The Antioxidant Cyclic 3-Hydroxymelatonin Promotes the Growth and Flowering of Arabidopsis thaliana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Escherichia coli Expression and Purification of Recombinant Arabidopsis thaliana Melatonin 3-Hydroxylase (M3H)
2.2. Measurement of In Vitro M3H Activity and Enzyme Kinetics
2.3. Quantification of Cyclic 3-Hydroxymelatonin (3-OHM) from Arabidopsis
2.4. Measurement of Radical Scavenging Activity
2.5. Arabidopsis Plants and Growth Conditions
2.6. Determination of Flowering Time
2.7. Subcellular Localization Analysis of Arabidopsis thaliana Melatonin 3-Hydroxylase (AtM3H)
2.8. RNA Extraction and Gene Expression Analysis
2.9. Bacterial Growth Assay
2.10. Salt Tolerance Assay
2.11. Statistical Analysis
3. Results
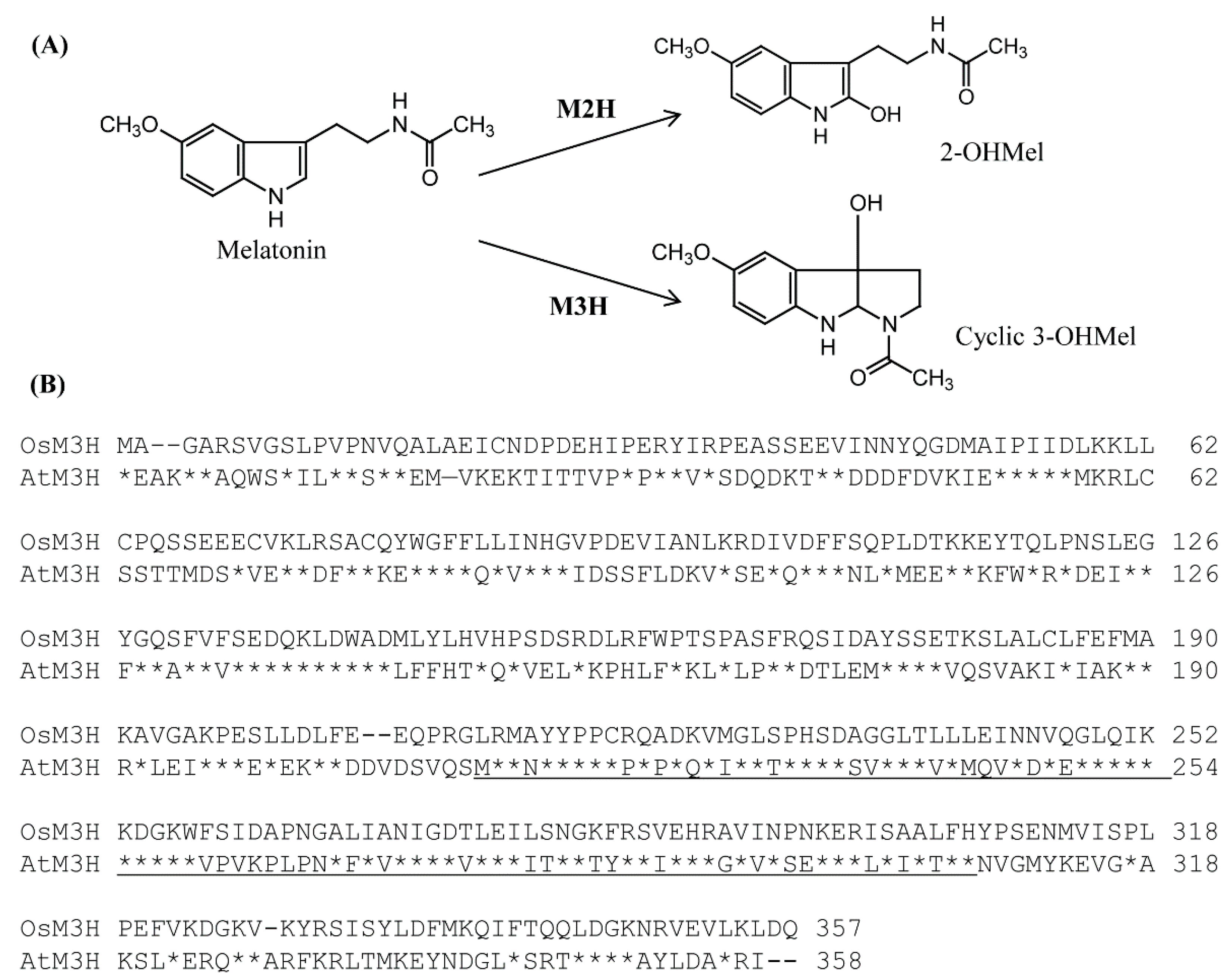
3.1. Sequence Analysis of Arabidopsis and Rice M3H
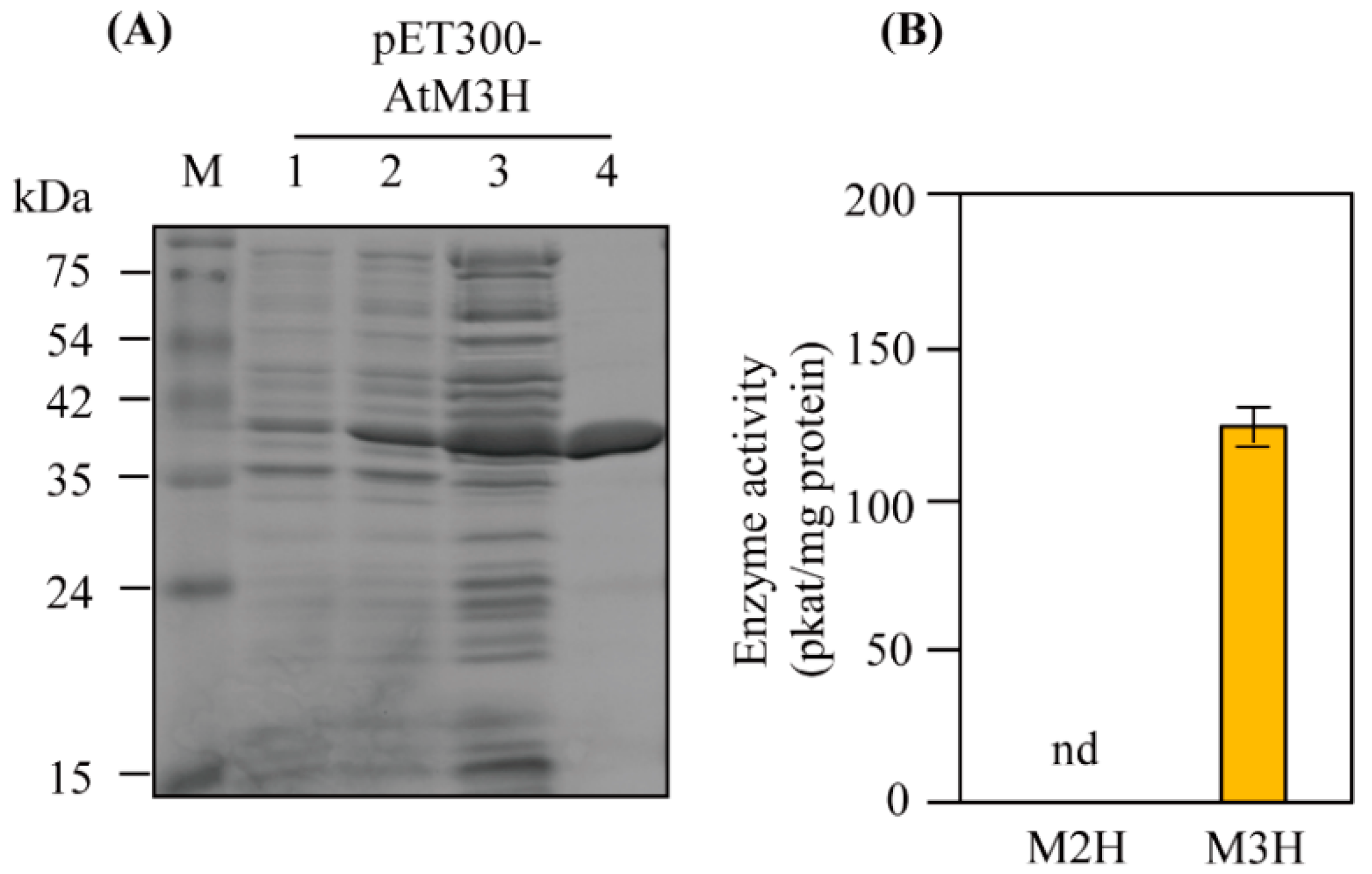
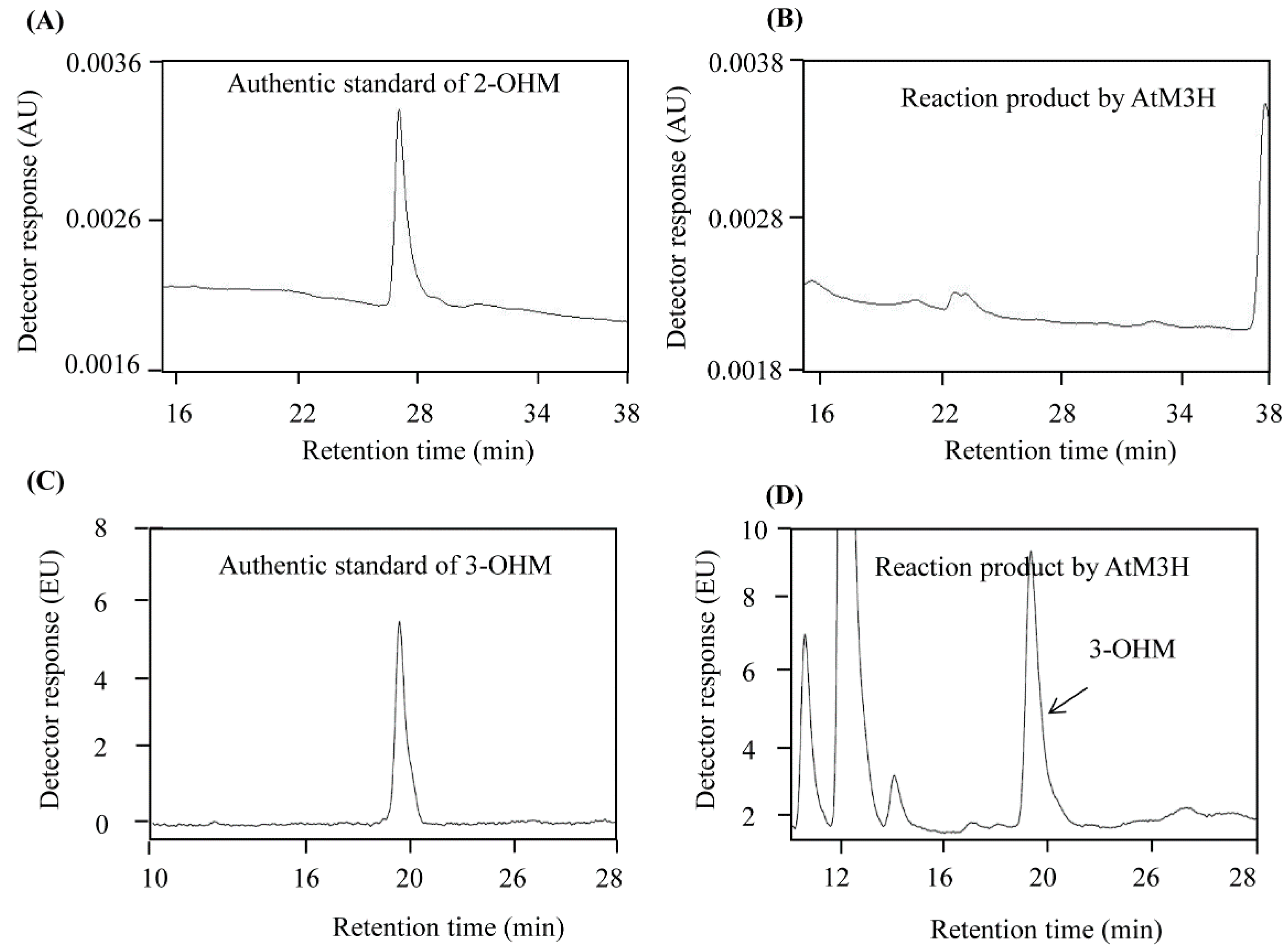
3.2. M3H Activity of Recombinant AtM3H
3.3. Characteristics of Recombinant AtM3H
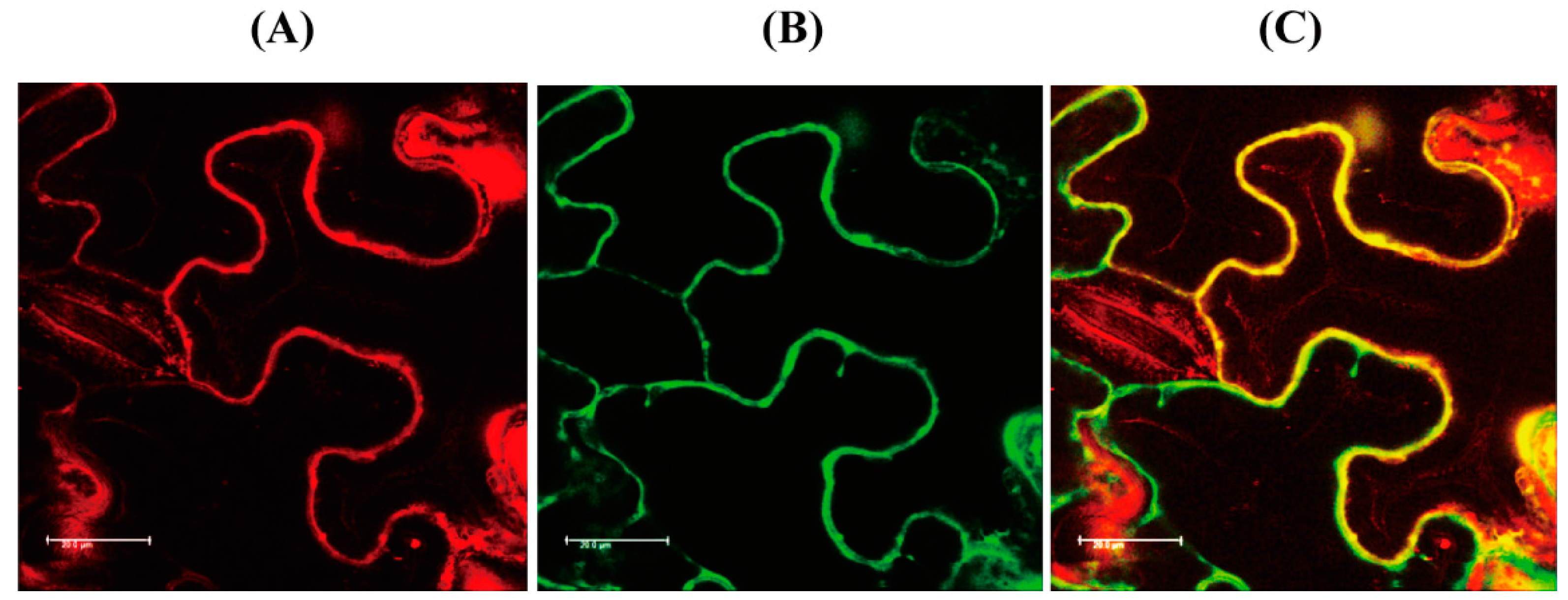
3.4. Subcellular Localization of AtM3H
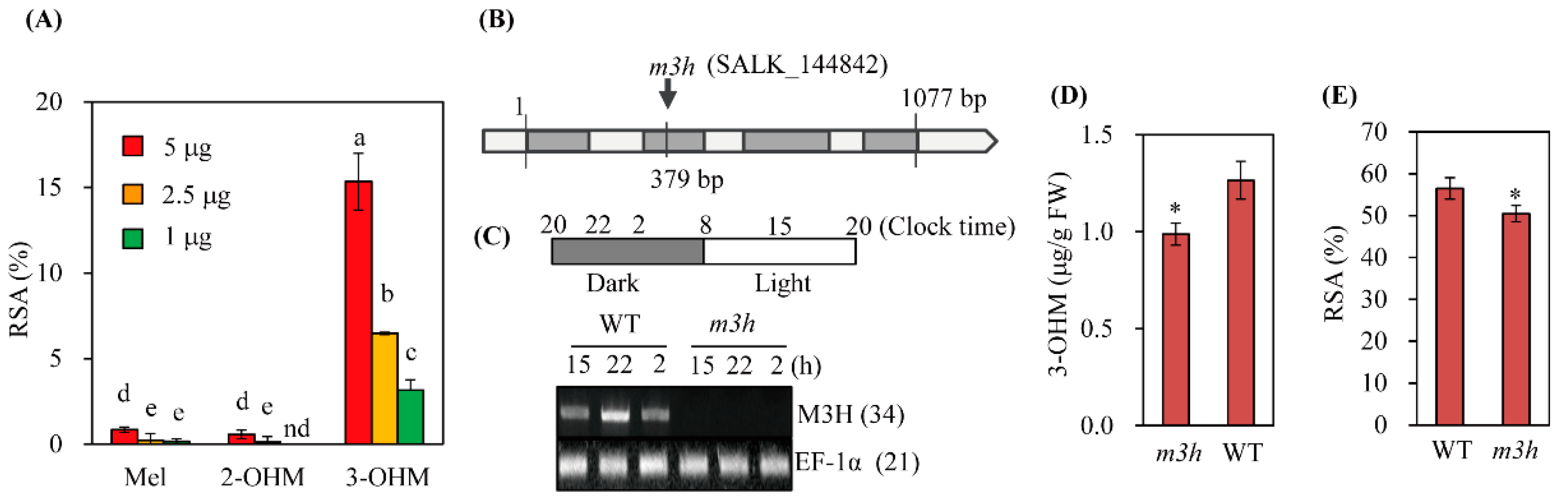
3.5. The Knockout Mutant of M3H (m3h) had Low Antioxidant Activity
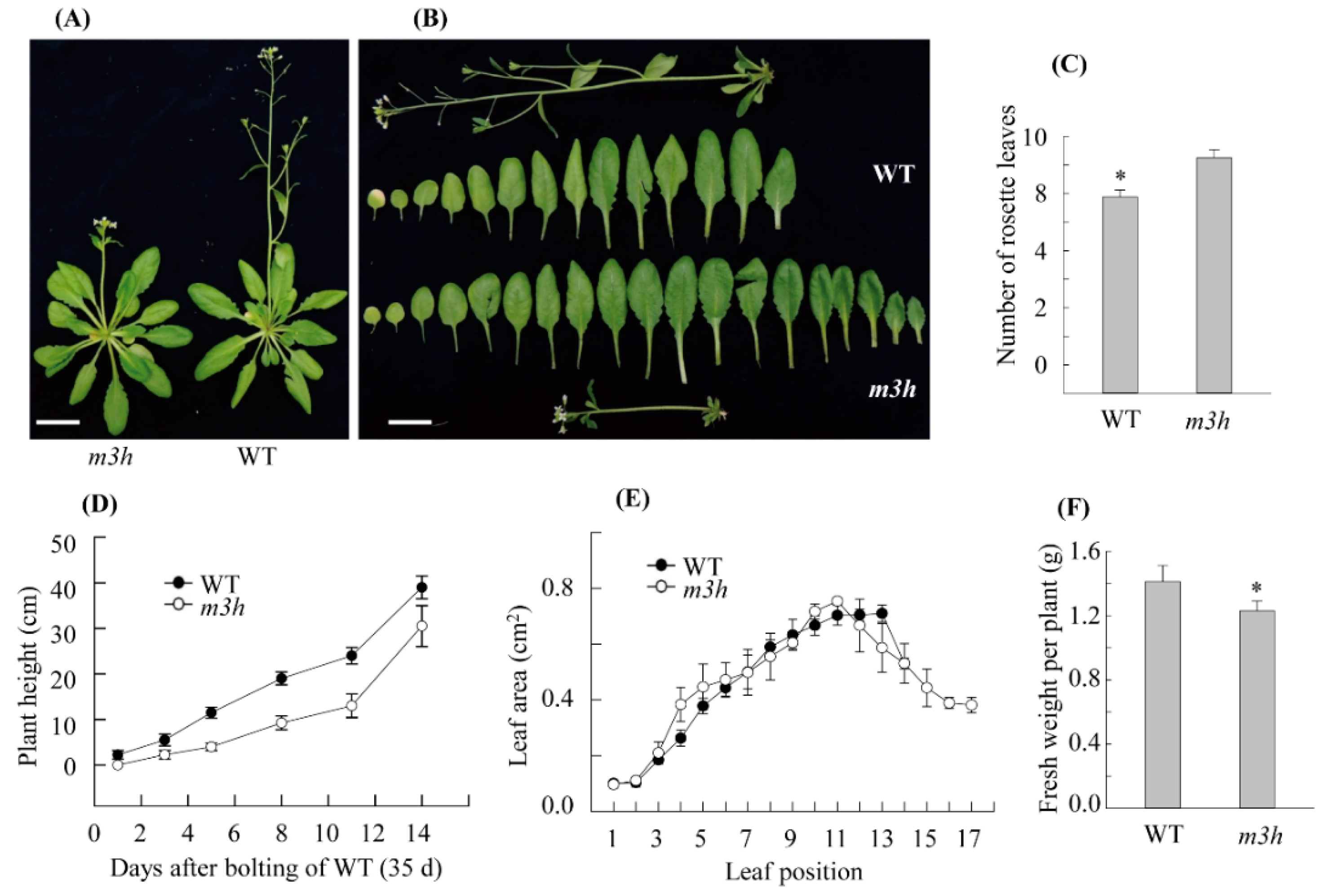
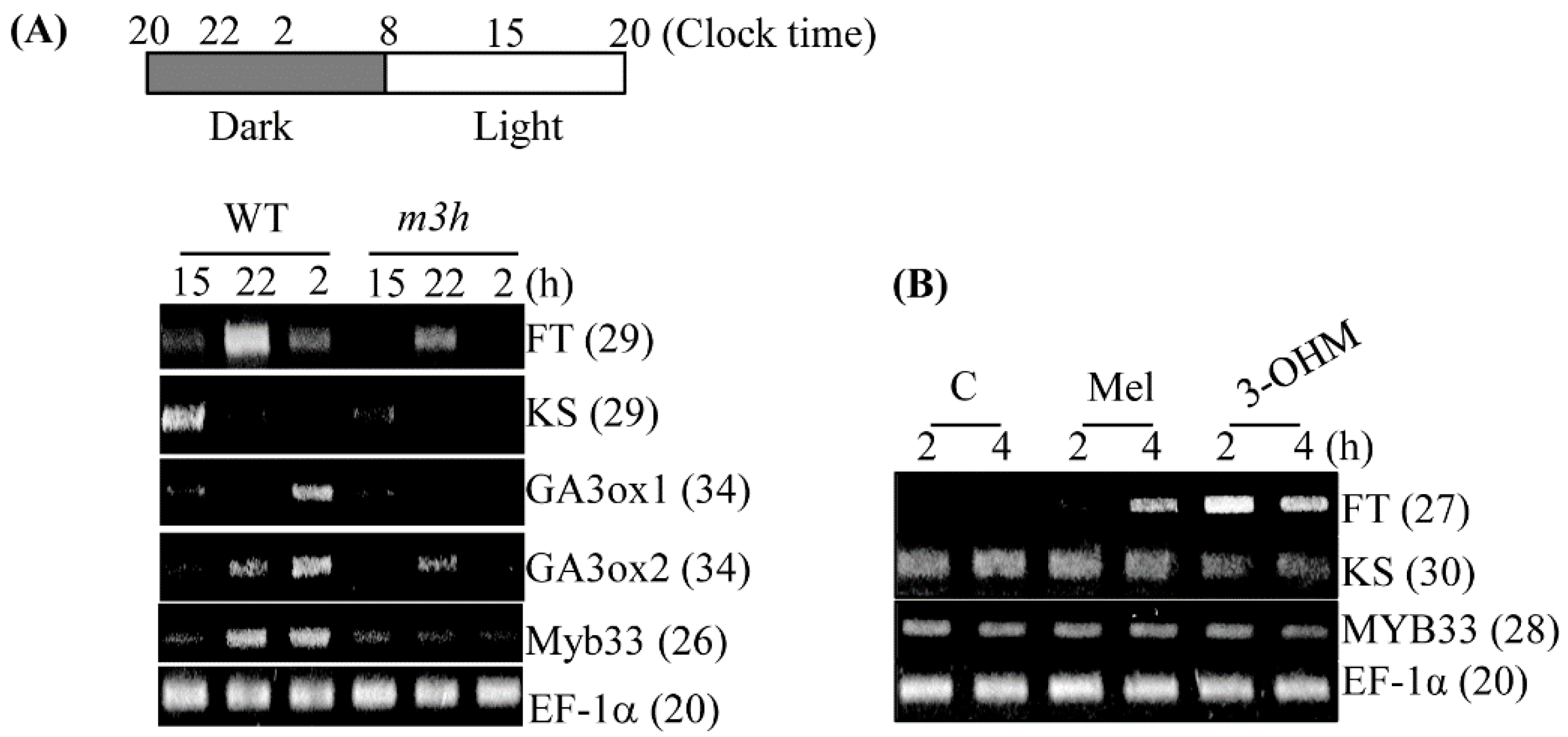
3.6. The m3h had Delayed Flowering and Retarded Growth
3.7. Stress Tolerance of m3h
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dobrikova, A.G. Signaling molecules in plants: Exogenous application. Acta Sci. Agric. 2017, 1, 38–41. [Google Scholar]
- Wei, J.; Li, D.X.; Zhang, J.R.; Shan, C.; Rengel, Z.; Song, Z.B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Back, K. The phytomelatonin receptor (PMRT1) Arabidopsis Cand2 is not a bona fide G protein-coupled melatonin receptor. Melatonin Res. 2020, 3, 177–186. [Google Scholar] [CrossRef]
