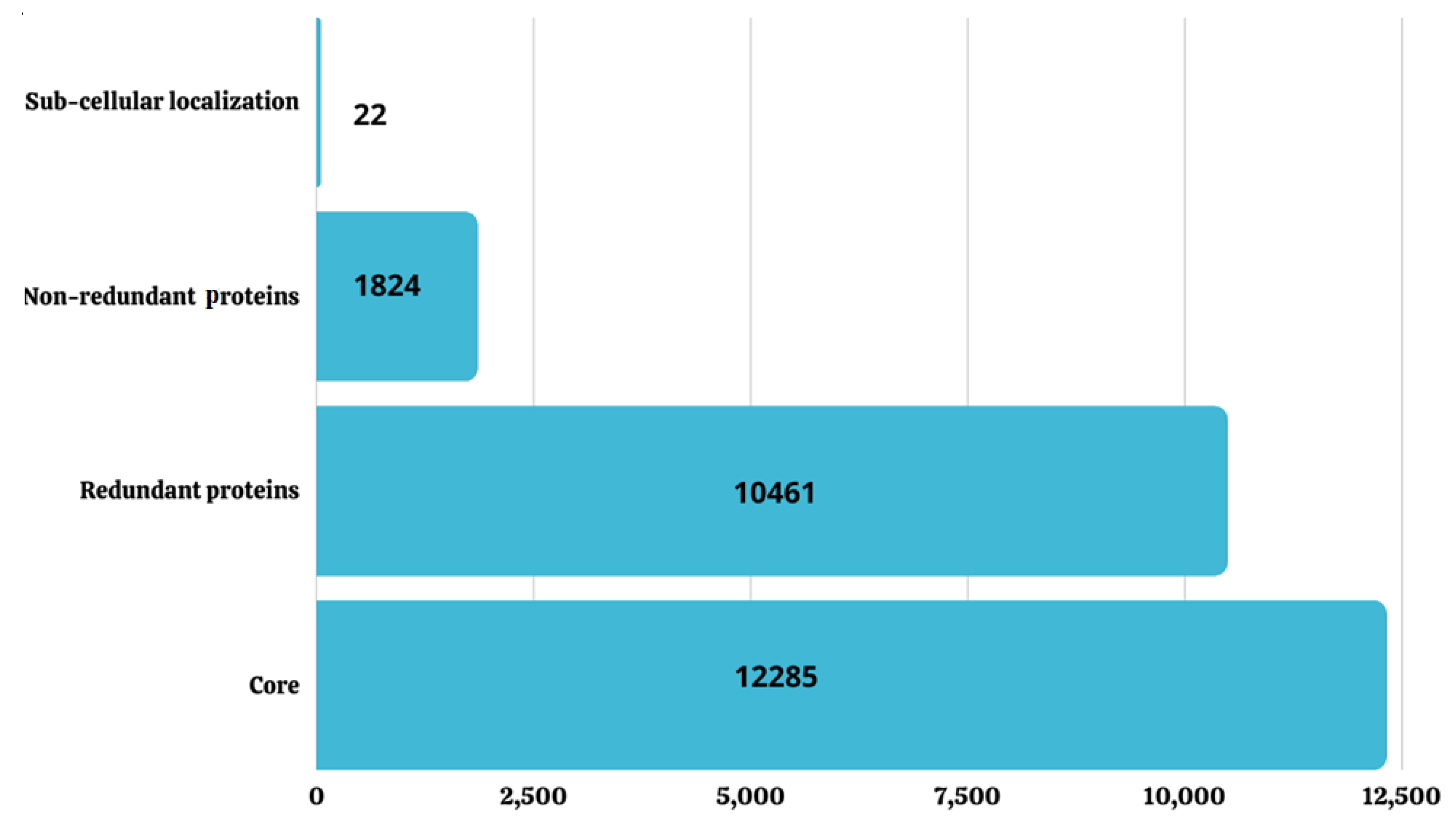
3.2. Pan-Genome Analysis
3.3. CD-HIT Analysis
3.4. Sub-Cellular Localization
3.5. Virulence Check
3.6. Transmembrane Helices and Physiochemical Properties Analysis
3.7. Homology Check against Probiotics
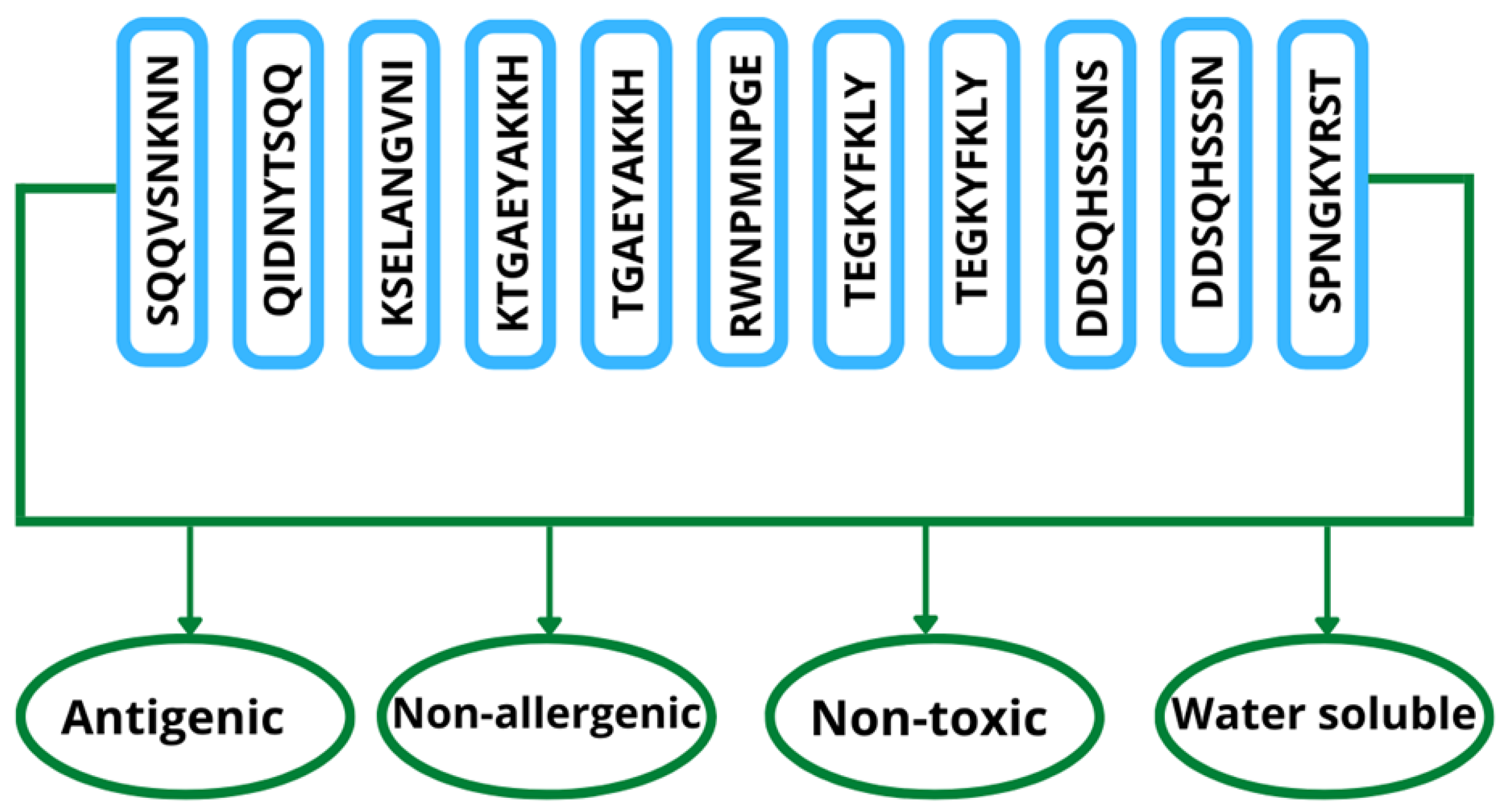
3.8. B and T Cells Epitope Prediction
3.9. Epitope Prioritization
3.10. Multi-Epitope Vaccine Construct Processing
3.11. Codon Optimization
3.12. Disulfide Engineering
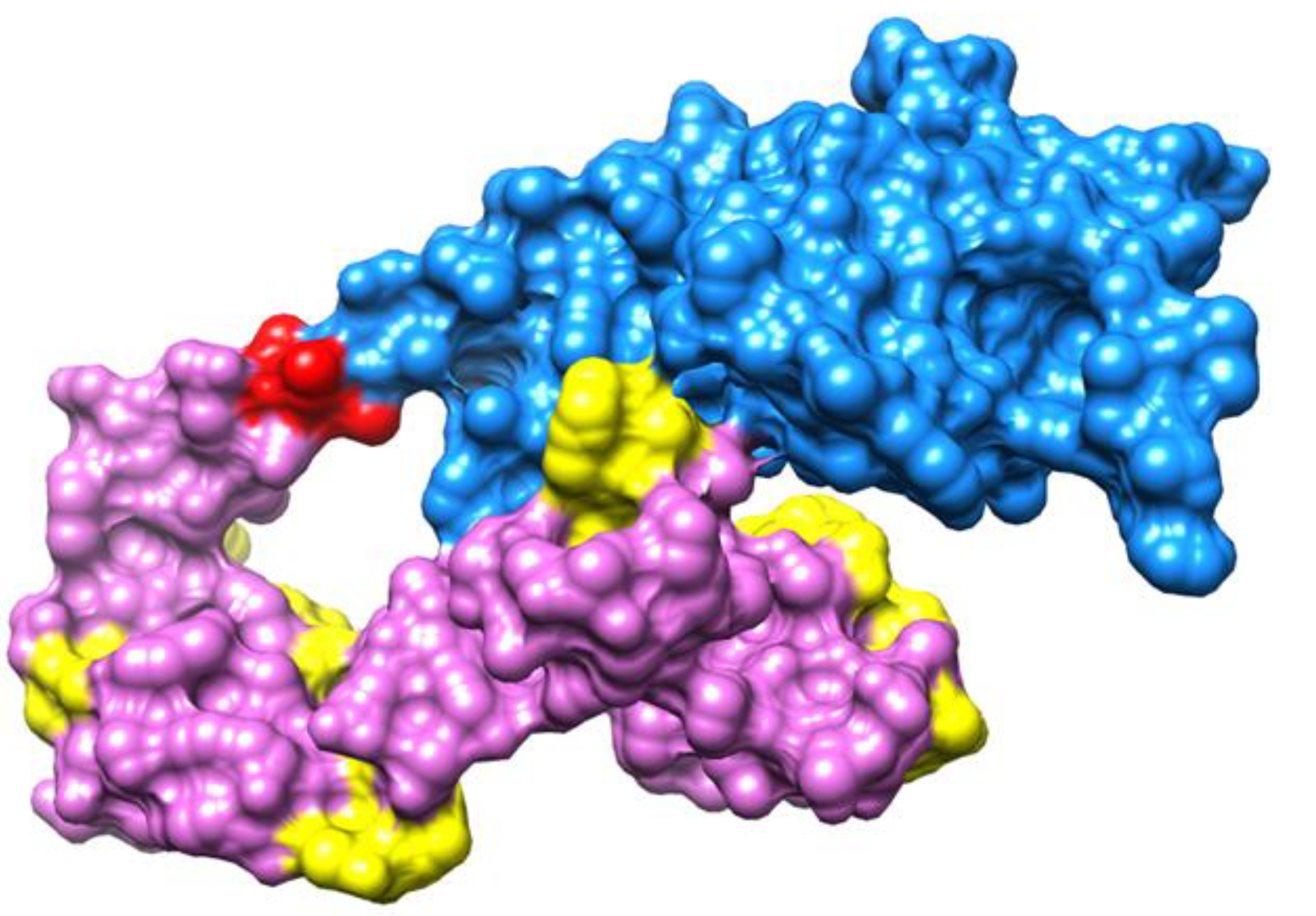
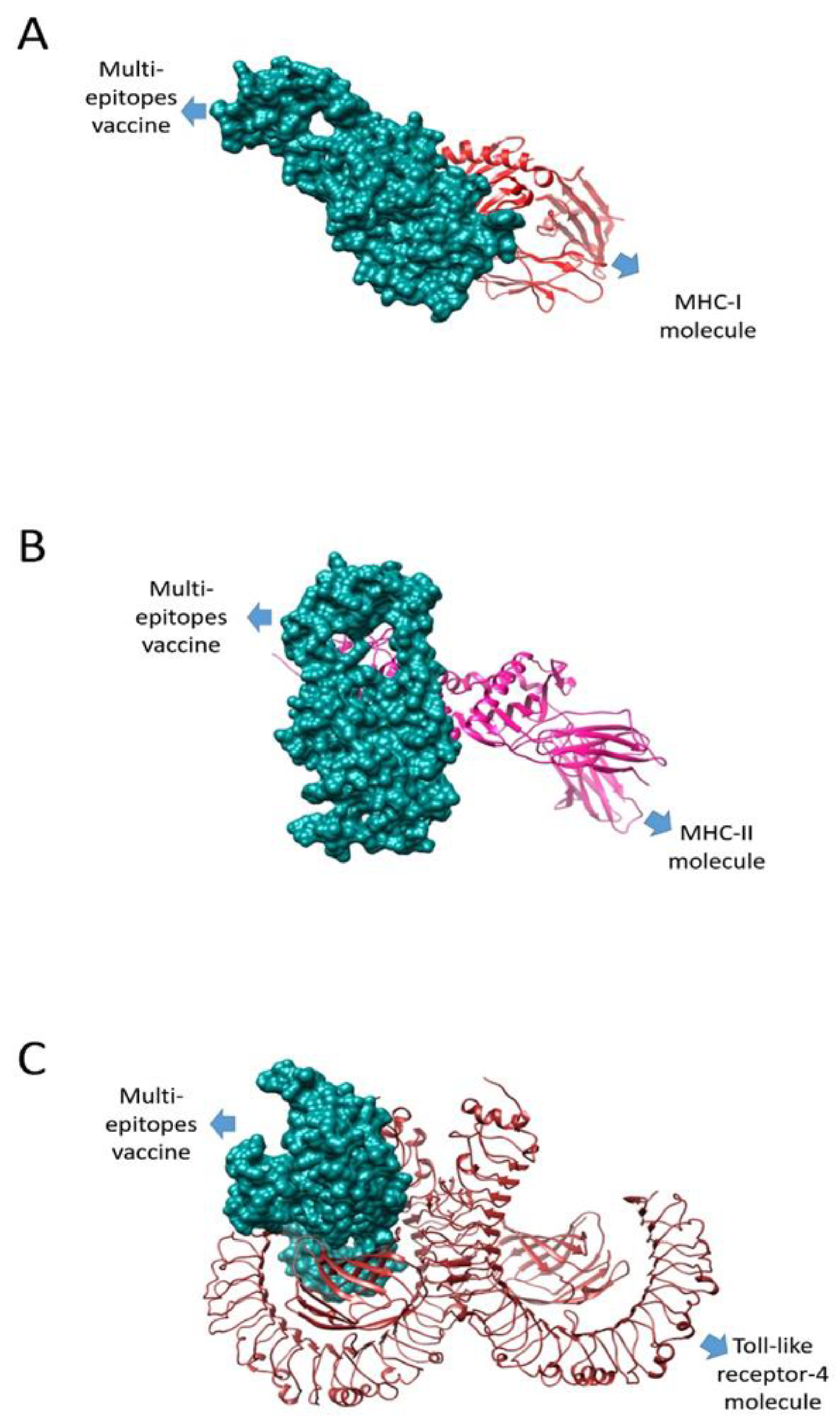
3.13. Molecular Docking
3.14. Docking Refinement
3.15. MD Simulation
3.16. Binding Free Energies Calculations
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Abdalla, N.M.; Haimour, W.O.; Osman, A.A.; Sarhan, M.A.; Musaa, H.A. Antibiotics sensitivity profile towards Staphylococcus hominis in Assir region of Saudi Arabia. J. Sci. Res. 2013, 5, 171–183. [Google Scholar] [CrossRef] [Green Version]
- Mendoza-Olazarán, S.; Morfin-Otero, R.; Rodriguez-Noriega, E.; Llaca-Diaz, J.; Flores-Treviño, S.; González-González, G.M.; Villarreal-Treviño, L.; Garza-González, E. Microbiological and molecular characterization of Staphylococcus hominis isolates from blood. PLoS ONE 2013, 8, e61161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voineagu, L.; Braga, V.; Botnarciuc, M.; Barbu, A.; Tataru, M. Emergence of Staphylococcus hominis Strains in General Infections. ARS Med. Tomitana 2012, 18, 80–82. [Google Scholar] [CrossRef] [Green Version]
- Abbas, G.; Zafar, I.; Ahmad, S.; Azam, S.S. Immunoinformatics design of a novel multi-epitope peptide vaccine to combat multi-drug resistant infections caused by Vibrio vulnificus. Eur. J. Pharm. Sci. 2020, 142, 105160; Cunha, B.A.; Esrick, M.D.; LaRusso, M. Staphylococcus hominis native mitral valve bacterial endocarditis (SBE) in a patient with hypertrophic obstructive cardiomyopathy. Heart Lung 2007, 36, 380–382. [Google Scholar] [CrossRef]
- Szczuka, E.; Krzymińska, S.; Bogucka, N.; Kaznowski, A. Multifactorial mechanisms of the pathogenesis of methicillin-resistant Staphylococcus hominis isolated from bloodstream infections. Antonie Van Leeuwenhoek 2018, 111, 1259–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, N.H.; Baruah, F.K.; Grover, R.K. Staphylococcus hominis subsp. novobiosepticus, an emerging multidrug-resistant bacterium, as a causative agent of septicaemia in cancer patients. Indian J. Med. Res. 2017, 146, 420. [Google Scholar] [PubMed]
- Sunbul, M.; Demirag, M.K.; Yilmaz, O.; Yilmaz, H.; Ozturk, R.; Leblebicioglu, H. Pacemaker lead endocarditis caused by Staphylococcus hominis. Pacing Clin. Electrophysiol. 2006, 29, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, A.-M.; Sollid, J.U.E. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob. Agents Chemother. 2007, 51, 1671–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donati, C.; Rappuoli, R. Reverse vaccinology in the 21st century: Improvements over the original design. Ann. N. Y. Acad. Sci. 2013, 1285, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Masignani, V.; Pizza, M.; Moxon, E.R. The development of a vaccine against meningococcus B using reverse vaccinology. Front. Immunol. 2019, 10, 751. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.; Shahid, F.; Khan, A.; Bhatti, S.; Ahmad, S.; Naz, A.; Almatroudi, A.; Qamar, M.T.u. Pan-vaccinomics approach towards a universal vaccine candidate against WHO priority pathogens to address growing global antibiotic resistance. Comput. Biol. Med. 2021, 136, 104705. [Google Scholar] [CrossRef] [PubMed]
- Al-Megrin, W.A.I.; Karkashan, A.; Alnuqaydan, A.M.; Alkhayl, F.F.A.; Alrumaihi, F.; Almatroudi, A.; Allemailem, K.S. Design of a Multi-Epitopes Based Chimeric Vaccine against Enterobacter cloacae Using Pan-Genome and Reverse Vaccinology Approaches. Vaccines 2022, 10, 886. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.J.; Kan, C.N.E.; Luo, C.; Kazlauskas, R.J. Consensus Finder web tool to predict stabilizing substitutions in proteins. Methods Enzymol. 2020, 643, 129–148. [Google Scholar] [PubMed]
- Asad, Y.; Ahmad, S.; Rungrotmongkol, T.; Ranaghan, K.E.; Azam, S.S. Immuno-informatics driven proteome-wide investigation revealed novel peptide-based vaccine targets against emerging multiple drug resistant Providencia stuartii. J. Mol. Graph. Model. 2018, 80, 238–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, S.; Ahmad, S.; Azam, S.S. Protein localized at the inner and outer membrane, periplasmic spaces and secretory proteins were selected for the next phase. The protein with multiple and unknown localization were discarded. Eur. J. Pharm. Sci. 2020, 146. [Google Scholar] [CrossRef]
