Antibody Response after SARS-CoV-2 Infection with the Delta and Omicron Variant
Abstract
:1. Introduction
2. Materials and Methods
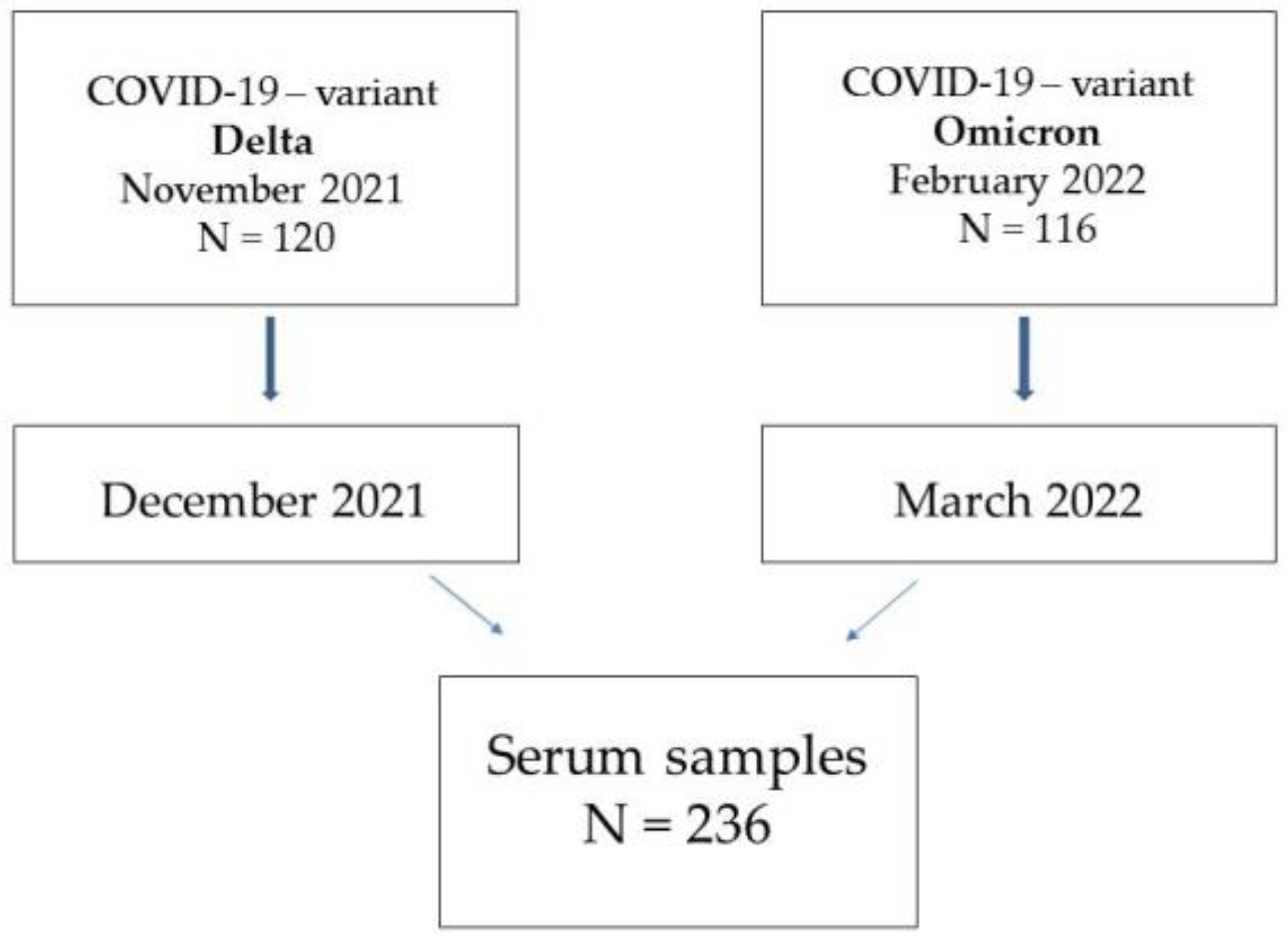
2.1. Study Design
2.2. Nasopharyngeal Swabs
2.2.1. Detection of SARS CoV-2
2.2.2. Detection of SARS CoV-2 Variants
2.3. Serum Samples Collection
Detection of SARS-CoV-2 Antibody
2.4. Statistical Analysis
2.5. Ethics
3. Results
Characteristics of the Studied Population
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Weekly Epidemiological Update on COVID-19—17 August 2022. Edition 105. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---17-August-2022 (accessed on 20 August 2022).