- Tan, D.X.; Reiter, R.J. An evolutionary view of melatonin synthesis and metabolism related to its biological functions in plant. J. Exp. Bot. 2020, 71, 4677–4689. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Mitogen-activated protein kinase pathways are required for melatonin-mediated defense responses in plants. J. Pineal Res. 2016, 60, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Back, K. Melatonin induction and its role in high light stress tolerance in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12504. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin regulates chloroplast protein quality control via a mitogen-activated protein kinase signaling pathway. Antioxidants 2021, 10, 511. [Google Scholar] [CrossRef]
- Yang, Q.; Peng, Z.; Ma, W.; Zhang, S.; Hou, S.; Wei, J.; Dong, S.; Yu, X.; Song, Y.; Gao, W.; et al. Melatonin functions in priming of stomatal immunity in Panax notoginseng and Arabidopsis thaliana. Plant Physiol. 2021, 187, 2837–2851. [Google Scholar] [CrossRef]
- Shi, H.; Chen, Y.; Tan, D.X.; Reiter, R.J.; Chan, Z.; He, C. Melatonin induces nitric oxide and the potential mechanisms relate to innate immunity against bacterial pathogen infection in Arabidopsis. J. Pineal Res. 2015, 59, 102–108. [Google Scholar] [CrossRef]
- Madigan, A.P.; Harris, C.; Bedon, F.; Franks, A.E.; Plummer, K.M. High doses of melatonin confer abiotic stress tolerance to phytopathogenic fungi grown in vitro. Melatonin Res. 2020, 3, 187–193. [Google Scholar] [CrossRef]