- Qamar, M.T.u.; Ahmad, S.; Fatima, I.; Ahmad, F.; Shahid, F.; Naz, A.; Abbasi, S.W.; Khan, A.; Mirza, M.U.; Ashfaq, U.A.; et al. Designing multi-epitope vaccine against Staphylococcus aureus by employing subtractive proteomics, reverse vaccinology and immuno-informatics approaches. Comput. Biol. Med. 2021, 132, 104389. [Google Scholar] [CrossRef] [PubMed]
- Bawa, C.; Dixit, P.; Preeti, P.; Rawal, K.; Huria, H. Evaluation of DnaJ Chaperone as a Potential Vaccine Candidate Using Computational Tools. 2022. Available online: https://osf.io/gh8ew (accessed on 31 August 2022).
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Membrane proteins. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Ullah, A.; Ahmad, S.; Ismail, S.; Afsheen, Z.; Khurram, M.; Qamar, M.u.; AlSuhaymi, N.; Alsugoor, M.H.; Allemailem, K.S. Towards a novel multi-epitopes chimeric vaccine for simulating strong immune responses and protection against morganella morganii. Int. J. Environ. Res. Public Health 2021, 18, 10961. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Kurgan, L. Sequence similarity searching. Curr. Protoc. Protein Sci. 2019, 95, e71. [Google Scholar] [CrossRef] [Green Version]
- Ras-Carmona, A.; Pelaez-Prestel, H.F.; Lafuente, E.M.; Reche, P.A. BCEPS: A web server to predict linear B cell epitopes with enhanced immunogenicity and cross-reactivity. Cells 2021, 10, 2744. [Google Scholar] [CrossRef] [PubMed]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune epitope database—Analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef] [PubMed]
- Bordbar, A.; Bagheri, K.P.; Ebrahimi, S.; Parvizi, P. Bioinformatics analyses of immunogenic T-cell epitopes of LeIF and PpSP15 proteins from Leishmania major and sand fly saliva used as model antigens for the design of a multi-epitope vaccine to control leishmaniasis. Infect. Genet. Evol. 2020, 80, 104189. [Google Scholar] [CrossRef] [PubMed]
- Soltan, M.A.; Eldeen, M.A.; Elbassiouny, N.; Mohamed, I.; El-Damasy, D.A.; Fayad, E.; Ali, O.A.A.; Raafat, N.; Eid, R.A.; Al-Karmalawy, A.A. Proteome based approach defines candidates for designing a multitope vaccine against the Nipah virus. Int. J. Mol. Sci. 2021, 22, 9330. [Google Scholar] [CrossRef] [PubMed]
- Naveed, M.; Yaseen, A.R.; Khalid, H.; Ali, U.; Rabaan, A.A.; Garout, M.; Halwani, M.A.; al Mutair, A.; Alhumaid, S.; al Alawi, Z.; et al. Execution and Design of an Anti HPIV-1 Vaccine with Multiple Epitopes Triggering Innate and Adaptive Immune Responses: An Immunoinformatic Approach. Vaccines 2022, 10, 869. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kumar, P.; Saumya, K.U.; Kapuganti, S.K.; Bhardwaj, T.; Giri, R. Exploring the SARS-CoV-2 structural proteins for multi-epitope vaccine development: An in-silico approach. Expert Rev. Vaccines 2020, 19, 887–898. [Google Scholar] [CrossRef]
- Sharma, N.; Naorem, L.D.; Jain, S.; Raghava, G.P.S. ToxinPred2: An improved method for predicting toxicity of proteins. Brief. Bioinform. 2022, 23, bbac174. [Google Scholar] [CrossRef]
- Lim, H.X.; Lim, J.; Jazayeri, S.D.; Poppema, S.; Poh, C.L. Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomed. J. 2021, 44, 18–30. [Google Scholar] [CrossRef]
- Chauhan, V.; Rungta, T.; Goyal, K.; Singh, M.P. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci. Rep. 2019, 9, 2517. [Google Scholar] [CrossRef] [Green Version]
- Das, N.C.; Patra, R.; Gupta, P.S.S.; Ghosh, P.; Bhattacharya, M.; Rana, M.K.; Mukherjee, S. Designing of a novel multi-epitope peptide based vaccine against Brugia malayi: An in silico approach. Infect. Genet. Evol. 2021, 87, 104633. [Google Scholar] [CrossRef]
- Shehzad, A.; Sumartono, C.; Nugraha, J.; Susilowati, H.; Wijaya, A.Y.; Ishfaq, H.; Ahmad, M.K.; Tyasningsih, W.; Rantam, F.A. Development of a multi-epitope spike glycoprotein vaccine to combat SARS-CoV-2 using the bioinformatics approach. J. Pharm. Pharmacogn. Res. 2022, 10, 445–458. [Google Scholar] [CrossRef]
- Ghosh, P.; Bhakta, S.; Bhattacharya, M.; Sharma, A.R.; Sharma, G.; Lee, S.-S.; Chakraborty, C. A novel multi-epitopic peptide vaccine candidate against Helicobacter pylori: In-silico identification, design, cloning and validation through molecular dynamics. Int. J. Pept. Res. Ther. 2021, 27, 1149–1166. [Google Scholar] [CrossRef]