- Huang, Y.; Yang, C.; Xu, X.-F.; Xu, W.; Liu, S.-W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Arbeitman, C.R.; Rojas, P.; Ojeda-May, P.; Garcia, M.E. The SARS-CoV-2 spike protein is vulnerable to moderate electric fields. Nat. Commun. 2021, 12, 5407. [Google Scholar] [CrossRef] [PubMed]
- Perez-Gomez, R. The Development of SARS-CoV-2 Variants: The Gene Makes the Disease. J. Dev. Biol. 2021, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Cosar, B.; Karagulleoglu, Z.Y.; Unal, S.; Ince, A.T.; Uncuoglu, D.B.; Tuncer, G.; Kilinc, B.R.; Ozkan, Y.E.; Ozkoc, H.C.; Demir, I.N.; et al. SARS-CoV-2 Mutations and their Viral Variants. Cytokine Growth Factor Rev. 2022, 63, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef]
- Choi, J.Y.; Smith, D.M. SARS-CoV-2 Variants of Concern. Yonsei Med. J. 2021, 62, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Govindarajulu, M.; Parise, R.S.; Neel, L.; Shankar, T.; Patel, S.; Lowery, P.; Smith, F.; Dhanasekaran, D.; Moore, T. Emerging SARS-CoV-2 Variants: A Review of Its Mutations, Its Implications and Vaccine Efficacy. Vaccines 2021, 9, 1195. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wu, Q.; Song, P.; You, C. The Variation of SARS-CoV-2 and Advanced Research on Current Vaccines. Front. Med. 2022, 8, 806641. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Hong, W.; Pan, X.; Lu, G.; Wei, X. Severe acute respiratory syndrome coronavirus 2 Omicron variant: Characteristics and prevention. Med. Comm. 2021, 2, 838–845. [Google Scholar]
- Kannan, S.; Shaik Syed Ali, P.; Sheeza, A. Omicron (B.1.1.529)-variant of concern-molecular profile and epidemiology: A mini review. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 8019–8022. [Google Scholar] [PubMed]
- Araf, Y.; Akter, F.; Tang, Y.-D.; Fatemi, R.; Parvez, M.S.A.; Zheng, C.; Hossain, M.G. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022, 94, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef] [Green Version]
- Iketani, S.; Liu, L.; Guo, Y.; Liu, L.; Chan, J.F.W.; Huang, Y.; Wang, M.; Luo, Y.; Yu, J.; Chu, H. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef]
- Rahimi, F.; Abadi, A.T.B. Implications of the SARS-CoV-2 subvariants BA.4 and BA.5. Int. J. Surg. 2022, 104, 106806. [Google Scholar] [CrossRef]
