The NIN-Like Protein OsNLP2 Negatively Regulates Ferroptotic Cell Death and Immune Responses to Magnaporthe oryzae in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. ΔOsnlp2 Mutant Genotyping
2.3. Fungal Cultures and Growth Conditions
2.4. Fungal Inoculation and Infection Evaluation
2.5. Cloning and Plasmid Construction
2.6. Agrobacterium Transformation, OsNLP2 Subcellular Localization, and Transient OsNLP2 Expression Assay
2.7. Complementation of OsNLP2 into ΔOsnlp2 Mutant
2.8. RT-PCR and Real-Time qRT-PCR Analyses
2.9. ROS Detection and Quantification
2.10. Prussian Blue Staining for Ferric Ion Detection
2.11. Lipid Peroxidation Assay
2.12. Treatment with Deferoxamine, Ferrostatin-1, Cytochalasin A, and Diphenyleneiodonium
3. Results
3.1. Identification of OsNLP2, ΔOsnlp2 Mutant, and OsNLP2 Complementation Plants
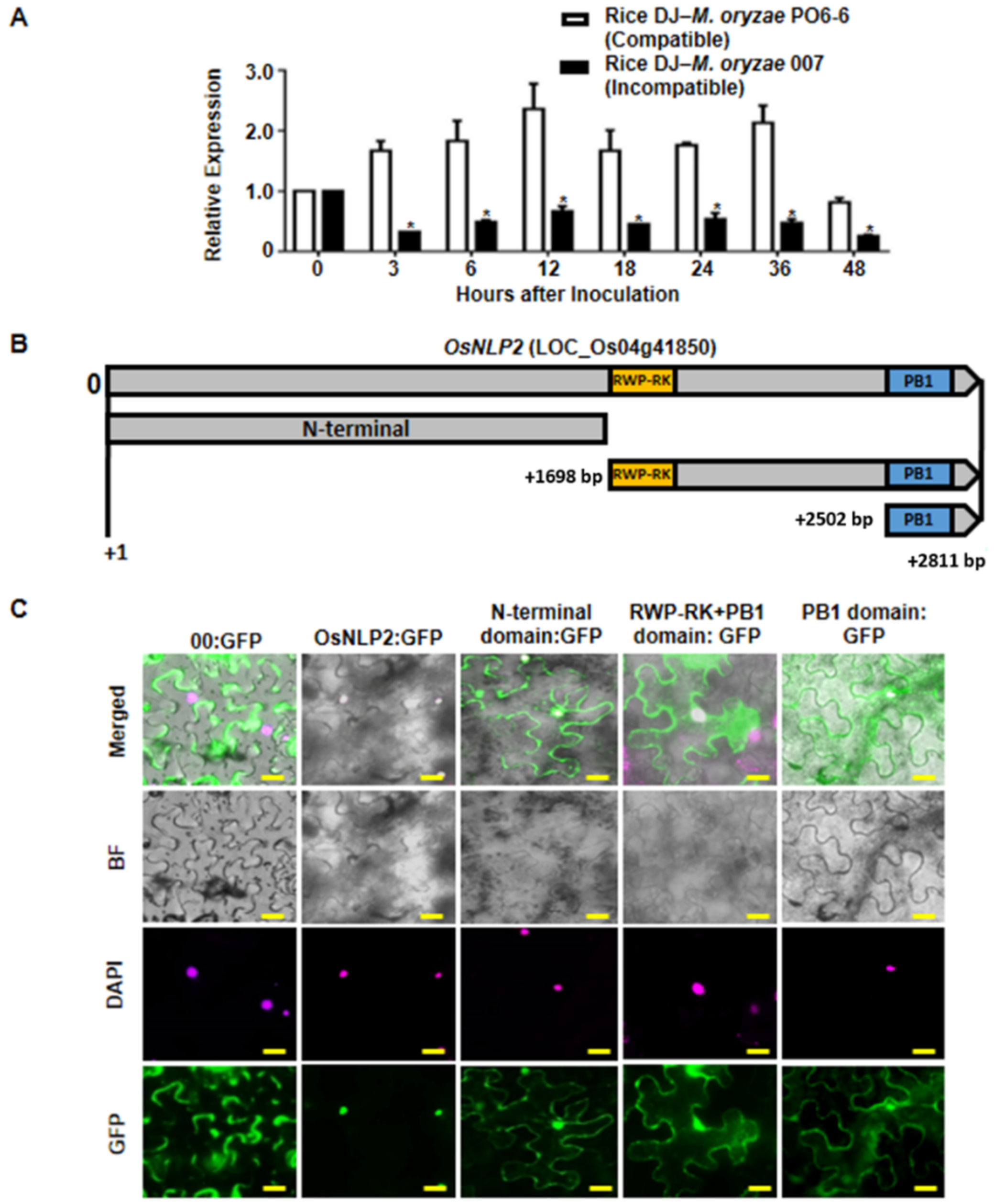
3.2. OsNLP2 Expression Patterns in Rice–M. oryzae Interactions
3.3. Subcellular Localization of OsNLP2
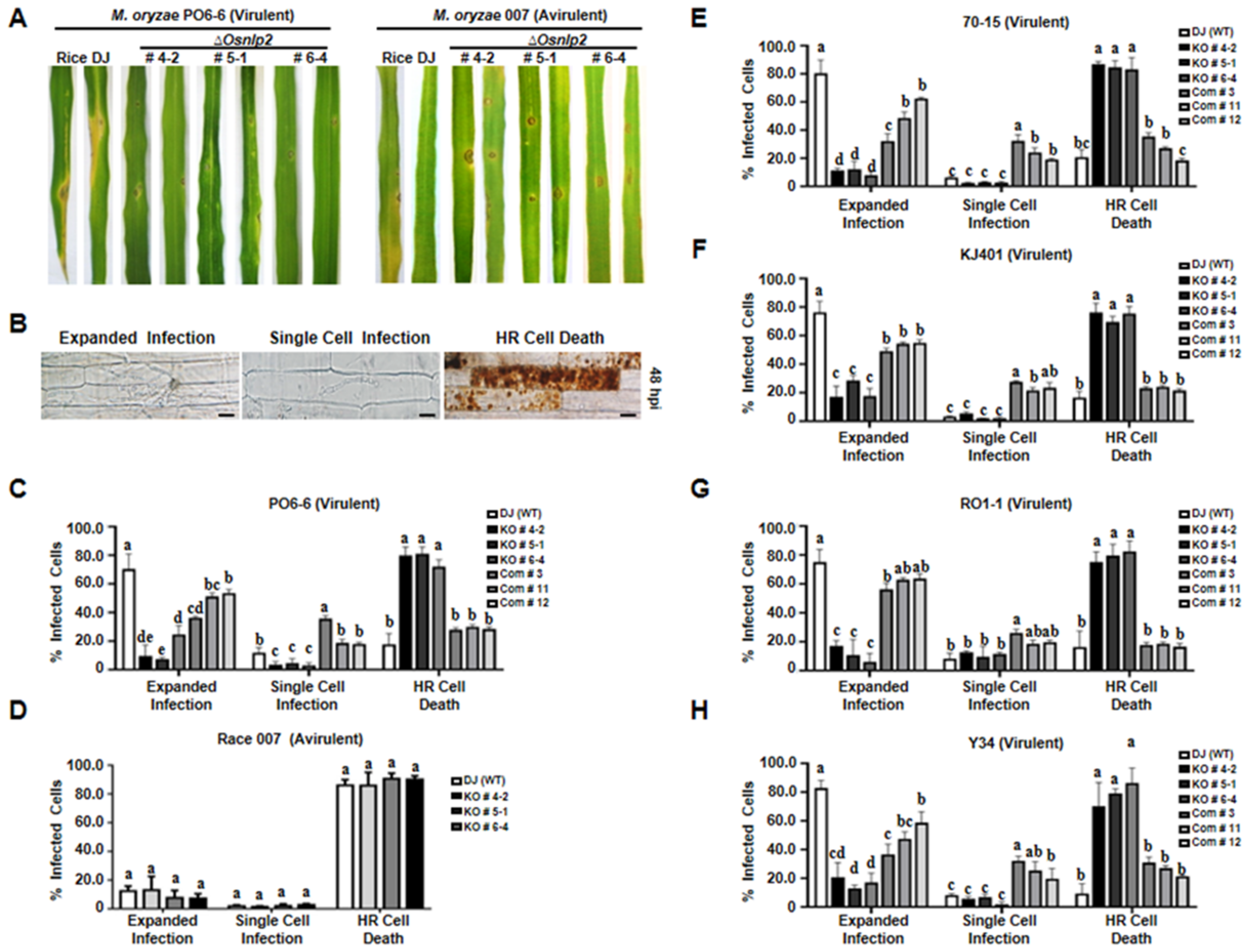
3.4. ΔOsNLP2 Mutation Triggers Hypersensitive Resistance Reactions against Different M. oryzae Strains Infection
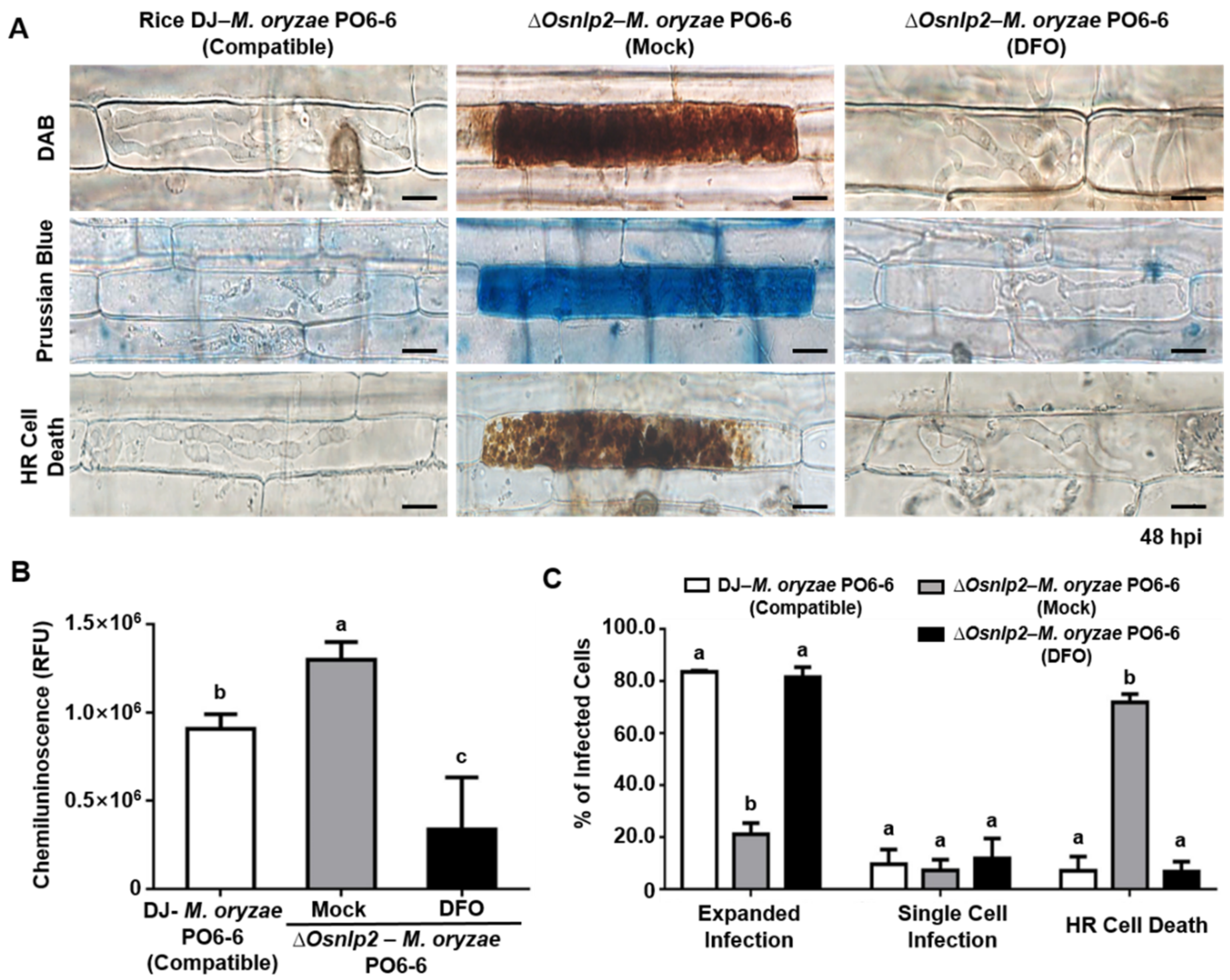
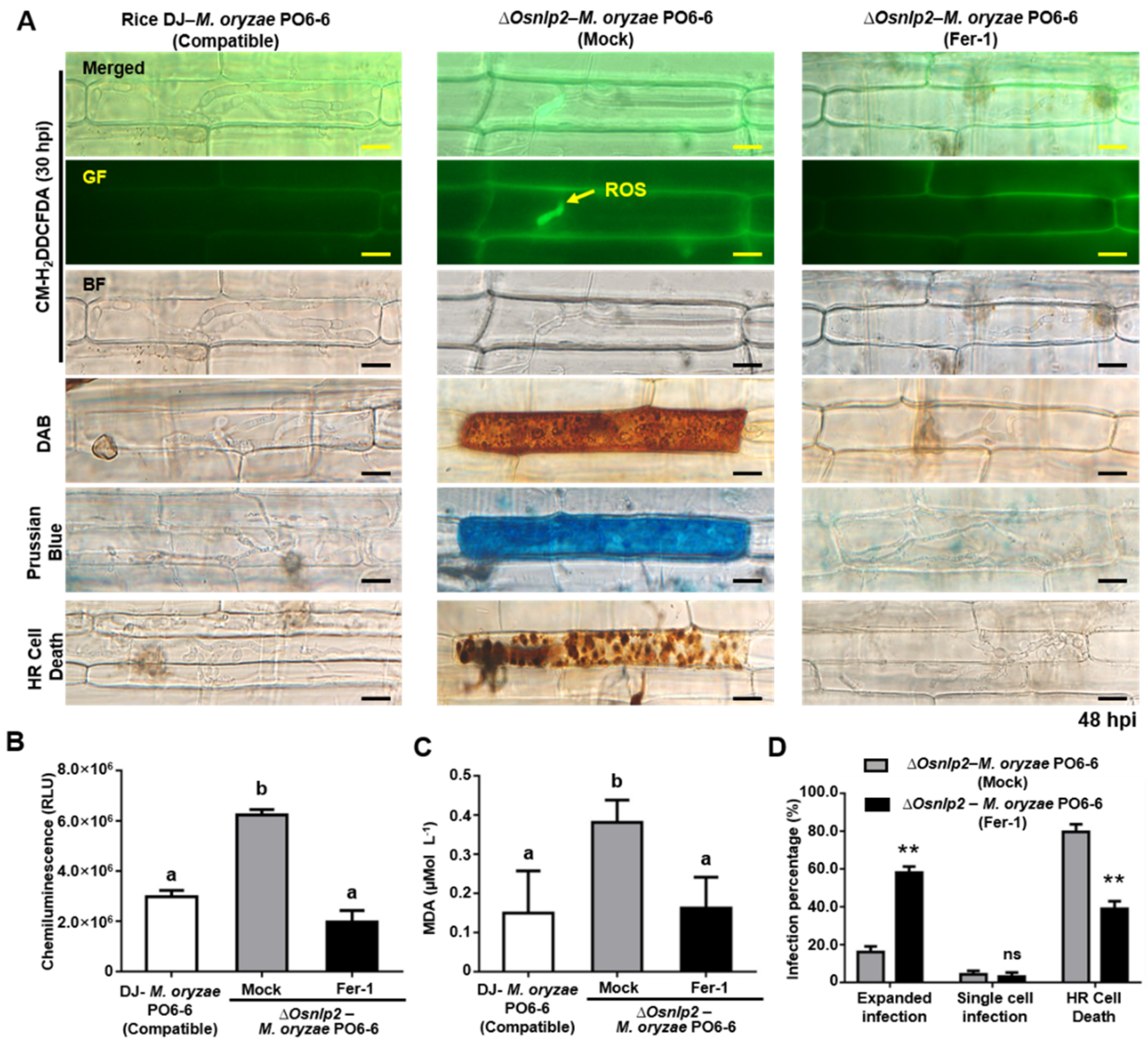
3.5. ROS-and Iron-Dependent Ferroptotic Cell Death in ΔOsnlp2 Mutants in Response to M. oryzae
3.6. Iron Chelator DFO and Ferroptosis Inhibitor Fer-1 Suppress ROS-and Iron-Dependent Ferroptotic Cell Death in ΔOsnlp2 Mutants in Response to M. oryzae
3.7. The Actin Polymerization Inhibitor Cyt A and NADPH Oxidase Inhibitor Diphenyleneiodonium (DPI) Suppress ROS and Ferric Ion Accumulation and HR Cell Death in ΔOsnlp2 Leaf Sheaths in Response to M. oryzae
3.8. Time-Course Expression of Defense-Related Genes in ΔOsnlp2 Mutant Cells during M. oryzae Infection
4. Discussion
4.1. Subcellular Localization and Functional Analysis of OsNLP2 Domains
4.2. OsNLP2 Expression Positively Regulates Susceptible Disease Development