- Mahmud, S.; Rafi, M.; Paul, G.K.; Promi, M.M.; Shimu, M.; Sultana, S.; Biswas, S.; Emran, T.B.; Dhama, K.; Alyami, S.A.; et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci. Rep. 2021, 11, 15431. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, M.; Alshammari, A.; Alasmari, A.F.; Alharbi, S.M.; Qamar, M.u.; Ullah, A.; Ahmad, S.; Irfan, M.; Khalil, A.A.K. Designing of a Recombinant Multi-Epitopes Based Vaccine against Enterococcus mundtii Using Bioinformatics and Immunoinformatics Approaches. Int. J. Environ. Res. Public Health 2022, 19, 3729. [Google Scholar] [CrossRef] [PubMed]
- Guest, J.D.; Vreven, T.; Zhou, J.; Moal, I.; Jeliazkov, J.R.; Gray, J.J.; Weng, Z.; Pierce, B.G. An expanded benchmark for antibody-antigen docking and affinity prediction reveals insights into antibody recognition determinants. Structure 2021, 29, 606–621. [Google Scholar] [CrossRef] [PubMed]
- Kar, T.; Narsaria, U.; Basak, S.; Deb, D.; Castiglione, F.; Mueller, D.M.; Srivastava, A.P. A candidate multi-epitope vaccine against SARS-CoV-2. Sci. Rep. 2020, 10, 10895. [Google Scholar] [CrossRef] [PubMed]
- Jalal, K.; Abu-Izneid, T.; Khan, K.; Abbas, M.; Hayat, A.; Bawazeer, S.; Uddin, R. Identification of vaccine and drug targets in Shigella dysenteriae sd197 using reverse vaccinology approach. Sci. Rep. 2022, 12, 251. [Google Scholar] [CrossRef] [PubMed]
- Srividya, D.; Mohan, A.H.S.; Rao, S.N. Expression and purification of codon-optimized cre recombinase in E. coli. Protein Expr. Purif. 2020, 167, 105546. [Google Scholar]
- Deng, H.; Yu, S.; Guo, Y.; Gu, L.; Wang, G.; Ren, Z.; Li, Y.; Li, K.; Li, R. Development of a multivalent enterovirus subunit vaccine based on immunoinformatic design principles for the prevention of HFMD. Vaccine 2020, 38, 3671–3681. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; et al. Amber 2021; University of California: San Francisco, CA, USA, 2021; Available online: https://ambermd.org/doc12/Amber21.pdf (accessed on 15 June 2022).
- Yelk, J.D. Molecular Dynamics Investigations of Duplex Columnar Liquid Crystal Phases of Nucleoside Triphosphates; University of Colorado at Boulder: Boulder, CO, USA, 2019; Available online: https://www.proquest.com/openview/3e239cbed4452aec18ef34dc4d491ffc/1?pq-origsite=gscholar&cbl=18750&diss=y (accessed on 15 June 2022).
- Hengphasatporn, K.; Kungwan, N.; Rungrotmongkol, T. Binding pattern and susceptibility of epigallocatechin gallate against envelope protein homodimer of Zika virus: A molecular dynamics study. J. Mol. Liq. 2019, 274, 140–147. [Google Scholar] [CrossRef]
- Rai, N.; Tiwari, S.P.; Maginn, E.J. Force field development for actinyl ions via quantum mechanical calculations: An approach to account for many body solvation effects. J. Phys. Chem. B 2012, 116, 10885–10897. [Google Scholar] [CrossRef]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef] [Green Version]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed]
- Freschi, L.; Vincent, A.T.; Jeukens, J.; Emond-Rheault, J.-G.; Kukavica-Ibrulj, I.; Dupont, M.-J.; Charette, S.J.; Boyle, B.; Levesque, R.C. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol. Evol. 2019, 11, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonkheer, E.M.; Brankovics, B.; Houwers, I.M.; van der Wolf, J.M.; Bonants, P.J.M.; Vreeburg, R.A.M.; Bollema, R.; de Haan, J.R.; Berke, L.; Smit, S.; et al. The Pectobacterium pangenome, with a focus on Pectobacterium brasiliense, shows a robust core and extensive exchange of genes from a shared gene pool. BMC Genom. 2021, 22, 265. [Google Scholar] [CrossRef] [PubMed]
- Van, T.T.H.; Lacey, J.A.; Vezina, B.; Phung, C.; Anwar, A.; Scott, P.C.; Moore, R.J. Survival mechanisms of Campylobacter hepaticus identified by genomic analysis and comparative transcriptomic analysis of in vivo and in vitro derived bacteria. Front. Microbiol. 2019, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, C.G.P.; Fitzpatrick, D.A. Pan-genome analyses of model fungal species. Microb. Genom. 2019, 5, e000243. [Google Scholar] [CrossRef]
- Gul, S.; Ahmad, S.; Ullah, A.; Ismail, S.; Khurram, M.; Qamar, M.u.; Hakami, A.R.; Alkhathami, A.G.; Alrumaihi, F.; Allemailem, K.S. Designing a recombinant vaccine against Providencia rettgeri using immunoinformatics approach. Vaccines 2022, 10, 189. [Google Scholar] [CrossRef]