- Ali, M.; Lamin-Samu, A.T.; Muhammad, I.; Farghal, M.; Khattak, A.M.; Jan, I.; Haq, S.U.; Khan, A.; Gong, Z.-H.; Lu, G. Melatonin mitigates the infection of Colletotrichum gloeosporioides via modulation of the chitinase gene and antioxidant activity in Capsicum annuum L. Antioxidants 2021, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, L.; Gu, P.; Zhan, X.; Zhang, Y.; Hou, C.; Wu, Z.; Wu, Y.F.; Wang, Q.C. Exogenous application of melatonin improves plant resistance to virus infection. Plant Pathol. 2019, 68, 1287–1295. [Google Scholar] [CrossRef]
- Bajwa, V.S.; Shukla, M.R.; Sherif, S.M.; Murch, S.J.; Saxena, P.K. Role of melatonin in alleviating cold stress in Arabidopsis thaliana. J. Pineal Res. 2014, 56, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, R.; Pareek, S.; Domínguez-Avila, J.A.; Gonzalez-Aguilar, G.A.; Valero, D.; Serrano, M. An exogenous pre-storage melatonin alleviates chilling injury in some mango fruit cultivars, by acting on the enzymatic and non-enzymatic antioxidant system. Antioxidants 2022, 11, 384. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Li, C.; Wei, Z.; Liang, D.; Ma, F. Long-term exogenous application of melatonin delays drought-induced leaf senescence in apple. J. Pineal Res. 2013, 54, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Wang, G.Y.; Muhammad, I.; Chi, Y.X.; Zeeshan, M.; Nasar, J.; Zhou, X.B. Interactive effects of melatonin and nitrogen improve drought tolerance of maize seedlings by regulating growth and physiological attributes. Antioxidants 2022, 11, 359. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zheng, G.; Li, W.; Wang, Y.; Hu, B.; Wang, H.; Wu, H.; Qian, Y.; Zhu, X.G.; Tan, D.X.; et al. Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J. Pineal Res. 2015, 59, 91–101. [Google Scholar] [CrossRef]