4.3. ΔOsnlp2 Mutants Exhibit ROS-and Iron-Dependent Ferroptotic HR Cell Death Responses to M. oryzae Infection
4.4. DFO, Fer-1, Cyt A, and DPI Inhibit ROS-and Iron-Dependent Ferroptotic Cell Death in ΔOsnlp2 Mutants
4.5. Broad-Spectrum, Nonspecific Resistance of ΔOsnlp2 Mutants to Different M. oryzae Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.S.; Hwang, I.S.; Hwang, B.K. Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 2012, 24, 1675–1690. [Google Scholar] [CrossRef] [PubMed]
- Jwa, N.S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar] [CrossRef] [PubMed]
- Dangol, S.; Chen, Y.; Hwang, B.K.; Jwa, N.S. Iron- and reactive oxygen species-dependent ferroptotic cell death in rice-Magnaporthe oryzae interactions. Plant Cell 2019, 31, 189–209. [Google Scholar] [CrossRef] [PubMed]
- Dangol, S.; Nguyen, N.K.; Singh, R.; Chen, Y.; Wang, J.; Lee, H.G.; Hwang, B.K.; Jwa, N.S. Mitogen-activated protein kinase OsMEK2 and OsMPK1 signaling is required for ferroptotic cell death in rice-Magnaporthe oryzae interactions. Front. Plant Sci. 2021, 12, 710794. [Google Scholar] [CrossRef]
- Meng, Q.; Gupta, R.; Min, C.W.; Kwon, S.W.; Wang, Y.; Je, B.I.; Kim, Y.J.; Jeon, J.S.; Agrawal, G.K.; Rakwal, R.; et al. Proteomics of rice-Magnaporthe oryzae interaction: What have we learned so far? Front. Plant Sci. 2019, 10, 1383. [Google Scholar] [CrossRef]
- Sánchez-Sanuy, F.; Peris-Peris, C.; Tomiyama, S.; Okada, K.; Hsing, Y.I.; San Segundo, B.; Campo, S. Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus. BMC Plant Biol. 2019, 19, 563. [Google Scholar] [CrossRef]
- Chen, Y.; Dangol, S.; Wang, J.; Jwa, N.S. Focal accumulation of ROS can block Pyricularia oryzae effector BAS4-expression and prevent infection in rice. Int. J. Mol. Sci. 2020, 21, 6196. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Cao, J.Y.; Dixon, S.J. Mechanisms of ferroptosis. Cell Mol. Life Sci. 2016, 73, 2195–2209. [Google Scholar] [CrossRef] [Green Version]