- Carruthers, M.; Yurchenko, A.A.; Augley, J.J.; Adams, C.E.; Herzyk, P.; Elmer, K.R. De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model salmonid fish species. BMC Genom. 2018, 19, 32. [Google Scholar]
- Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Combating tigecycline resistant Acinetobacter baumannii: A leap forward towards multi-epitope based vaccine discovery. Eur. J. Pharm. Sci. 2019, 132, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pachathundikandi, S.K.; Tegtmeyer, N.; Backert, S. Signal transduction of Helicobacter pylori during interaction with host cell protein receptors of epithelial and immune cells. Gut Microbes 2013, 4, 454–474. [Google Scholar] [CrossRef] [Green Version]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalsass, M.; Brozzi, A.; Medini, D.; Rappuoli, R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 2019, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Heng, J.; Dai, S.; Shaw, N.; Zhou, B.; Huang, B.; He, Z.; Wang, Y.; Jiang, T.; Li, X.; et al. An efficient strategy for high throughput screening of recombinant integral membrane protein expression and stability. Protein Expr. Purif. 2011, 78, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Steed, H.; Macfarlane, S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 2008, 104, 305–344. [Google Scholar] [CrossRef]
- Maiuolo, J.; Musolino, V.; Gliozzi, M.; Carresi, C.; Scarano, F.; Nucera, S.; Scicchitano, M.; Oppedisano, F.; Bosco, F.; Macri, R.; et al. Involvement of the Intestinal Microbiota in the Appearance of Multiple Sclerosis: Aloe vera and Citrus bergamia as Potential Candidates for Intestinal Health. Nutrients 2022, 14, 2711. [Google Scholar] [CrossRef] [PubMed]
- Rivera, A.; Siracusa, M.C.; Yap, G.S.; Gause, W.C. Innate cell communication kick-starts pathogen-specific immunity. Nat. Immunol. 2016, 17, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.R.; Garg, S.; Brenner, M.B. Antigen presentation by CD1: Lipids, T cells, and NKT cells in microbial immunity. Adv. Immunol. 2009, 102, 1–94. [Google Scholar] [PubMed]
- Sinha, J.K.; Bhattacharya, S. A Text Book of Immunology; Academic publishers: Cambridge, MA, USA, 2006. [Google Scholar]
- Kapingidza, A.B.; Kowal, K.; Chruszcz, M. Antigen–Antibody complexes. In Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and Other Body Fluid Proteins; Springer: Cham, Switzerland, 2020; pp. 465–497. [Google Scholar]
- Purcell, A.W.; McCluskey, J.; Rossjohn, J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007, 6, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Khan, S.; Saleem, S.; Nizam-Uddin, N.; Mohammad, A.; Khan, T.; Ahmad, S.; Arshad, M.; Ali, S.S.; Suleman, M.; et al. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med. 2021, 133, 104420. [Google Scholar] [CrossRef]
- Aguttu, C.; Okech, B.A.; Mukisa, A.; Lubega, G.W. Screening and characterization of hypothetical proteins of Plasmodium falciparum as novel vaccine candidates in the fight against malaria using reverse vaccinology. J. Genet. Eng. Biotechnol. 2021, 19, 103. [Google Scholar] [CrossRef]
- Binz, H.; Wigzell, H. Shared idiotypic determinants on B and T lymphocytes reactive against the same antigenic determinants. I. Demonstration of similar or identical idiotypes on IgG molecules and T-cell receptors with specificity for the same alloantigens. J. Exp. Med. 1975, 142, 197–211. [Google Scholar] [CrossRef] [Green Version]
- Zaman, M.; Toth, I. Immunostimulation by synthetic lipopeptide-based vaccine candidates: Structure-activity relationships. Front. Immunol. 2013, 4, 318. [Google Scholar] [CrossRef] [PubMed]
- Qamar, M.u.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.; Fatima, I.; Shahid, F.; Chen, L.-L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar]
- Aldakheel, F.M.; Abrar, A.; Munir, S.; Aslam, S.; Allemailem, K.S.; Khurshid, M.; Ashfaq, U.A. Proteome-wide mapping and reverse vaccinology approaches to design a multi-epitope vaccine against clostridium perfringens. Vaccines 2021, 9, 1079. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule ofDrosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Bošnački, D.; Eikelder, H.M.M.t.; Steijaert, M.N.; de Vink, E.P. Stochastic analysis of amino acid substitution in protein synthesis. In International Conference on Computational Methods in Systems Biology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 367–386. [Google Scholar]
