The Epidemiological Features of the SARS-CoV-2 Omicron Subvariant BA.5 and Its Evasion of the Neutralizing Activity of Vaccination and Prior Infection
Abstract
:1. Introduction
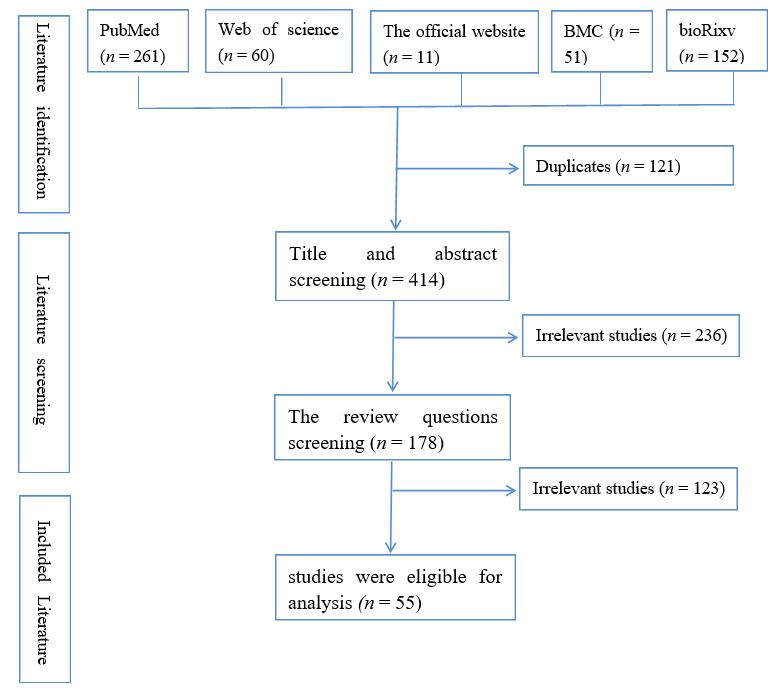
2. Methods
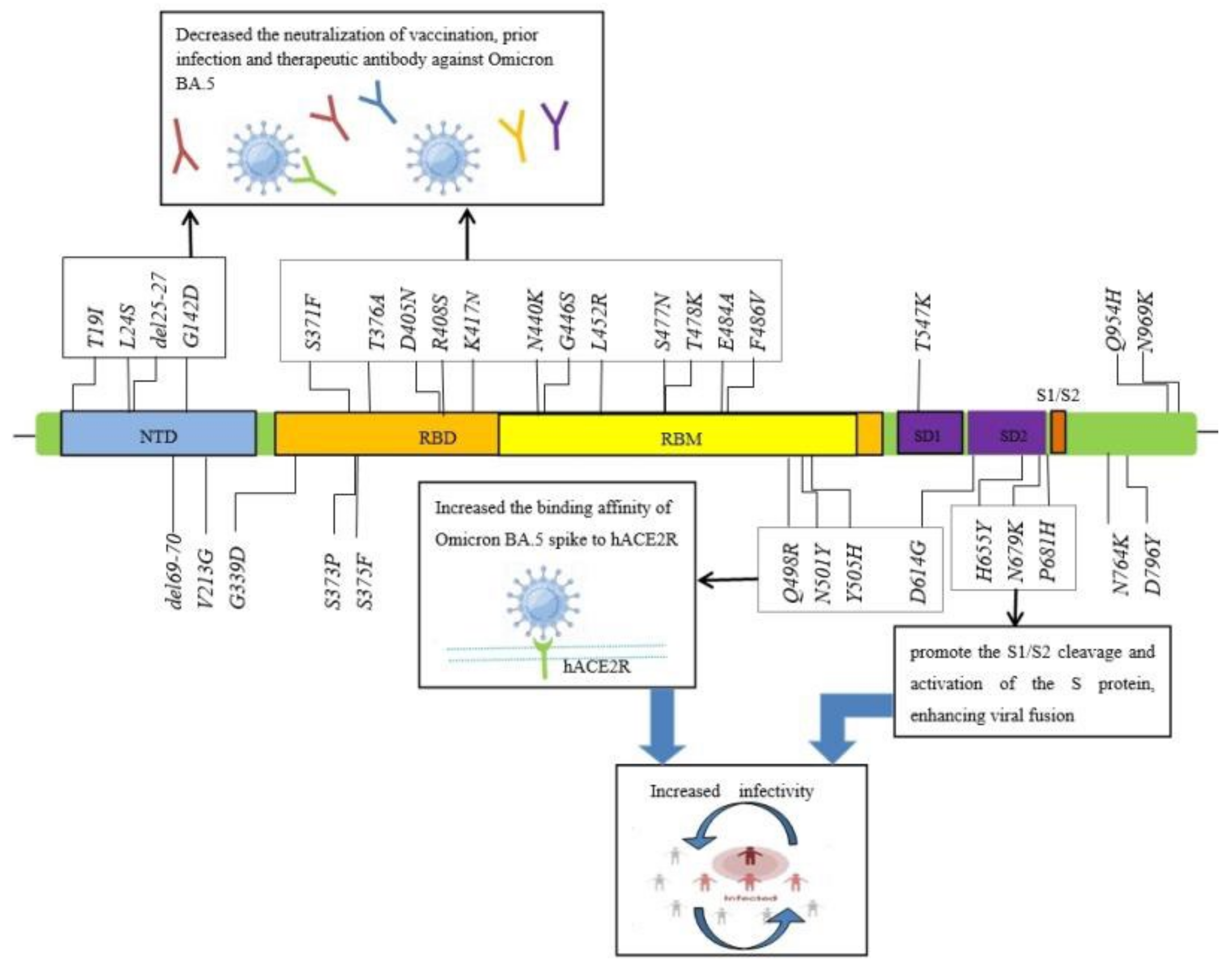
3. The Characteristics of SARS-CoV-2 Omicron BA.5 Spike Mutations
4. The Virological Characteristics of the SARS-CoV-2 Omicron BA.5
5. SARS-CoV-2 Omicron BA.5 Spread Faster in Many Countries and Has a Low Risk of Severe Clinical Outcomes
6. Omicron BA.5 Exhibits Stronger Neutralization Evasion Than BA.2 against nAbs Elicited by Vaccination and Prior Infection and Some Antiviral Drugs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Technical Report. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. 2021. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 17 May 2022).
- World Health Organization (WHO). Statement on Omicron Sublineage BA.2; WHO: Geneva, Switzerland, 2022; Available online: https://www.who.int/news/item/22-02-2022-statement-on-omicron-sublineage-ba.2 (accessed on 22 February 2022).
- Ai, J.W.; Wang, X.; Zhao, X.Y.; Zhang, Y.; Jiang, Y.C.; Li, M.H.; Cui, Y.; Chen, Y.; Qiao, R.; Li, L.; et al. Antibody Resistance of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2 and BA.3 Sub-lineages. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Moyo, S.; Amoako, D.G.; Baxter, C.; et al. Continued Emergence and Evolution of Omicron in South Africa: New BA.4 and BA.5 lineages. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.D.; Sun, Y.H.; Xu, H.H.; Ye, Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022, 4, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Makoni, M. South Africa responds to new SARS-CoV-2 variant. Lancet 2021, 397, 267. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- Singh, J.; Rahman, S.A.; Ehtesham, N.Z.; Hira, S.; Hasnain, S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021, 27, 1131–1133. [Google Scholar] [CrossRef]
- Tian, D.D.; Sun, Y.H.; Zhou, J.M.; Ye, Q. The global epidemic of SARS-CoV-2 variants and their mutational immune escape. J. Med. Virol. 2021, 94, 847–857. [Google Scholar] [CrossRef]
- Chen, J.; Qiu, Y.; Wang, R.; Wei, G.W. Persistent Laplacian Projected Omicron BA.4 and BA.5 to Become New Dominating Variants. medRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Desingu, P.A.; Nagarajan, K. The emergence of Omicron lineages BA.4 and BA.5, and the global spreading trend. J. Med. Virol. 2022, 94, 5077–5079. [Google Scholar] [CrossRef]
- Callaway, E. What Omicron’s BA.4 and BA.5 variants mean for the pandemic. Nature 2022, 606, 848–849. [Google Scholar] [CrossRef]