- Fenton, H.J.H. Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 1894, 65, 899–910. [Google Scholar] [CrossRef]
- Pierre, J.L.; Fontecave, M. Iron and activated oxygen species in biology: The basic chemistry. Biometals. 1999, 12, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Zaho, M.J.; Jung, L.; Tanielian, C.; Mechin, R. Kinetics of the competitive degradation of deoxyribose and other biomolecules by hydroxyl radicals produced by the Fenton reaction. Free Radic. Res. 1994, 20, 345–363. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef]
- Parker, D.; Beckmann, M.; Zubair, H.; Enot, D.P.; Caracuel-Rios, Z.; Overy, D.P.; Snowdon, S.; Talbot, N.J.; Draper, J. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009, 59, 723–737. [Google Scholar] [CrossRef]
- Singh, R.; Dangol, S.; Chen, Y.; Choi, J.; Cho, Y.S.; Lee, J.E.; Choi, M.O.; Jwa, N.S. Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-Malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol. Cells 2016, 39, 426–438. [Google Scholar] [CrossRef]
- Takahashi, S.; Kimura, S.; Kaya, H.; Iizuka, A.; Wong, H.L.; Shimamoto, K.; Kuchitsu, K. Reactive oxygen species production and activation mechanism of the rice NADPH oxidase OsRbohB. J. Biochem. 2012, 152, 37–43. [Google Scholar] [CrossRef]
- Mur, L.A.J.; Simpson, C.; Kumari, A.; Gupta, A.K.; Gupta, K.J. Moving nitrogen to the centre of plant defence against pathogens. Ann. Bot. 2017, 119, 703–709. [Google Scholar] [CrossRef]
- Gupta, K.J.; Brotman, Y.; Segu, S.; Zeier, T.; Zeier, J.; Persijn, S.T.; Cristescu, S.M.; Harren, F.J.; Bauwe, H.; Fernie, A.R.; et al. The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 2013, 64, 553–568. [Google Scholar] [CrossRef]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef] [Green Version]