- Moustafa-Farag, M.; Elkelish, A.; Dafea, M.; Khan, M.; Arnao, M.B.; Abdelhamid, M.T.; El-Ezz, A.A.; Almoneafy, A.; Mahmoud, A.; Awad, M.; et al. Role of melatonin in plant tolerance to soil stressors: Salinity, pH and heavy metals. Molecules 2020, 25, 5359. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, Y.; Ma, X.; Ouyang, Z.; Deng, L.; Shen, S.; Dong, X.; Du, N.; Dong, H.; Guo, Z.; et al. Melatonin alleviates copper toxicity via improving ROS metabolism and antioxidant defense response in tomato seedlings. Antioxidants 2022, 11, 758. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and reactive oxygen and nitrogen species: A model for the plant redox network. Melatonin Res. 2019, 2, 152–168. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.X.; Zhou, Z.; Cruz, M.H.C.; Fuentes-Broto, L.; Galano, A. Phytomelatonin: Assisting plants to survive and thrive. Molecules 2015, 20, 7396–7437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Chen, J.; Ma, Y.; Huang, M.; Qiu, T.; Bian, H.; Han, N.; Wang, J. Function, mechanism, and application of plant melatonin: An update with a focus on the cereal crop, barley (Hordeum vulgare L.). Antioxidants 2022, 11, 634. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin in the evolution of plants and other phototrophs. Melatonin Res. 2019, 2, 10–36. [Google Scholar] [CrossRef]
- Tan, D.X.; Manchester, L.C.; Liu, X.; Rosales-Corral, S.A.; Acuna-Castroviejo, D.; Reiter, R.J. Mitochondria and chloroplasts as the original sites of melatonin synthesis: A hypothesis related to melatonin’s primary function and evolution in eukaryotes. J. Pineal Res. 2013, 54, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Back, K. Melatonin metabolism, signaling and possible roles in plants. Plant J. 2021, 105, 376–391. [Google Scholar] [CrossRef]