- Shrestha, L.B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022, 32, e2381. [Google Scholar] [CrossRef]
- Cao, Y.L.; Yisimayi, A.; Jian, F.C.; Song, W.L.; Xiao, T.H.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef]
- Hachmann, N.P.; Miller, J.; Collier, A.I.; Ventura, J.D.; Yu, J.Y.; Rowe, M.; Bondzie, E.A.; Powers, O.; Surve, N.; Hall, K.; et al. Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022, 387, 86–88. [Google Scholar] [CrossRef]
- Muik, A.; Lui, B.G.; Bacher, M.; Wallisch, A.-K.; Toker, A.; Finlayson, A.; Krüger, K.; Ozhelvaci, O.; Grikscheit, K.; Hoehl, S.; et al. Omicron BA.2 breakthrough infection enhances cross-neutralization of BA.2.12.1 and BA.4/BA.5. Sci. Immunol. 2022, eade2283. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F.F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef]
- Planas, D.; Bruel, T.; Grzelak, L.; Benhassine, F.G.; Staropoli, S.; Porrot, F.; Planchais, C.; Buchrieser, J.; Rajah, M.M.; Bishop, E.; et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021, 27, 917–924. [Google Scholar] [CrossRef]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 592, 616–622. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Q.; Wei, P.C.; Chen, Z.Z.; Aviszus, K.; Yang, J.; Downing, W.; Peterson, S.; Jiang, C.; Liang, B.; et al. The basis of a more contagious 501Y.V1 variant of SARS-COV-2. bioRxiv 2021. preprint. [Google Scholar] [CrossRef]
- Shah, M.; Woo, H.G. Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front. Immunol. 2022, 12, 830527. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Q.; Ge, J.; Ren, W.L.; Zhang, R.; Lan, J.; Ju, B.; Su, B.; Yu, F.; Chen, P.; et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity 2021, 54, 1611–1621.e5. [Google Scholar] [CrossRef]
- Li, Q.Q.; Nie, J.H.; Wu, J.J.; Zhang, L.; Ding, R.X.; Wang, H.X.; Zhang, Y.; Li, T.; Liu, S.; Zhang, M.; et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 2021, 184, 2362–2371.e9. [Google Scholar] [CrossRef]
- Callaway, E. Delta coronavirus variant: Scientists brace for impact. Nature 2021, 595, 17–18. [Google Scholar] [CrossRef]
- Chung, H.Y.; Noh, J.Y.; Koo, B.S.; Hong, J.J.; Kim, H.K. SARS-CoV-2 mutations acquired during serial passage in human cell lines are consistent with several of those found in recent natural SARS-CoV-2 variants. Comput. Struct. Biothchnol. J. 2022, 2, 925–1934. [Google Scholar] [CrossRef]
- Chi, X.Y.; Yan, R.H.; Zhang, J.; Zhang, G.Y.; Zhang, Y.Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 36, 50–655. [Google Scholar] [CrossRef]
- Escalera, A.; Reiche, A.S.G.; Aslam, S.; Mena, I.; Laporte, M.; Pearl, R.L.; Fossati, A.; Rathnasinghe, R.; Alshammary, H.; van de Guchte, A.; et al. Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe 2022, 3, 73–387.e377. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, T.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 60, 300–306. [Google Scholar] [CrossRef]