- Chardin, C.; Girin, T.; Roudier, F.; Meyer, C.; Krapp, A. The plant RWP-RK transcription factors: Key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 2014, 65, 5577–5587. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Z.S.; Xia, J.Q.; Alfatih, A.; Song, Y.; Huang, Y.J.; Wan, G.Y.; Sun, L.Q.; Tang, H.; Liu, Y.; et al. Rice NIN-LIKE PROTEIN 4 plays a pivotal role in nitrogen use efficiency. Plant Biotechnol. J. 2021, 19, 448–461. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. The role of protein-protein interactions mediated by the PB1 domain of NLP transcription factors in nitrate-inducible gene expression. BMC Plant Biol. 2019, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, F.; Rizzo, P.; Rutten, T.; Altschmied, L.; Bäumlein, H. RWP-RK domain-containing transcription factors control cell differentiation during female gametophyte development in Arabidopsis. New Phytol. 2017, 213, 1909–1924. [Google Scholar] [CrossRef]
- Koszegi, D.; Johnston, A.J.; Rutten, T.; Czihal, A.; Altschmied, L.; Kumlehn, J.; Wüst, S.E.; Kirioukhova, O.; Gheyselinck, J.; Grossniklaus, U.; et al. Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 2011, 67, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, W.; Yang, Y.; Li, Z.; Li, N.; Qi, S.; Crawford, N.M.; Wang, Y. The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci. Rep. 2018, 8, 1487. [Google Scholar] [CrossRef]
- Alfatih, A.; Wu, J.; Zhang, Z.S.; Xia, J.Q.; Jan, S.U.; Yu, L.H.; Xiang, C.B. Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J. Exp. Bot. 2020, 71, 6032–6042. [Google Scholar] [CrossRef]
- Singh, R.; Lee, M.O.; Lee, J.E.; Choi, J.; Park, J.H.; Kim, E.H.; Yoo, R.H.; Cho, J.I.; Jeon, J.S.; Rakwal, R.; et al. Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 2012, 160, 477–487. [Google Scholar] [CrossRef]
- Jeon, J.S.; Lee, S.; Jung, K.H.; Jun, S.H.; Jeong, D.H.; Lee, J.; Kim, C.; Jang, S.; Yang, K.; Nam, J.; et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000, 22, 561–570. [Google Scholar] [CrossRef]