- Qamar, M.u.; Shahid, F.; Aslam, S.; Ashfaq, U.A.; Aslam, S.; Fatima, I.; Fareed, M.M.; Zohaib, A.; Chen, L.-L. Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2. Infect. Dis. Poverty 2020, 9, 1–14. [Google Scholar]
- Dombkowski, A.A.; Sultana, K.Z.; Craig, D.B. Protein disulfide engineering. FEBS Lett. 2014, 588, 206–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gongora-Benitez, M.; Tulla-Puche, J.; Albericio, F. Multifaceted roles of disulfide bonds. Peptides as therapeutics. Chem. Rev. 2014, 114, 901–926. [Google Scholar] [CrossRef]
- Safavi, A.; Kefayat, A.; Mahdevar, E.; Abiri, A.; Ghahremani, F. Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches. Vaccine 2020, 38, 7612–7628. [Google Scholar] [CrossRef] [PubMed]
- al Saba, A.; Adiba, M.; Saha, P.; Hosen, M.I.; Chakraborty, S.; Nabi, A.H.M.N. An in-depth in silico and immunoinformatics approach for designing a potential multi-epitope construct for the effective development of vaccine to combat against SARS-CoV-2 encompassing variants of concern and interest. Comput. Biol. Med. 2021, 136, 104703. [Google Scholar] [CrossRef] [PubMed]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef] [PubMed]
- Andrusier, N.; Mashiach, E.; Nussinov, R.; Wolfson, H.J. Principles of flexible protein–Protein docking. Proteins Struct. Funct. Bioinform. 2008, 73, 271–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, T.; Amato, I.Y. Basics of Molecular Simulation and its Application to Biomolecules. In Quantum Bio-Informatics II From Quantum Information to Bio-Informatics; World Scientific Publishing: Singapore, 2009; pp. 228–241. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [PubMed]
- Zhang, X.; Perez-Sanchez, H.; Lightstone, F.C. A comprehensive docking and MM/GBSA rescoring study of ligand recognition upon binding antithrombin. Curr. Top. Med. Chem. 2017, 17, 1631–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinnasamy, S.; Selvaraj, G.; Selvaraj, C.; Kaushik, A.C.; Kaliamurthi, S.; Khan, A.; Singh, S.K.; Wei, D.-Q. Combining in silico and in vitro approaches to identification of potent inhibitor against phospholipase A2 (PLA2). Int. J. Biol. Macromol. 2020, 144, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Gautham, M.; Spicer, N.; Chatterjee, S.; Goodman, C. What are the challenges for antibiotic stewardship at the community level? An analysis of the drivers of antibiotic provision by informal healthcare providers in rural India. Soc. Sci. Med. 2021, 275, 113813. [Google Scholar] [CrossRef] [PubMed]
- Jasovský, D.; Littmann, J.; Zorzet, A.; Cars, O. Antimicrobial resistance—A threat to the world’s sustainable development. Ups. J. Med. Sci. 2016, 121, 159–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micoli, F.; Bagnoli, F.; Rappuoli, R.; Serruto, D. The role of vaccines in combatting antimicrobial resistance. Nat. Rev. Microbiol. 2021, 19, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Gupta, R.K.; Deshpande, M.C.; Schwendeman, S.P. Biodegradable poly (lactic-co-glycolic acid) microparticles for injectable delivery of vaccine antigens. Adv. Drug Deliv. Rev. 2005, 57, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Shahid, F.; Zaheer, T.; Ashraf, S.T.; Shehroz, M.; Anwer, F.; Naz, A.; Ali, A. Chimeric vaccine designs against Acinetobacter baumannii using pan genome and reverse vaccinology approaches. Sci. Rep. 2021, 11, 13213. [Google Scholar] [CrossRef] [PubMed]
- Allemailem, K.S. A Comprehensive Computer Aided Vaccine Design Approach to Propose a Multi-Epitopes Subunit Vaccine against Genus Klebsiella Using Pan-Genomics, Reverse Vaccinology, and Biophysical Techniques. Vaccines 2021, 9, 1087. [Google Scholar] [CrossRef] [PubMed]
- Satyam, R.; Bhardwaj, T.; Jha, N.K.; Jha, S.K.; Nand, P. Toward a chimeric vaccine against multiple isolates of Mycobacteroides-An integrative approach. Life Sci. 2020, 250, 117541. [Google Scholar] [CrossRef] [PubMed]
- Dar, H.A.; Ismail, S.; Waheed, Y.; Ahmad, S.; Jamil, Z.; Aziz, H.; Hetta, H.F.; Muhammad, K. Designing a multi-epitope vaccine against Mycobacteroides abscessus by pangenome-reverse vaccinology. Sci. Rep. 2021, 11, 11197. [Google Scholar] [CrossRef] [PubMed]


| Organism Name | Strain | Size | GC% |
|---|---|---|---|
| S. hominis | FDAARGOS_575 | 2.25743 | 31.6637 |