- Sabir, D.K. Analysis of SARS-COV2 spike protein variants among Iraqi isolates. Gene Rep. 2022, 2, 01420. [Google Scholar] [CrossRef]
- Aoki, A.; Adachi, H.; Mori, Y.; Ito, M.; Sato, K.; Okuda, K.; Sakakibara, T.; Okamoto, Y.; Jinno, H. Discrimination of SARS-CoV-2 Omicron Sublineages BA.1 and BA.2 Using a High-Resolution Melting-Based Assay: A Pilot Study. Microbiol. Spectr. 2022, 10, e0136722. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Tada, T.; Dcosta, B.M.; Landau, N.R. Neutralization of SARS-CoV-2 Omicron BA.2 by Therapeutic Monoclonal Antibodies. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Nutalai, R.; Zhou, D.; Tuekprakhon, A.; Ginn, H.M.; Supasa, P.; Liu, C.; Huo, J.; Mentzer, A.J.; Duyvesteyn, H.M.E.; Dijokaite-Guraliuc, A.; et al. Potent cross-reactive antibodies following Omicron breakthrough in vaccines. Cell 2022, 185, 2116–2131.e18. [Google Scholar] [CrossRef] [PubMed]
- Touret, F.; Baronti, C.; Pastorino, B.; Villarroel, P.M.S.; Ninove, L.; Nougairède, A.; de Lamballerie, X. In vitro activity of therapeutic antibodies against SARS-CoV-2 Omicron BA.1, BA.2 and BA.5. Sci. Rep. 2022, 12, 12609. [Google Scholar] [CrossRef]
- Yamasoba, D.; Kosugi, Y.; Kimura, I.; Fujita, S.; Uriu, K.; Ito, J.; Sato, K. Sensitivity of novel SARS-CoV-2 Omicron subvariants, BA.2.11, BA.2.12.1, BA.4 and BA.5 to therapeutic monoclonal antibodies. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.C.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. SARS-CoV-2 Omicron BA.2.12.1, BA.4, and BA.5 subvariants evolved to extend antibody evasion. Nature 2022. preprint. [Google Scholar] [CrossRef]
- Qu, P.; Faraone, J.N.; Evans, J.P.; Zou, X.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Lozanski, G.; Mallampalli, R.K.; Saif, L.J.; et al. Differential evasion of Delta and Omicron immunity and enhanced fusogenicity of SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 subvariants. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J.; Uriu, K.; et al. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA.2.12.1, BA.4 and BA.5. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC). Epidemiological update: SARS-CoV-2 Omicron sublineages BA.4 and BA.5. Available online: https://www.ecdc.europa.eu/en/news-events (accessed on 13 May 2022).
- Brüssow, H. COVID-19: Omicron—The latest, the least virulent, but probably not the last variant of concern of SARS-CoV-2. Microb. Biotechnol. 2022, 15, 1927–1939. [Google Scholar] [CrossRef]