- Kim, S.R.; Jeon, J.S.; An, G. Development of an efficient inverse PCR method for isolating gene tags from T-DNA insertional mutants in rice. Methods Mol. Biol. 2011, 678, 139–146. [Google Scholar] [CrossRef]
- Dangol, S.; Singh, R.; Chen, Y.; Jwa, N.S. Visualization of multicolored in vivo organelle markers for co-localization studies in oryza sativa. Mol Cells. 2017, 40, 828–836. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, K.A.; Jensen, P.E.; Møller, I.M.; Schulz, A. Monitoring reactive oxygen species formation and localisation in living cells by use of the fluorescent probe CM-H2DCFDA and confocal laser microscopy. Physiol. Plant 2009, 136, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Rustérucci, C.; Aviv, D.H.; Holt, B.F.; Dangl, J.L.; Parker, J.E. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 2001, 13, 2211–2224. [Google Scholar] [CrossRef]
- Liu, X.; Bai, X.; Wang, X.; Chu, C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007, 164, 969–979. [Google Scholar] [CrossRef]
- Smith, M.A.; Harris, P.L.; Sayre, L.M.; Perry, G. Iron accumulation in Alzheimer disease is a source of redox-generated free radicals. Proc. Natl. Acad. Sci. USA 1997, 18, 9866–9868. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, R.G.; Gao, Y.J.; Zheng, S.Z.; Xu, P.; Zhang, S.Q.; Sun, D.Y. Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 2009, 149, 1773–1784. [Google Scholar] [CrossRef]
- Chi, M.H.; Park, S.Y.; Kim, S.; Lee, Y.H. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009, 5, e1000401. [Google Scholar] [CrossRef]
- Mutte, S.K.; Weijers, D. Deep evolutionary history of the Phox and Bem1 (PB1) domain across eukaryotes. Sci. Rep. 2020, 10, 3797. [Google Scholar] [CrossRef] [Green Version]
- Fryer, M.J.; Oxborough, K.; Mullineaux, P.M.; Baker, N.R. Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 2002, 53, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Mylonas, C.; Kouretas, D. Lipid peroxidation and tissue damage. In Vivo 1999, 13, 295–309. [Google Scholar] [PubMed]
