Unravelling the Differential Host Immuno-Inflammatory Responses to Staphylococcus aureus and Escherichia coli Infections in Sepsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients Selection
2.2. Blood Sample Collection
2.3. Blood Culture
2.4. Bacterial Identification by RT-PCR
2.5. Isolation of Neutrophils from Blood
2.6. Spiking
2.7. iNOS Expression
2.8. Total Nitrite Assay
2.9. NETs Release
2.10. Cytokines Estimation
2.11. Statistical Analysis after the Estimation of Total Nitrite Assay and Cytokine
3. Results
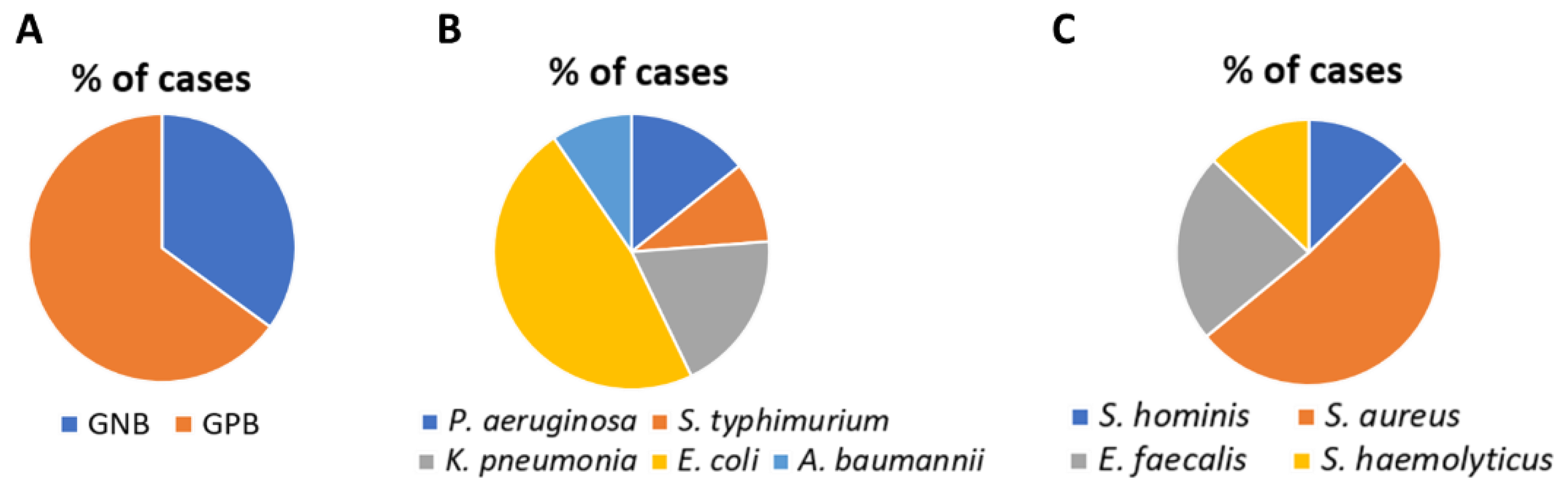
3.1. Detection of Pathogens by Blood Culture and RT-PCR
3.2. Expression of iNOS Generation and Estimation of Total Nitrite Content
3.3. NETs Formation and Estimation of Cytokines
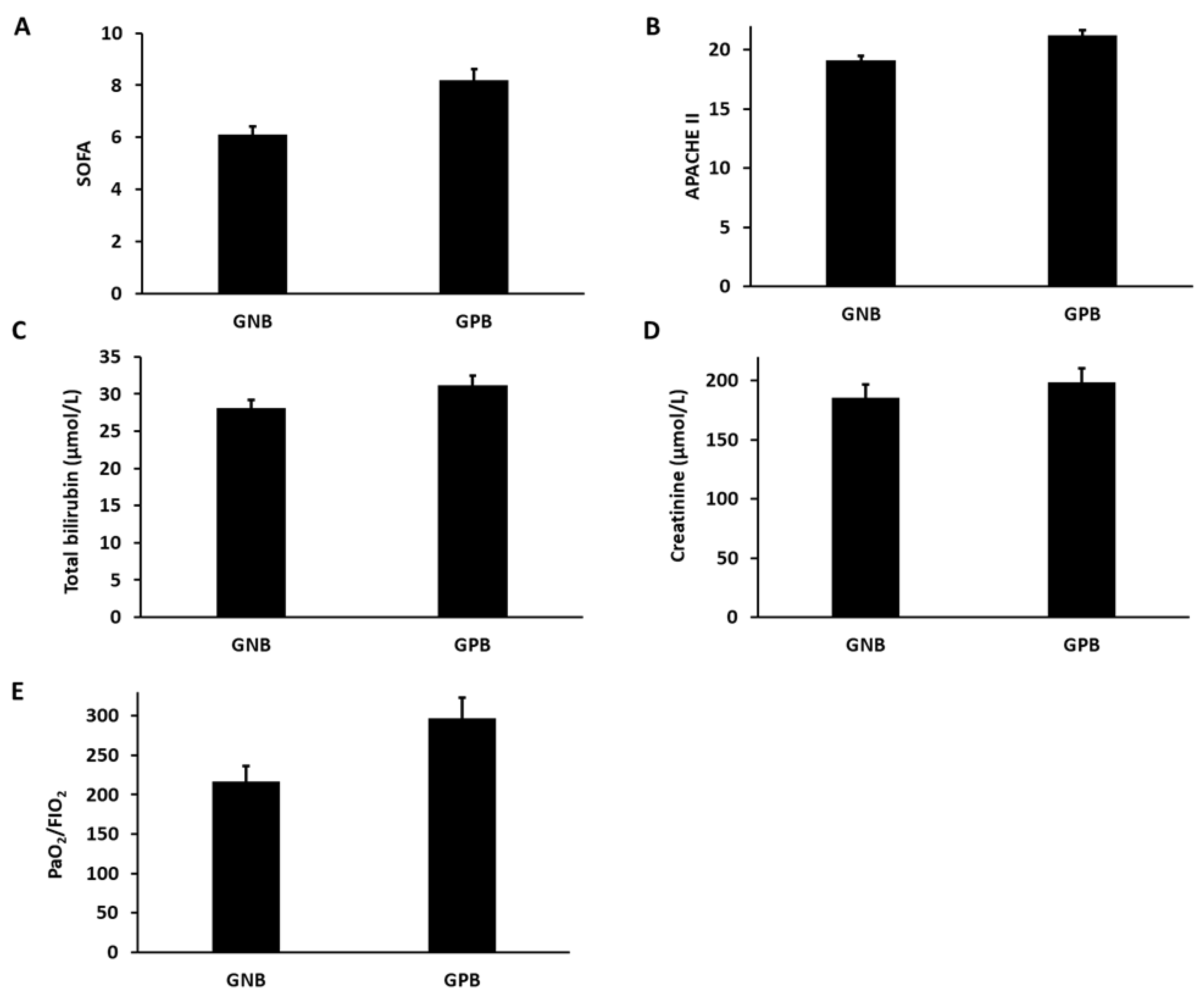
3.4. Estimation of Organ Dysfunction
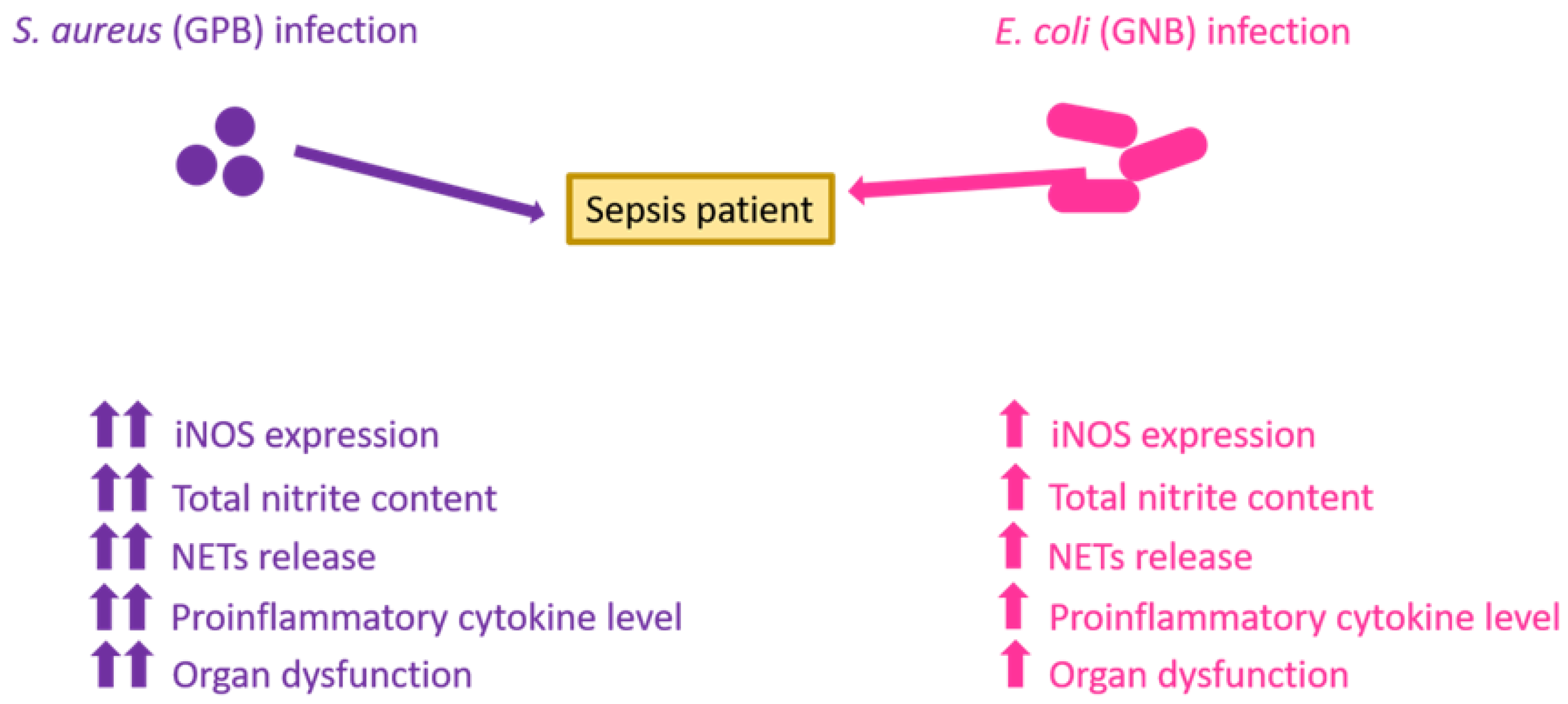
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dickson, K.; Lehmann, C. Inflammatory response to different toxins in experimental sepsis models. Int. J. Mol. Sci. 2019, 20, 4341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minasyan, H. Sepsis and septic shock: Pathogenesis and treatment perspectives. J. Crit. Care 2017, 40, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Tunjungputri, R.N.; van de Heijden, W.; Urbanus, R.T.; de Groot, P.G.; van der Ven, A.; de Mast, Q. Higher platelet reactivity and platelet-monocyte complex formation in Gram-positive sepsis compared to Gram-negative sepsis. Platelets 2017, 28, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Ogura, H.; Kushimoto, S.; Shiraishi, A.; Sugiyama, T.; Deshpande, G.A.; Uchida, M.; Nagata, I.; Saitoh, D.; Fujishima, S.; et al. Variations in infection sites and mortality rates among patients in intensive care units with severe sepsis and septic shock in Japan. J. Intensive Care 2019, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Pfalzgraff, A.; Weindl, G. Intracellular lipopolysaccharide sensing as a potential therapeutic target for sepsis. Trends Pharmacol. Sci. 2019, 40, 187–197. [Google Scholar] [CrossRef]