| FDAARGOS_746 | 2.37219 | 31.6522 | |
| 19A | 2.28122 | 31.6534 | |
| S34-1 | 2.25454 | 31.4135 | |
| FDAARGOS_747 | 2.24212 | 31.5385 | |
| FDAARGOS_762 | 2.25551 | 31.529 | |
| K1 | 2.25341 | 31.4 |
| Fasta Files | Core Genes | ACC Genes | Unique Genes | Exclusively Absent Genes |
|---|---|---|---|---|
| 19A protein.faa | 1755 | 211 | 101 | 13 |
| FDAARGOS_747.protein.faa | 1755 | 250 | 51 | 5 |
| FDAARGOS_762.protein.faa | 1755 | 232 | 49 | 9 |
| FDAARGOS_protein.faa | 1755 | 290 | 160 | 8 |
| FDAARGOS_proteins.faa | 1755 | 281 | 53 | 7 |
| K1.proteins.faa | 1755 | 237 | 74 | 58 |
| S34-1.proteins. faa | 1755 | 235 | 58 | 16 |
| Proteins | VFDB | |
|---|---|---|
| Extracellular | Bit Score | Sequence Identity |
| >core/1166/1/Org1_Gene1302 | 211 | 49% |
| >core/1180/1/Org1_Gene1448 | 169 | 45% |
| >core/1802/1/Org1_Gene1985 | 287 | 70% |
| >core/99/2/Org2_Gene1570 | 132 | 44% |
| >core/16/4/Org4_Gene1407 | 1569 | 58% |
| Extracellular Proteins | Transmembrane Helices | No. of Amino Acids | Molecular Weight | Theoretical PI | Instability Index | Aliphatic Index |
|---|---|---|---|---|---|---|
| >core/1166/1/Org1_Gene1302 | 1 | 282 | 32.28 | 8.37 | 17.03 | 76.49 |
| >core/1180/1/Org1_Gene1448 | 0 | 321 | 56.32 | 10.51 | 42.52 | 85.39 |
| >core/1679/1/Org1_Gene1002 | 0 | 200 | 22.73 | 4.99 | 30.44 | 81.45 |
| >core/1802/1/Org1_Gene1985 | 1 | 181 | 20.81 | 9 | 20.19 | 74.75 |
| >core/99/2/Org2_Gene1570 | 0 | 385 | 58.60 | 8.65 | 45.67 | 84.52 |
| >core/16/4/Org4_Gene1407 | 0 | 402 | 63.05 | 9.32 | 44.64 | 85.39 |
| B Cell Epitopes | B Cells Peptides |
|---|---|
| >core/1166/1/Org1_Gene1302 N-acetylglucosaminidase | QIFFKKVNEVEKVQHVNVTLDKAAAKQIDNYTSQQVSNKNNNAW RDASASEIKGAMDSSKFIDDDKQKYQFLDLSKY QGIDKNRIKRMLFDRPTLLKHTD |
| KSELANGVNIDGKK | |
| EDPIKTGAEYAKKHGWDT | |
| SHDDQNTLYSMRWNPMNPGEH | |
| KTEGKYFKLYVYKDDQ | |
| >core/1802/1/Org1_Gene1985 Thermonuclease family protein | HTGPFKDDSQHSSSNSTQIELKGK |
| TVKPNTPVQPY | |
| LAREKYFSPNGKYRST |
| T Cell Epitopes (MHC-I and MHC-II) | |||
|---|---|---|---|
| MHC-II | Percentile Score | MHC-I | Percentile Score |
| KNRIKRMLFDRPTLL | 0.21 | MLFDRPTLL | 0.01 |
| KNRIKRMLF | 0.94 | ||
| QKYQFLDLSKYQGID | 6.1 | YQFLDLSKY | 0.01 |
| DLSKYQGID | 71 | ||
| QKYQFLDLSK | 4.5 | ||
| IKGAMDSSKFIDDDK | 6.2 | KGAMDSSKF | 1.1 |
| SSKFIDDDK | 1.8 | ||
| GAMDSSKFI | 1.5 | ||
| NAWRDASASEIKGAM | 4.6 | SASEIKGAM | 0.23 |
| NAWRDASAS | 4.9 | ||
| QIDNYTSQQVSNKNN | 22 | YTSQQVSNK | 0.07 |
| SQQVSNKNN | 17 | ||
| QIDNYTSQQ | 2.8 | ||
| VQHVNVTLDKAAAKQ | 3.2 | VTLDKAAAK | 0.02 |
| VQHVNVTLDK | 1.8 | ||
| VQHVNVTLDK | 2.2 | ||
| QIFFKKVNEVEKVQH | 7.5 | IFFKKVNEV | 0.2 |
| KVNEVEKVQH | 1.1 | ||
| QIFFKKVNEV | 0.83 | ||
| KSELANGVNID | 7.2 | SELANGVNI | 0.14 |
| SELANGVNID | 4.9 | ||
| KSELANGVNI | 1.1 | ||
| SELANGVNIDGKK | 28 | SELANGVNI | 0.14 |
| NGVNIDGKK | 3.2 | ||
| DPIKTGAEYAKKH | 15 | DPIKTGAEY | 0.02 |
| KTGAEYAKKH | 3.3 | ||
| TGAEYAKKHGWDT | 31 | AEYAKKHGW | 0.01 |
| YAKKHGWDT | 2.1 | ||
| TGAEYAKKH | 4.4 | ||
| NTLYSMRWNPMNPGE | 3.3 | SMRWNPMNP | 1.6 |
| RWNPMNPGE | 3.2 | ||
| NTLYSMRWNP | 11 | ||
| SHDDQNTLYSMRWNP | 36 | DQNTLYSMR | 0.15 |
| TLYSMRWNP | 3.5 | ||
| SHDDQNTLY | 0.23 | ||
| KTEGKYFKLYVYKDD | 3.5 | TEGKYFKLY | 0.01 |
| YFKLYVYKDD | 48 | ||
| TEGKYFKLYVYKDDQ | 4.3 | TEGKYFKLY | 0.01 |
| KLYVYKDDQ | 15 | ||
| DDSQHSSSNSTQIEL | 14 | HSSSNSTQI | 0.63 |
| SSNSTQIEL | 0.76 | ||
| DDSQHSSSNS | 32 | ||
| QHSSSNSTQIELKGK | 18 | SSNSTQIELK | 0.06 |
| STQIELKGK | 0.18 | ||
| QHSSSNSTQI | 4.5 | ||
| HTGPFKDDSQHSSSN | 19 | GPFKDDSQH | 0.84 |
| DDSQHSSSN | 23 | ||
| HTGPFKDDSQ | 13 | ||
| TVKPNTPVQPY | 6 | KPNTPVQPY | 0.05 |
| TVKPNTPVQ | 0.74 | ||
| LAREKYFSPNGKYRS | 4.4 | KYFSPNGKY | 0.01 |
| YFSPNGKYRS | 3.8 | ||
| LAREKYFSP | 1.1 | ||
| AREKYFSPNGKYRST | 4.5 | KYFSPNGKY | 0.01 |
| SPNGKYRST | 0.2 | ||
| AREKYFSPNG | 13 | ||
| Pair of Residues | Chi3 | Energy |
|---|---|---|
| Leu4-Glu104 | 116.71 | 3.66 |
| Phe10-Asn25 | −88.2 | 5.92 |
| Leu13-Asp28 | 109.19 | 3.15 |
| Leu13-Asn35 | 80.16 | 3.35 |
| Thr27-Tyr33 | 92.9 | 4.18 |
| Thr36-Leu41 | 95.75 | 3.06 |
| Ile38-Leu41 | 114.86 | 3.82 |
| Thr49-Ala53 | −99.93 | 4.39 |
| Glu50-His78 | −99.21 | 2.56 |
| Met58-Ala67 | −88.37 | 3.6 |
| Pro74-Gln77 | −103.37 | 2.03 |
| Ala96-Lys102 | −80.09 | 2.7 |
| Ala96-Ala119 | −106.52 | 1.15 |
| Leu106-Trp109 | 93.83 | 3.1 |
| Trp109-Lys112 | 79.05 | 0.99 |
| Pro140-Thr149 | −84.69 | 5.82 |