- Grotto, D.; Maria, L.S.; Valentini, J.; Paniz, C.; Schmitt, G.; Garcia, S.C.; Pomblum, V.J.; Rocha, J.B.T.; Farina, M. Importance of the lipid peroxidation biomarkers and methodological aspects for malondialdehyde quantification. Quimica. Nova. 2009, 32, 169–174. [Google Scholar] [CrossRef]
- Nakashita, H.; Yoshioka, K.; Takayama, M.; Kuga, R.; Midoh, N.; Usami, R.; Horikoshi, K.; Yoneyama, K.; Yamaguchi, I. Characterization of PBZ1, a probenazole-inducible gene, in suspension-cultured rice cells. Biosci. Biotechnol. Biochem. 2001, 65, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Uehlein, N.; Kaldenhoff, R.; Guo, S.; Zhu, Y.; Kai, L. Aquaporin PIP2;1 affects water transport and root growth in rice (Oryza sativa L.). Plant Physiol. Biochem. 2019, 139, 152–160. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 2016, 67, 1663–1676. [Google Scholar] [CrossRef]
- Yant, L.; Mathieu, J.; Dinh, T.T.; Ott, F.; Lanz, C.; Wollmann, H.; Chen, X.; Schmid, M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 2010, 22, 2156–2170. [Google Scholar] [CrossRef]
- Seo, E.; Choi, D. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief Funct. Genomics 2015, 14, 260–267. [Google Scholar] [CrossRef] [Green Version]
- Ho, Y.S.; Burden, L.M.; Hurley, J.H. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000, 19, 5288–5299. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Emergence of a new step towards understanding the molecular mechanisms underlying nitrate-regulated gene expression. J. Exp. Bot. 2014, 65, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Solomon, P.S.; Tan, K.C.; Oliver, R.P. The nutrient supply of pathogenic fungi; a fertile field for study. Mol. Plant Pathol. 2003, 4, 203–210. [Google Scholar] [CrossRef]