- Mallat, J.; Leone, S.; Cascella, M.; Fiore, M. Should endotoxin be a research priority in Gram-negative sepsis and septic shock? Expert Rev. Clin. Pharmacol. 2019, 12, 697–699. [Google Scholar] [CrossRef] [Green Version]
- Danielski, L.G.; Giustina, A.D.; Bonfante, S.; Barichello, T.; Petronilho, F. The NLRP3 inflammasome and its role in sepsis development. Inflammation 2020, 43, 24–31. [Google Scholar] [CrossRef]
- Chen, F.; Zou, L.; Williams, B.; Chao, W. Targeting toll-like receptors in sepsis: From bench to clinical trials. Antioxid. Redox Signal. 2021, 35, 1324–1339. [Google Scholar] [CrossRef]
- Leung, A.K.; Genga, K.R.; Topchiy, E.; Cirstea, M.; Shimada, T.; Fjell, C.; Russell, J.A.; Boyd, J.H.; Walley, K.R. Reduced Proprotein convertase subtilisin/kexin 9 (PCSK9) function increases lipoteichoic acid clearance and improves outcomes in Gram positive septic shock patients. Sci. Rep. 2019, 9, 10588. [Google Scholar] [CrossRef] [Green Version]
- Bühler, T.; Boos, N.; Leuppi-Taegtmeyer, A.B.; Berger, C.T. Febrile illness and bicytopenia within hours after tick-borne encephalitis booster vaccination. NPJ Vaccines 2019, 4, 52. [Google Scholar] [CrossRef] [Green Version]
- Chousterman, B.G.; Swirski, F.K.; Weber, G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017, 39, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Cron, R.Q.; Behrens, E.M. (Eds.) Cytokine Storm Syndrome; Springer Nature: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Cabrera, C.P.; Manson, J.; Shepherd, J.M.; Torrance, H.D.; Watson, D.; Longhi, M.P.; Hoti, M.; Patel, M.B.; O’Dwyer, M.; Nourshargh, S.; et al. Signatures of inflammation and impending multiple organ dysfunction in the hyperacute phase of trauma: A prospective cohort study. PLoS Med. 2017, 14, e1002352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margraf, A.; Ludwig, N.; Zarbock, A.; Rossaint, J. Systemic inflammatory response syndrome after surgery: Mechanisms and protection. Anesth. Analg. 2020, 131, 1693–1707. [Google Scholar] [CrossRef]