- Lee, K.; Choi, G.H.; Back, K. Cadmium-induced melatonin synthesis in rice requires light, hydrogen peroxide, and nitric oxide: Key regulatory roles for tryptophan decarboxylase and caffeic acid O-methyltransferase. J. Pineal Res. 2017, 63, e12441. [Google Scholar] [CrossRef]
- Byeon, Y.; Back, K. Molecular cloning of melatonin 2-hydroxylase responsible for 2-hydroxymelatonin production in rice (Oryza sativa). J. Pineal Res. 2015, 58, 343–351. [Google Scholar] [CrossRef]
- Lee, H.J.; Back, K. 2-Hydroxymelatonin confers tolerance against combined cold and drought stress in tobacco, tomato, and cucumber as a potent anti-stress compound in the evolution of land plants. Melatonin Res. 2019, 2, 35–46. [Google Scholar] [CrossRef]
- Shah, A.A.; Ahmed, S.; Ali, A.; Yasin, N.A. 2-Hydroxymelatonin mitigates cadmium stress in Cucumis sativus seedlings: Modulation of antioxidant enzymes and polyamines. Chemosphere 2020, 243, 125308. [Google Scholar] [CrossRef]
- Lee, K.; Zawadzka, A.; Czarnocki, Z.; Reiter, R.J.; Back, K. Molecular cloning of melatonin 3-hydroxylase and its production of cyclic 3-hydroxymelatonin in rice (Oryza sativa). J. Pineal Res. 2016, 61, 470–478. [Google Scholar] [CrossRef]
- Choi, G.H.; Back, K. Cyclic 3-hydroxymelatonin exhibits diurnal rhythm and cyclic 3-hydroxymelatonin overproduction increases secondary tillers in rice by upregulating MOC1 expression. Melatonin Res. 2019, 2, 120–138. [Google Scholar] [CrossRef]