- Tavernier, V.; Cadiou, S.; Pageau, K.; Laugé, R.; Reisdorf-Cren, M.; Langin, T.; Masclaux-Daubresse, C. The plant nitrogen mobilization promoted by Colletotrichum lindemuthianum in Phaseolus leaves depends on fungus pathogenicity. J. Exp. Bot. 2007, 58, 3351–3360. [Google Scholar] [CrossRef]
- Greenberg, J.T.; Yao, N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol. 2004, 6, 201–211. [Google Scholar] [CrossRef]
- Choi, H.W.; Kim, D.S.; Kim, N.H.; Jung, H.W.; Ham, J.H.; Hwang, B.K. Xanthomonas filamentous hemagglutinin-like protein Fha1 interacts with pepper hypersensitive-induced reaction protein CaHIR1 and functions as a virulence factor in host plants. MPMI 2013, 26, 1441–1454. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Zilka, O.; Shah, R.; Li, B.; Friedmann Angeli, J.P.; Griesser, M.; Conrad, M.; Pratt, D.A. On the mechanism of cytoprotection by Ferrostatin-1 and Liproxstatin-1 and the role of lipid peroxidation in ferroptotic cell death. ACS Cent. Sci. 2017, 3, 232–243. [Google Scholar] [CrossRef]
- Cooper, J.A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987, 105, 1473–1478. [Google Scholar] [CrossRef] [Green Version]
- Mathur, J.; Spielhofer, P.; Kost, B.; Chua, N. The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 1999, 126, 5559–5568. [Google Scholar] [CrossRef]
- Yun, B.W.; Atkinson, H.A.; Gaborit, C.; Greenland, A.; Read, N.D.; Pallas, J.A.; Loake, G.J. Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 2003, 34, 768–777. [Google Scholar] [CrossRef]
- Gross, P.; Julius, C.; Schmelzer, E.; Hahlbrock, K. Translocation of cytoplasm and nucleus to fungal penetration sites is associated with depolymerization of microtubules and defence gene activation in infected, cultured parsley cells. EMBO J. 1993, 12, 1735–1744. [Google Scholar] [CrossRef]