- Sonar, S.A.; Lal, G. The iNOS activity during an immune response controls the CNS pathology in experimental autoimmune encephalomyelitis. Front. Immunol. 2019, 10, 710. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Gupta, E.; Kaushik, S.; Kumar Srivastava, V.; Mehta, S.; Jyoti, A. Evaluation of oxidative stress and antioxidant status: Correlation with the severity of sepsis. Scand. J. Immunol. 2018, 87, e12653. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Gupta, E.; Srivastava, V.K.; Kaushik, S.; Saxena, J.; Goyal, L.K.; Mehta, S.; Jyoti, A. Nitrosative stress and cytokines are linked with the severity of sepsis and organ dysfunction. Br. J. Biomed. Sci. 2019, 76, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gupta, E.; Kaushik, S.; Kumar Srivastava, V.; Mehta, S.; Jyoti, A. Quantification of NETs formation in neutrophil and its correlation with the severity of sepsis and organ dysfunction. Clin. Chim. Acta 2019, 495, 606–610. [Google Scholar] [CrossRef]
- Gül, F.; Arslantaş, M.K.; Cinel, İ.; Kumar, A. Changing definitions of sepsis. Turk. J. Anaesthesiol. Reanim. 2017, 45, 129. [Google Scholar] [CrossRef]
- Dong, M.; Fisher, C.; Añez, G.; Rios, M.; Nakhasi, H.L.; Hobson, J.P.; Beanan, M.; Hockman, D.; Grigorenko, E.; Duncan, R. Standardized methods to generate mock (spiked) clinical specimens by spiking blood or plasma with cultured pathogens. J. Appl. Microbiol. 2016, 120, 1119–1129. [Google Scholar] [CrossRef] [Green Version]
- Keshari, R.S.; Jyoti, A.; Dubey, M.; Kothari, N.; Kohli, M.; Bogra, J.; Barthwal, M.K.; Dikshit, M. Cytokines induced neutrophil extracellular traps formation: Implication for the inflammatory disease condition. PLoS ONE 2012, 7, e48111. [Google Scholar] [CrossRef] [Green Version]
- Wan, T.; Zhao, Y.; Fan, F.; Hu, R.; Jin, X. Dexamethasone inhibits S. aureus-induced neutrophil extracellular pathogen-killing mechanism, possibly through toll-like receptor regulation. Front. Immunol. 2017, 8, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; McCann, K.; Tripathi, J.K.; Chauhan, P.; Zerbe, C.; Mishra, B.B.; Holland, S.M.; Sharma, J. Tamoxifen restores extracellular trap formation in neutrophils from patients with chronic granulomatous disease in a reactive oxygen species–independent manner. J. Allergy Clin. Immunol. 2019, 144, 597–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, J.K.; Sharma, A.; Sukumaran, P.; Sun, Y.; Mishra, B.B.; Singh, B.B.; Sharma, J. Oxidant sensor cation channel TRPM2 regulates neutrophil extracellular trap formation and protects against pneumoseptic bacterial infection. FASEB J. 2018, 32, 6848–6859. [Google Scholar] [CrossRef]