| Gly143-Asp146 | −78.71 | 2.98 |
| Asn147-Ser150 | 78.25 | 2.72 |
| Tyr148-Pro169 | −99.43 | 5.6 |
| Ser150-Gly153 | 99.22 | 0.78 |
| Pro154-Gly168 | −91.74 | 0.15 |
| Gly157-Glu160 | 80.42 | 4.03 |
| Ala162-Val165 | −62.14 | 6.82 |
| Gly170-Lys173 | −106.59 | 3.22 |
| Gly175-Ala179 | 120.42 | 9.13 |
| Tyr192-Gly199 | 90.13 | 5.39 |
| Phe221-Tyr224 | 101.86 | 2.6 |
| Lys222-Gln233 | −85.19 | 2.76 |
| Gly225-Ser245 | 67.24 | 5.62 |
| Gly227-Asp230 | −95.1 | 2.68 |
| Asp231-Pro243 | −77.12 | 3.04 |
| Solution No. | Score | Area | Atomic Contact Energy | Transformation |
|---|---|---|---|---|
| 1 | 19,690 | 2856.90 | 243.71 | −0.03 0.30 −0.09 52.34 47.37 58.64 |
| 2 | 19,200 | 3027.60 | 270.89 | 0.75 0.42 0.94 57.44 55.30 16.13 |
| 3 | 18,192 | 3065.80 | 157.31 | −3.09 −0.32 −0.24 62.27 28.26 48.61 |
| 4 | 17,630 | 3437.00 | 489.66 | 0.64 −0.12 −1.90 30.94 44.61 52.59 |
| 5 | 17,448 | 2402.00 | 332.04 | −2.53 0.48 2.20 47.52 38.06 57.76 |
| 6 | 17,354 | 2529.80 | 356.97 | −2.47 0.59 1.94 48.65 35.91 57.87 |
| 7 | 17,298 | 2026.20 | 211.10 | −2.53 0.44 2.46 46.83 39.76 57.70 |
| 8 | 16,830 | 2582.40 | 421.25 | −1.32 1.24 −0.69 16.14 44.00 37.89 |
| 9 | 16,788 | 2898.30 | 123.86 | 2.57 −0.79 −1.91 7.67 22.75 27.87 |
| 10 | 16,722 | 2468.90 | 80.82 | 0.36 −0.87 1.68 50.92 37.24 65.79 |
| 11 | 15,864 | 2500.60 | 400.30 | 2.90 1.04 −1.53 39.11 −3.56 38.58 |
| 12 | 15,804 | 2384.30 | 235.58 | −0.21 0.41 −0.16 48.41 47.33 59.09 |
| 13 | 15,752 | 2633.60 | 211.87 | 1.34 −0.24 −3.02 25.94 15.16 18.01 |
| 14 | 15,648 | 2207.40 | 349.69 | 2.57 −0.63 −2.05 5.24 25.83 31.22 |
| 15 | 15,482 | 2210.30 | 339.58 | 0.90 0.63 2.53 10.72 23.61 32.81 |
| 16 | 15,454 | 2190.40 | 430.68 | 1.85 −0.76 −2.43 47.55 26.12 8.92 |
| 17 | 15,452 | 2184.50 | 448.16 | −0.33 −0.92 0.49 51.82 12.75 19.05 |
| 18 | 15,436 | 2974.60 | 139.08 | 0.22 −0.06 −2.19 57.28 10.49 57.29 |
| 19 | 15,404 | 3022.70 | 446.40 | −0.15 −0.29 0.67 13.83 38.25 47.47 |
| 20 | 15,390 | 3328.10 | −35.46 | 2.70 −0.46 −1.70 46.00 55.26 63.86 |
| Solution No. | Score | Area | Atomic Contact Energy | Transformation |
|---|---|---|---|---|
| 1 | 19,030 | 2568.20 | 178.30 | −2.52 0.46 0.61 103.23 100.92 −12.98 |
| 2 | 19,012 | 2616.00 | 174.36 | 3.12 −0.94 −3.01 118.54 31.94 −4.33 |
| 3 | 18,762 | 3157.30 | 339.60 | −0.95 −1.48 −0.11 102.18 52.19 −15.79 |
| 4 | 17,928 | 2860.90 | 498.89 | 2.13 −0.68 2.42 115.91 101.86 1.25 |
| 5 | 17,924 | 3188.70 | −118.16 | −2.22 −0.86 −0.00 121.14 24.49 10.29 |
| 6 | 17,584 | 2759.50 | −36.31 | 2.22 0.44 2.53 142.80 64.79 15.68 |
| 7 | 17,258 | 2463.10 | 365.25 | 2.36 1.27 −2.48 85.84 67.41 3.01 |
| 8 | 16,858 | 2782.20 | 189.39 | −1.26 −0.73 −0.94 146.96 58.95 −5.46 |
| 9 | 16,806 | 2721.60 | 333.73 | 2.88 −0.76 −3.13 118.27 32.94 −6.90 |
| 10 | 16,788 | 2957.30 | −44.01 | −3.09 −1.10 −3.03 117.35 34.09 −6.06 |
| 11 | 16,616 | 2588.80 | 367.97 | −2.23 0.84 0.94 72.25 63.69 10.94 |
| 12 | 16,606 | 2181.70 | 29.05 | 0.23 0.32 −2.44 94.64 88.42 −23.96 |
| 13 | 16,602 | 2646.70 | 223.64 | 0.41 0.79 0.94 122.16 92.19 −6.03 |
| 14 | 16,598 | 2646.80 | 289.18 | −1.62 0.07 0.15 72.26 77.33 −6.13 |
| 15 | 16,542 | 2996.90 | 199.69 | −0.30 −0.95 −2.67 82.55 81.78 14.62 |
| 16 | 16,534 | 2063.40 | 3.75 | 3.06 0.75 1.28 139.48 75.91 −15.37 |
| 17 | 16,532 | 2808.00 | 17.66 | −1.36 −0.55 −0.95 149.27 54.23 −4.98 |
| 18 | 16,338 | 3494.90 | 200.24 | 2.42 −1.51 −3.10 99.91 52.71 −12.04 |
| 19 | 16,330 | 2461.90 | 116.63 | 0.88 0.49 2.74 89.56 69.78 −12.43 |
| 20 | 16,262 | 2662.00 | 81.94 | 1.13 0.51 2.68 89.25 66.06 −9.25 |
| Solution No. | Score | Area | Atomic Contact Energy | Transformation |
|---|---|---|---|---|
| 1 | 20,864 | 2763.80 | 319.39 | −2.98 0.85 1.45 −48.02 20.31 −23.51 |
| 2 | 19,780 | 2391.10 | 482.45 | −2.50 −0.35 2.08 −16.75 42.27 −16.53 |
| 3 | 19,694 | 3221.60 | 338.75 | 0.84 −1.45 2.63 −0.69 18.65 −54.86 |
| 4 | 19,676 | 2551.00 | 315.79 | −2.61 0.04 −1.92 9.24 32.35 −58.78 |
| 5 | 19,228 | 2566.00 | 228.48 | −0.75 −0.01 −2.79 −33.15 41.12 −15.34 |
| 6 | 18,084 | 3343.40 | 246.14 | 0.32 −1.46 2.08 1.33 18.82 −51.37 |
| 7 | 18,058 | 4052.50 | 372.52 | 3.13 0.68 1.53 −43.62 21.45 −22.30 |
| 8 | 17,838 | 2669.80 | 222.59 | −2.80 0.05 −1.83 −31.47 42.41 −10.22 |
| 9 | 17,764 | 2476.80 | 177.68 | 3.11 0.80 1.69 −50.10 18.45 −24.37 |
| 10 | 17,748 | 3277.60 | −178.96 | −1.03 −0.37 −1.37 17.61 33.74 −54.26 |