- Wu, S.; Tian, C.; Liu, P.; Guo, D.; Zheng, W.; Huang, X.; Zhang, Y.; Liu, L. Effects of SARS-CoV-2 mutations on protein structures and intraviral protein–protein interactions. J. Med. Virol. 2021, 93, 2132–2140. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. COVID-19 Vaccine Breakthrough Infections Reported to CDC—United States, January 1–April 30, 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 792–793. [Google Scholar] [CrossRef]
- Hirsh, J.; Htay, T.; Bhalla, S.; Nguyen, V.; Cervantes, J. Breakthrough SARS-CoV-2 infections after COVID-19 immunization. J. Investig. Med. 2022, 70, 1429–1432. [Google Scholar] [CrossRef]
- Lavezzo, E.; Pacenti, M.; Manuto, L.; Boldrin, C.; Cattai, M.; Grazioli, M.; Bianca, F.; Sartori, M.; Caldart, F.; Castelli, G.; et al. Neutralising reactivity against SARS-CoV-2 Delta and Omicron variants by vaccination and infection history. Genome. Med. 2022, 14, 61. [Google Scholar] [CrossRef]
- Lassaunière, R.; Polacek, C.; Frische, A.; Boding, L.; Sækmose, S.G.; Rasmussen, M.; Fomsgaard, A. Neutralizing Antibodies Against the SARS-CoV-2 Omicron Variant (BA.1) 1 to 18 Weeks After the Second and Third Doses of the BNT162b2 mRNA Vaccine. JAMA Netw. Open 2022, 5, e2212073. [Google Scholar] [CrossRef]
- Kuhlmann, C.; Mayer, C.K.; Claassen, M.; Maponga, T.; Burgers, W.A.; Keeton, R.; Riou, C.; Sutherland, A.D.; Suliman, T.; Shaw, M.L.; et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 2022, 399, 625–626. [Google Scholar] [CrossRef]
- Dimeglio, C.; Migueres, M.; Mansuy, J.M.; Saivin, S.; Miedougé, M.; Chapuy-Regaud, S.; Izopet, J. Antibody titers and breakthrough infections with Omicron SARS-CoV-2. J. Infect. 2022, 84, e13–e15. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Karim, F.; Cele, S.; Reedoy, K.; San, J.E.; Lustig, G.; Tegally, H.; Rosenberg, Y.; Bernstein, M.; Jule, Z.; et al. Omicron infection enhances Delta antibody immunity in vaccinated persons. Nature 2022, 607, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Then, E.; Lucas, C.; Monteiro, V.S.; Miric, M.; Brache, V.; Cochon, L.; Vogels, C.B.F.; Malik, A.A.; De la Cruz, E.; Jorge, A.; et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022, 28, 481–485. [Google Scholar] [CrossRef]
- Collie, S.; Champion, J.; Moultrie, H.; Bekker, L.-G.; Gray, G. Effectiveness of BNT162b2 Vaccine against Omicron Variant in South Africa. N. Engl. J. Med. 2022, 386, 494–496. [Google Scholar] [CrossRef]