- Idelevich, E.A.; Storck, L.M.; Sparbier, K.; Drews, O.; Kostrzewa, M.; Becker, K. Rapid direct susceptibility testing from positive blood cultures by the matrix-assisted laser desorption ionization–time of flight mass spectrometry-based direct-on-target microdroplet growth assay. J. Clin. Microbiol. 2018, 56, e00913-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessle, C.C.; Andersson, B.; Wold, A.E. Gram-positive and Gram-negative bacteria elicit different patterns of pro-inflammatory cytokines in human monocytes. Cytokine 2005, 30, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, M.; Yu, Y.; Pan, S.; Liu, Y. Antimicrobial susceptibility among gram-positive and gram-negative blood-borne pathogens collected between 2012–2016 as part of the Tigecycline Evaluation and Surveillance Trial. Antimicrob. Resist. Infect. Control 2018, 7, 152. [Google Scholar] [CrossRef]
- Gao, L.; Liu, X.; Zhang, D.; Xu, F.; Chen, Q.; Hong, Y.; Feng, G.; Shi, Q.; Yang, B.; Xu, L. Early diagnosis of bacterial infection in patients with septicopyemia by laboratory analysis of PCT, CRP and IL-6. Exp. Ther. Med. 2017, 13, 3479–3483. [Google Scholar] [CrossRef] [Green Version]
- Thomas-Rüddel, D.O.; Poidinger, B.; Kott, M.; Weiss, M.; Reinhart, K.; Bloos, F. Influence of pathogen and focus of infection on procalcitonin values in sepsis patients with bacteremia or candidemia. Crit. Care 2018, 22, 128. [Google Scholar] [CrossRef] [Green Version]
- Kwiecinski, J.M.; Horswill, A.R. Staphylococcus aureus bloodstream infections: Pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol. 2020, 53, 51–60. [Google Scholar] [CrossRef]
- Lee, E.J.; Park, J.S.; Lee, Y.Y.; Kim, D.Y.; Kang, J.L.; Kim, H.S. Anti-inflammatory and anti-oxidant mechanisms of an MMP-8 inhibitor in lipoteichoic acid-stimulated rat primary astrocytes: Involvement of NF-κB, Nrf2, and PPAR-γ signaling pathways. J. Neuroinflamm. 2018, 15, 326. [Google Scholar] [CrossRef] [Green Version]