- Seki, M.; Narusaka, M.; Kamiya, A.; Ishida, J.; Satou, M.; Sakurai, T.; Nakajima, M.; Enju, A.; Akiyama, K.; Oono, Y.; et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science 2002, 296, 141–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, K.; Kim, Y.S.; Park, S.; Back, K. Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 2009, 150, 1380–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.X.; Hardeland, R.; Manchester, L.C.; Galano, A.; Reiter, R.J. Cyclic-3-hydroxymelatonin (C3HOM), a potent antioxidant, scavenges free radicles and suppresses oxidative reactions. Curr. Med. Chem. 2014, 21, 1557–1565. [Google Scholar] [CrossRef]
- Galano, A.; Tan, D.X.; Reiter, R.J. Cyclic 3-hydroxymelatonin, a key metabolite enhancing the peroxyl radical scavenging activity of melatonin. RSC Adv. 2014, 4, 5220–5227. [Google Scholar] [CrossRef]
- Pérez-González, A.; Galano, A.; Alvarez-Idaboy, J.R.; Tan, D.X.; Reiter, R.J. Radical-trapping and preventive antioxidant effects of 2-hydroxymelatoin and 4-hydroxymelatonin: Contributions to the melatonin protection against oxidative stress. Biochem. Biophys. Acta 2017, 1861, 2206–2217. [Google Scholar] [CrossRef]
- Lee, H.Y.; Byeon, Y.; Tan, D.X.; Reiter, R.J.; Back, K. Arabidopsis serotonin N-acetyltransferase knockout mutant plants exhibit decreased melatonin and salicylic acid levels resulting in susceptibility to an avirulent pathogen. J. Pineal Res. 2015, 58, 291–299. [Google Scholar] [CrossRef]
- Hardeland, R. Taxon- and site-specific melatonin catabolism. Molecules 2017, 22, 2015. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.X.; Manchester, L.C.; Mascio, P.; Martinez, G.R.; Prado, F.M.; Reiter, R.J. Novel rhythms of N1-acetyl-N2-formyl-5-methoxykynuramine and its precursor melatonin in water hyacinth: Importance for phytoremediation. FASEB J. 2007, 21, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Tan, D.X.; Reiter, R.J.; Back, K. Predominance of 2-hydroxymelatonin over melatonin in plants. J. Pineal Res. 2015, 59, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yue, Q.; Xiang, G.; Bian, F.; Yao, Y. Melatonin promotes ripening of grape berry via increasing the levels of ABA, H2O2, and particularly ethylene. Hortic. Res. 2018, 5, 41. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Back, K. 2-Hydroxymelatonin promotes the resistance of rice plant to multiple simultaneous abiotic stresses (combined cold and drought). J. Pineal Res. 2016, 61, 303–316. [Google Scholar] [CrossRef]
- Shah, A.A.; Ahmed, S.; Yasin, N.A. 2-Hydroxymelatonin induced nutritional orchestration in Cucumis sativus under cadmium toxicity: Modulation of non-enzymatic antioxidants and gene expression. Int. J. Phytoremediat. 2020, 22, 497–570. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. 2-Hydroxymelatonin, rather than melatonin, is responsible for RBOH-dependent reactive oxygen species production leading to premature senescence in plants. Antioxidant 2021, 10, 1782. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. 2-Hydroxymelatonin promotes seed germination by increasing reactive oxygen species production and gibberellin synthesis in Arabidopsis thaliana. Antioxidant 2022, 11, 737. [Google Scholar] [CrossRef]
- Choi, G.H.; Back, K. Suppression of melatonin 2-hydroxylase increases melatonin production leading to the enhanced abiotic stress tolerance against cadmium, senescence, salt, and tunicamycin in rice plants. Biomolecules 2019, 9, 589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotchoni, S.O.; Larrimore, K.E.; Mukherjee, M.; Kempinski, C.F.; Barth, C. Alterations in the endogenous ascorbic acid content affect flowering time in Arabidopsis. Plant Physiol. 