- Lu, L.; Mok, B.W.Y.; Chen, L.L.; Chan, J.M.C.; Tsang, O.T.Y.; Lam, B.H.S.; Chuang, V.W.M.; Chu, A.W.H.; Chan, W.M.; Ip, J.D.; et al. Neutralization of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant by Sera From BNT162b2 or CoronaVac Vaccine Recipients. Clin. Infect. Dis. 2022, 75, e822–e826. [Google Scholar] [CrossRef] [PubMed]
- Stærke, N.B.; Reekie, J.; Nielsen, H.; Benfield, T.; Wiese, L.; Knudsen, L.S.; Iversen, M.B.; Iversen, K.; Fogh, K.; Bodilsen, J. Levels of SARS-CoV-2 antibodies among fully vaccinated individuals with Delta or Omicron variant breakthrough infections. Nat. Commun. 2022, 13, 4466. [Google Scholar] [CrossRef]
- Montesinos, I.; Dahma, H.; Wolff, F.; Dauby, N.; Dalunoy, S.; Wuyts, M.; Detemmerman, C.; Duterme, C.; Vandenberg, O.; Martin, C.; et al. Neutralizing antibody responses following natural SARS-CoV-2 infection: Dynamics and correlation with commercial serologic tests. J. Clin. Virol. 2021, 144, 104988. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, I.; Gruson, D.; Kabamba, B.; Dahma, H.; Van den Wijngaert, S.; Reza, S.; Carbone, V.; Vandenberg, O.; Gulbis, B.; Wolff, F.; et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J. Clin. Virol. 2020, 128, 104413. [Google Scholar] [CrossRef]
- Desingu, P.A.; Nagarajan, K.; Dhama, K. Emergence of Omicron third lineage BA.3 and its importance. J. Med. Virol. 2022, 94, 1808–1810. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J. Med. Virol. 2022, 94, 1728–1733. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Rissmann, M.; Kok, A.; Rosu, M.E.; Schipper, D.; Breugem, T.I.; van den Doel, P.B.; Chandler, F.; Bestebroer, T.; de Wit, M. Antigenic cartography of SARS-CoV-2 reveals that Omicron BA.1 and BA.2 are antigenically distinct. Sci. Immunol. 2022, 7, eabq4450. [Google Scholar] [CrossRef]
- Rahimi, F.; Abadi, A.T. The Omicron subvariant BA.2: Birth of a new challenge during the COVID-19 pandemic. Int. J. Surg. 2022, 99, 106261. [Google Scholar] [CrossRef]
- Tatsi, E.-B.; Filippatos, F.; Michos, A. SARS-CoV-2 variants and effectiveness of vaccines: A review of current evidence. Epidemiol. Infect. 2021, 149, e237. [Google Scholar] [CrossRef]
- Lazarevic, I.; Pravica, V.; Miljanovic, D.; Cupic, M. Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses 2021, 13, 1192. [Google Scholar] [CrossRef] [PubMed]
- Boehm, E.; Kronig, I.; Neher, R.A.; Eckerle, I.; Vetter, P.; Kaiser, L.; Geneva Centre for Emerging Viral Diseases. SARS-CoV-2 variants: The pandemics within the pandemic. Clin. Microbiol. Infect. 2021, 27, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | N | % | |