- Skalamera, D.; Heath, M.C. Changes in the cytoskeleton accompanying infection-induced nuclear movements and the hypersensitive response in plant cells invaded by rust fungi. Plant J. 1998, 16, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Trush, M.A. Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem. Biophys. Res. Commun. 1998, 253, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, J. Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 2002, 215, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.; Brosché, M.; Kangasjärvi, J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 2003, 8, 335–342. [Google Scholar] [CrossRef]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef]
- Liu, G.; Greenshields, D.L.; Sammynaiken, R.; Hirji, R.N.; Selvaraj, G.; Wei, Y. Targeted alterations in iron homeostasis underlie plant defense responses. J. Cell Sci. 2007, 120, 596–605. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Deng, H.; Wang, S. Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 2017, 90, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 2017, 170, 114–126.e115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wang, J.; Nguyen, N.K.; Hwang, B.K.; Jwa, N.S. The NIN-Like Protein OsNLP2 Negatively Regulates Ferroptotic Cell Death and Immune Responses to Magnaporthe oryzae in Rice. Antioxidants 2022, 11, 1795. https://doi.org/10.3390/antiox11091795
Chen Y, Wang J, Nguyen NK, Hwang BK, Jwa NS. The NIN-Like Protein OsNLP2 Negatively Regulates Ferroptotic Cell Death and Immune Responses to Magnaporthe oryzae in Rice. Antioxidants. 2022; 11(9):1795. https://doi.org/10.3390/antiox11091795
Chicago/Turabian StyleChen, Yafei, Juan Wang, Nam Khoa Nguyen, Byung Kook Hwang, and Nam Soo Jwa. 2022. "The NIN-Like Protein OsNLP2 Negatively Regulates Ferroptotic Cell Death and Immune Responses to Magnaporthe oryzae in Rice" Antioxidants 11, no. 9: 1795. https://doi.org/10.3390/antiox11091795
APA StyleChen, Y., Wang, J., Nguyen, N. K., Hwang, B. K., & Jwa, N. S. (2022). The NIN-Like Protein OsNLP2 Negatively Regulates Ferroptotic Cell Death and Immune Responses to Magnaporthe oryzae in Rice. Antioxidants, 11(9), 1795. https://doi.org/10.3390/antiox11091795