2009, 149, 803–815. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Hwang, O.J.; Back, K. Phytomelatonin as a signaling molecule for protein quality control via chaperone, autophagy, and ubiquitin–proteasome systems in plants. J. Exp. Bot. 2022. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Cano, A.; Hernández-Ruiz, J. Phytomelatonin: An unexpected molecule with amazing performance in plants. J. Exp. Bot. 2022. [Google Scholar] [CrossRef]
- Lee, K.; Back, K. Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62, e12392. [Google Scholar] [CrossRef]
- Huangfu, L.; Chen, R.; Lu, Y.; Zhang, E.; Miao, J.; Zuo, Z.; Zhao, Y.; Zhu, M.; Zhang, Z.; Li, P.; et al. OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol. J. 2022, 20, 1122–1139. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.Y.; Back, K. The Antioxidant Cyclic 3-Hydroxymelatonin Promotes the Growth and Flowering of Arabidopsis thaliana. Antioxidants 2022, 11, 1157. https://doi.org/10.3390/antiox11061157
Lee HY, Back K. The Antioxidant Cyclic 3-Hydroxymelatonin Promotes the Growth and Flowering of Arabidopsis thaliana. Antioxidants. 2022; 11(6):1157. https://doi.org/10.3390/antiox11061157
Chicago/Turabian StyleLee, Hyoung Yool, and Kyoungwhan Back. 2022. "The Antioxidant Cyclic 3-Hydroxymelatonin Promotes the Growth and Flowering of Arabidopsis thaliana" Antioxidants 11, no. 6: 1157. https://doi.org/10.3390/antiox11061157
APA StyleLee, H. Y., & Back, K. (2022). The Antioxidant Cyclic 3-Hydroxymelatonin Promotes the Growth and Flowering of Arabidopsis thaliana. Antioxidants, 11(6), 1157. https://doi.org/10.3390/antiox11061157