|---|---|---|---|
| Sex | Female | 118 | 50.0 |
| Male | 118 | 50.0 | |
| Age | 40–50 | 118 | 50.0 |
| 70–85 | 118 | 50.0 | |
| Past COVID-19 | Delta | 120 | 50.8 |
| Omicron | 116 | 49.2 |
| Patients’ Group | NCP | RBD | S2 |
|---|---|---|---|
| COVID-19 Delta | 640.9 (300.7–989.3) | 886.0 (210.1–1000.1) | 746.8 (190.7–1000.0) |
| COVID-19 Omicron | 438.1 (210.0–690.7) | 345.8 (190.1–512.1) | 345.1 (180.1–740.5) |
| p | 10−6 * | 10−6 * | 10−6 * |
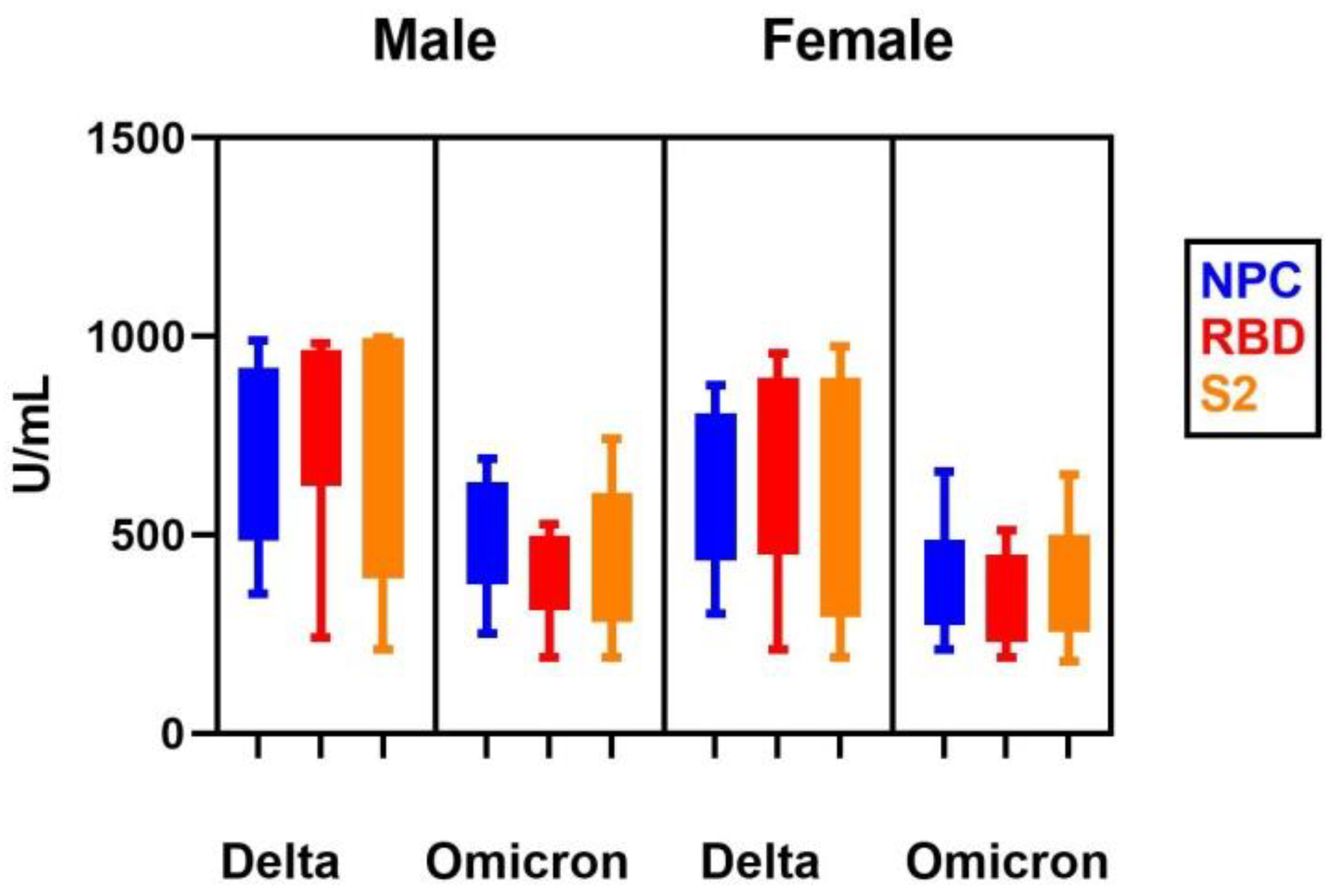
| Patients’ Group | NCP | RBD | S2 | |||
|---|---|---|---|---|---|---|
| Delta | Omicron | Delta | Omicron | Delta | Omicron | |
| Male | ||||||
| 40–50 | 717.3 (350.7–989.3) | 485.4 (250.8–690.7) | 948.7 (240.9–1005.5) | 456.2 (190.5–525.7) | 852.8 (210.7–1000.0) | 361.4 (190.6–740.5) |
| p | 0.001 * | 0.001 * | 0.001 * | |||
| 70–85 | 667.3 (350.7–989.3) | 485.4 (250.8–690.7) | 842.7 (240.9–982.3) | 456.2 (190.3–526.1) | 723.5 (210.7–997.1) | 361.5 (190.5–740.3) |
| p | 0.001 * | 0.001 * | 0.001 * | |||
| Female | ||||||
| 40–50 | 497.5 (350.7–989.3) | 390.7 (210.0–659.0) | 842.1 (210.1–1005.0) | 345.8 (190.1–512.1) | 723.4 (210.7–1000.0) | 314.8 (180.5–650.6) |
| p | >0.05 | 0.001 * | 0.001 * | |||
| 70–85 | 439.7 (300.9–878.7) | 390.6 (210.0–658.1) | 790.0 (210.4–956.4) | 345.5 (190.0–510.1) | 606.7 (190.7–975.8) | 340.0 (180.5–650.3) |
| p | >0.05 | 0.001 * | 0.001 * | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Błaszczuk, A.; Michalski, A.; Sikora, D.; Malm, M.; Drop, B.; Polz-Dacewicz, M. Antibody Response after SARS-CoV-2 Infection with the Delta and Omicron Variant. Vaccines 2022, 10, 1728. https://doi.org/10.3390/vaccines10101728
Błaszczuk A, Michalski A, Sikora D, Malm M, Drop B, Polz-Dacewicz M. Antibody Response after SARS-CoV-2 Infection with the Delta and Omicron Variant. Vaccines. 2022; 10(10):1728. https://doi.org/10.3390/vaccines10101728
Chicago/Turabian StyleBłaszczuk, Agata, Aleksander Michalski, Dominika Sikora, Maria Malm, Bartłomiej Drop, and Małgorzata Polz-Dacewicz. 2022. "Antibody Response after SARS-CoV-2 Infection with the Delta and Omicron Variant" Vaccines 10, no. 10: 1728. https://doi.org/10.3390/vaccines10101728
APA StyleBłaszczuk, A., Michalski, A., Sikora, D., Malm, M., Drop, B., & Polz-Dacewicz, M. (2022). Antibody Response after SARS-CoV-2 Infection with the Delta and Omicron Variant. Vaccines, 10(10), 1728. https://doi.org/10.3390/vaccines10101728