| 11 | 17,652 | 2927.20 | 7.50 | 0.63 0.97 0.47 −55.98 24.76 −26.27 |
| 12 | 17,566 | 3376.60 | 435.51 | −0.34 0.53 −1.72 −58.27 11.71 −52.97 |
| 13 | 17,424 | 2759.50 | 95.73 | −0.28 −0.02 0.43 −15.69 49.62 −18.02 |
| 14 | 17,340 | 3144.90 | 319.63 | −2.21 1.12 −2.60 −48.43 35.44 −21.42 |
| 15 | 17,340 | 2747.30 | 93.26 | 2.49 0.83 −2.57 −57.86 24.72 −0.77 |
| 16 | 17,322 | 2663.00 | 183.48 | −2.28 0.63 −0.33 −68.26 −18.01 4.10 |
| 17 | 17,226 | 2159.20 | 218.29 | 0.42 −0.86 1.59 35.96 −6.27 −64.19 |
| 18 | 17,218 | 2911.20 | 375.82 | −1.15 −0.50 −0.61 16.58 15.77 −70.72 |
| 19 | 17,040 | 2249.90 | 421.54 | 0.38 −0.73 −1.75 −44.40 47.72 −11.97 |
| 20 | 17,006 | 2235.50 | 218.89 | −2.60 0.40 1.70 −72.01 −1.85 5.10 |
| Solution Number | Global Energy | Attractive van der Waals | Repulsive van der Waals | Atomic Contact Energy | Hydrogen Bonds Energy |
|---|---|---|---|---|---|
| 8 | −4.84 | −3.77 | 0.34 | −1.23 | −0.99 |
| 9 | 6.04 | −0.55 | 0.00 | 0.99 | 0.00 |
| 5 | 9.41 | −3.75 | 0.00 | 2.70 | −0.32 |
| 4 | 9.55 | −3.40 | 1.80 | 3.71 | −0.46 |
| 2 | 10.22 | −0.00 | 0.00 | −0.00 | 0.00 |
| 7 | 12.30 | −10.85 | 3.92 | 1.55 | 0.00 |
| 6 | 18.35 | −3.40 | 1.18 | 2.00 | 0.00 |
| 10 | 24.67 | −6.10 | 0.97 | 1.15 | 0.00 |
| 3 | 28.63 | −36.11 | 16.03 | 10.96 | −2.12 |
| 1 | 31.21 | −14.77 | 29.21 | 5.98 | −3.36 |
| Solution Number | Global Energy | Attractive van der Waals | Repulsive van der Waals | Atomic Contact Energy | Hydrogen Bonds Energy |
|---|---|---|---|---|---|
| 9 | 2.55 | −4.82 | 0.80 | 1.65 | 0.00 |
| 5 | 4.36 | −1.93 | 1.65 | −0.43 | 0.00 |
| 10 | 10.58 | −3.28 | 1.32 | 3.89 | −0.34 |
| 8 | 15.30 | −32.77 | 30.61 | 13.93 | −2.23 |
| 2 | 16.03 | −7.26 | 2.03 | 5.34 | −0.42 |
| 6 | 28.39 | −9.07 | 24.19 | 6.31 | −0.46 |
| 3 | 55.64 | −8.79 | 10.16 | 13.06 | 0.00 |
| 4 | 444.29 | −32.75 | 578.96 | 18.68 | −4.13 |
| 1 | 922.72 | −28.54 | 1153.02 | 19.35 | −4.68 |
| 7 | 1040.22 | −28.82 | 1310.23 | 15.62 | −5.01 |
| Solution Number | Global Energy | Attractive van der Waals | Repulsive van der Waals | Atomic Contact Energy | Hydrogen Bonds Energy |
|---|---|---|---|---|---|
| 8 | −36.67 | −32.83 | 33.92 | 2.84 | −6.88 |
| 1 | −3.76 | −25.41 | 6.33 | 10.26 | −2.29 |
| 4 | 2.59 | −1.90 | 0.30 | 0.62 | 0.00 |
| 2 | 20.09 | −49.11 | 52.92 | 16.08 | −7.74 |
| 5 | 364.56 | −27.61 | 498.29 | 1.45 | −2.59 |
| 9 | 511.26 | −63.30 | 728.16 | 0.06 | −3.96 |
| 3 | 776.89 | −38.99 | 1019.72 | 14.40 | −4.25 |
| 6 | 887.29 | −40.59 | 1162.81 | 10.78 | −2.95 |
| 7 | 1292.51 | −36.49 | 1632.36 | 20.70 | −1.52 |
| 10 | 7376.28 | −56.57 | 9336.35 | −11.09 | −6.70 |
| Energy Parameter | TLR-4–Vaccine Complex | MHC-I–Vaccine Complex | MHC-II–Vaccine Complex |
|---|---|---|---|
| MM/GBSA | |||
| VDWAALS | −56.68 | −51.38 | −66.62 |
| EEL | −33.65 | −32.55 | −25.87 |
| Delta G gas | −90.33 | −83.93 | −92.49 |
| Delta G solv | 15.86 | 21.68 | 20.54 |
| Delta Total | −74.47 | −62.25 | −71.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawaz, M.; Ullah, A.; Al-Harbi, A.I.; Haq, M.U.; Hameed, A.R.; Ahmad, S.; Aziz, A.; Raziq, K.; Khan, S.; Irfan, M.; et al. Genome-Based Multi-Antigenic Epitopes Vaccine Construct Designing against Staphylococcus hominis Using Reverse Vaccinology and Biophysical Approaches. Vaccines 2022, 10, 1729. https://doi.org/10.3390/vaccines10101729
Nawaz M, Ullah A, Al-Harbi AI, Haq MU, Hameed AR, Ahmad S, Aziz A, Raziq K, Khan S, Irfan M, et al. Genome-Based Multi-Antigenic Epitopes Vaccine Construct Designing against Staphylococcus hominis Using Reverse Vaccinology and Biophysical Approaches. Vaccines. 2022; 10(10):1729. https://doi.org/10.3390/vaccines10101729
Chicago/Turabian StyleNawaz, Mahreen, Asad Ullah, Alhanouf I. Al-Harbi, Mahboob Ul Haq, Alaa R. Hameed, Sajjad Ahmad, Aamir Aziz, Khadija Raziq, Saifullah Khan, Muhammad Irfan, and et al. 2022. "Genome-Based Multi-Antigenic Epitopes Vaccine Construct Designing against Staphylococcus hominis Using Reverse Vaccinology and Biophysical Approaches" Vaccines 10, no. 10: 1729. https://doi.org/10.3390/vaccines10101729
APA StyleNawaz, M., Ullah, A., Al-Harbi, A. I., Haq, M. U., Hameed, A. R., Ahmad, S., Aziz, A., Raziq, K., Khan, S., Irfan, M., & Muhammad, R. (2022). Genome-Based Multi-Antigenic Epitopes Vaccine Construct Designing against Staphylococcus hominis Using Reverse Vaccinology and Biophysical Approaches. Vaccines, 10(10), 1729. https://doi.org/10.3390/vaccines10101729