- Yamasoba, D.; Kimura, I.; Nasser, H.; Morioka, Y.; Nao, N.; Ito, J.; Uriu, K.; Tsuda, M.; Zahradnik, J.; Shirakawa, K.; et al. Virological characteristics of SARS-CoV-2 BA.2 variant. bioRxiv 2022. [Google Scholar] [CrossRef]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moltrie, H.; Groome, M.; Gyamfi Amoako, D.; Everatt, J.; Bhiman, J.; Scheepers, C.; et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: A data linkage study. Lancet 2022, 399, 437–446. [Google Scholar] [CrossRef]
- Sheikh, A.; Kerr, S.; Woolhouse, M.; McMenamin, J.; Robertson, C. Collaborators EAVE II. Severity of Omicron variant of concern and effectiveness of vaccine boosters against symptomatic disease in Scotland (EAVE II): A national cohort study with nested test-negative design. Lancet Infect. Dis. 2022, 22, 959–966. [Google Scholar] [CrossRef]
- Lauring, A.S.; Tenforde, M.W.; Chappell, J.D.; Gaglani, M.; Ginde, A.A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.; Casey, J.D.; et al. Clinical severity of, and effectiveness of mRNA vaccines against, Covid-19 from Omicron, Delta, and Alpha SARS-CoV-2 variants in the United States: Prospective observational study. BMJ 2022, 376, e069761. [Google Scholar] [CrossRef]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hisley, W.; Lopez Bernal, J.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalization and death associated with SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef]
- Davie, M.A.; Morden, E.; Rosseau, P.; Arendse, J.; Bam, J.L.; Boloko, L.; Cloete, K.; Cohen, C.; Chetty, N.; Dane, P.; et al. Outcomes of laboratory-confirmed SARS-CoV-2 infection during resurgence driven by Omicron lineages BA.4 and BA.5 compared with previous waves in the Western Cape Province, South Africa. medRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Schulz, S.R.; Cossmann, A.; Stankov, M.V.; Jack, H.M.; Behrens, G.M.N.; Pohlmann, S.; Hoffmann, M.; et al. Augmented neutralisation resistance of emerging omicron subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect. Dis. 2022, 22, 1117–1118. [Google Scholar] [CrossRef]
- Yamasoba, D.; Kosugi, Y.; Kimura, I.; Fujita, S.; Uriu, K.; Ito, J.; Sato, K. Neutralization sensitivity of SARS-CoV-2 omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect. Dis. 2022, 22, 942–943. [Google Scholar] [CrossRef]
- Takashita, E.; Yamayoshi, S.; Simon, V.; Bakel, H.V.; Sordillo, E.M.; Pekosz, A.; Fukushi, S.; Suzuki, T.; Maeda, K.; Halfmann, P.; et al. Efficacy of Antibodies and Antiviral Drugs against Omicron BA.2.12.1, BA.4, and BA.5 Subvariants. N. Engl. J. Med. 2022, 387, 468–470. [Google Scholar] [CrossRef]