- Spiller, F.; Formiga, R.O.; da Silva Coimbra, J.F.; Alves-Filho, J.C.; Cunha, T.M.; Cunha, F.Q. Targeting nitric oxide as a key modulator of sepsis, arthritis and pain. Nitric Oxide 2019, 89, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.L.; Suzuki, K. The pattern recognition receptors and lipopolysaccharides (LPS)-induced systemic inflammation. Int. J. Res. Stud. Med. Health Sci. 2017, 2, 1–7. [Google Scholar]
- Tallam, A.; Perumal, T.M.; Antony, P.M.; Jäger, C.; Fritz, J.V.; Vallar, L.; Balling, R.; Del Sol, A.; Michelucci, A. Gene regulatory network inference of immunoresponsive gene 1 (IRG1) identifies interferon regulatory factor 1 (IRF1) as its transcriptional regulator in mammalian macrophages. PLoS ONE 2016, 1, e0149050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogdan, C. Nitric oxide synthase in innate and adaptive immunity: An update. Trends Immunol. 2015, 36, 161–178. [Google Scholar] [CrossRef]
- Winkler, M.S.; Kluge, S.; Holzmann, M.; Moritzm, E.; Robbe, L.; Bauer, A.; Zahrte, C.; Priefler, M.; Schwedhelm, E.; Böger, R.H.; et al. Markers of nitric oxide are associated with sepsis severity: An observational study. Crit. Care 2017, 21, 189. [Google Scholar] [CrossRef] [Green Version]
- Shaker, N.B.; Ali, K.O. Role of IL-6 and IL-10 as biomarkers in diagnosis of bacteremia in hemodialysis patients. Al-Kufa Univ. J. Biol. 2019, 11, 34–41. [Google Scholar]
- Jorch, S.K.; Kubes, P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 2017, 23, 279–287. [Google Scholar] [CrossRef]
- Chaudhry, H.; Zhou, J.; Zhong, Y.I.; Ali, M.M.; McGuire, F.; Nagarkatti, P.S.; Nagarkatti, M. Role of cytokines as a double-edged sword in sepsis. In Vivo 2013, 27, 669–684. [Google Scholar]
- Black, L.P.; Puskarich, M.A.; Henson, M.; Miller, T.; Reddy, S.T.; Fernandez, R.; Guirgis, F.W. Quantitative and qualitative assessments of cholesterol association with bacterial infection type in sepsis and septic shock. J. Intensive Care Med. 2021, 36, 808–817. [Google Scholar] [CrossRef]
- Surbatovic, M.; Popovic, N.; Vojvodic, D.; Milosevic, I.; Acimovic, G.; Stojicic, M.; Veljovic, M.; Jevdjic, J.; Djordjevic, D.; Radakovic, S. Cytokine profile in severe Gram-positive and Gram-negative abdominal sepsis. Sci. Rep. 2015, 5, 11355. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Gupta, E.; Kaushik, S.; Kumar Srivastava, V.; Mehta, S.; Jyoti, A. Functional role of iNOS-Rac2 interaction in neutrophil extracellular traps (NETs) induced cytotoxicity in sepsis. Clin. Chim. Acta 2021, 513, 43–49. [Google Scholar] [CrossRef] [PubMed]



| Parameters | Value |
|---|---|
| Total number of patients | 60 |
| Age (years) | 49.11 ± 12.35 |
| Male/female ratio | 37/23 |
| Body Temperature (°C) | 38.62 ± 1.28 |
| Heart rate (beats/min) | 112.45 ± 9.7 |
| Respiratory rate (breaths/min) | 27.3 ± 4.2 |
| Mean arterial pressure (mm Hg) | 86.13 ± 13.22 |
| SOFA score | 7.1 ± 1.23 |
| APACHE II score | 20.1 ± 4.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, E.; Kumar, S.; Srivastava, V.K.; Saxena, J.; Siddiqui, A.J.; Mehta, S.; Kaushik, S.; Jyoti, A. Unravelling the Differential Host Immuno-Inflammatory Responses to Staphylococcus aureus and Escherichia coli Infections in Sepsis. Vaccines 2022, 10, 1648. https://doi.org/10.3390/vaccines10101648
Gupta E, Kumar S, Srivastava VK, Saxena J, Siddiqui AJ, Mehta S, Kaushik S, Jyoti A. Unravelling the Differential Host Immuno-Inflammatory Responses to Staphylococcus aureus and Escherichia coli Infections in Sepsis. Vaccines. 2022; 10(10):1648. https://doi.org/10.3390/vaccines10101648
Chicago/Turabian StyleGupta, Ena, Sanni Kumar, Vijay Kumar Srivastava, Juhi Saxena, Arif Jamal Siddiqui, Sudhir Mehta, Sanket Kaushik, and Anupam Jyoti. 2022. "Unravelling the Differential Host Immuno-Inflammatory Responses to Staphylococcus aureus and Escherichia coli Infections in Sepsis" Vaccines 10, no. 10: 1648. https://doi.org/10.3390/vaccines10101648
APA StyleGupta, E., Kumar, S., Srivastava, V. K., Saxena, J., Siddiqui, A. J., Mehta, S., Kaushik, S., & Jyoti, A. (2022). Unravelling the Differential Host Immuno-Inflammatory Responses to Staphylococcus aureus and Escherichia coli Infections in Sepsis. Vaccines, 10(10), 1648. https://doi.org/10.3390/vaccines10101648