- Xie, X.P.; Zoul, J.; Liu, M.R.; Ren, P.; Shi, P.Y. Neutralization of SARS-CoV-2 Omicron sublineages by 4 doses of mRNA vaccine. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Bhiman, J.N.; Richardson, S.R.; Lambson, B.E.; Kgagudi, P.; Mzindle, N.; Kaldine, H.; Crowther, C.; Gray, G.; Bekker, L.-G. Novavax Trial Clinical Lead Author Group; et al. Novavax NVX-COV2373 triggers potent neutralization of Omicron sublineages. bioRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Xia, S.; Wang, L.J.; Zhu, Y.; Lu, L.; Jiang, S.B. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct. Target. Ther. 2022, 7, 241. [Google Scholar] [CrossRef]
- Tuekprakhon, A.; Nutalai, R.; Guraliuc, A.D.; Zhou, D.M.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.E.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e13. [Google Scholar] [CrossRef]

| WHO Label | Omicron |
|---|---|
| Pango lineage | BA.5 |
| Next strain | 22B |
| GISAID clade | GRA |
| Higher mutated strain in the spike protein | T19I, L24S, del25-27, del69-70, D142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, G446S, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, T547K D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Earliest detected | South Africa, Jan-2022 |
| Higher infectivity | BA.5 dominated in April (72%) and in May (92%) in South Africa #. BA.5 dominated in August (88.8%) and in September (83.1%) in USA *. |
| Lower risk of hospitalization, ICU admission, and mortality | Adjusted hazard ratio [aHR] 1.12; 95% confidence interval [CI]: 0.93; 1.34 [49]. |
| Immune escape | BA.5 could further escape from the immunity induced by vaccination and BA.1 or BA.2 prior infections. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, D.; Nie, W.; Sun, Y.; Ye, Q. The Epidemiological Features of the SARS-CoV-2 Omicron Subvariant BA.5 and Its Evasion of the Neutralizing Activity of Vaccination and Prior Infection. Vaccines 2022, 10, 1699. https://doi.org/10.3390/vaccines10101699
Tian D, Nie W, Sun Y, Ye Q. The Epidemiological Features of the SARS-CoV-2 Omicron Subvariant BA.5 and Its Evasion of the Neutralizing Activity of Vaccination and Prior Infection. Vaccines. 2022; 10(10):1699. https://doi.org/10.3390/vaccines10101699
Chicago/Turabian StyleTian, Dandan, Wenjian Nie, Yanhong Sun, and Qing Ye. 2022. "The Epidemiological Features of the SARS-CoV-2 Omicron Subvariant BA.5 and Its Evasion of the Neutralizing Activity of Vaccination and Prior Infection" Vaccines 10, no. 10: 1699. https://doi.org/10.3390/vaccines10101699
APA StyleTian, D., Nie, W., Sun, Y., & Ye, Q. (2022). The Epidemiological Features of the SARS-CoV-2 Omicron Subvariant BA.5 and Its Evasion of the Neutralizing Activity of Vaccination and Prior Infection. Vaccines, 10(10), 1699. https://doi.org/10.3390/vaccines10101699





