A Multifactorial Regulation of Glutathione Metabolism behind Salt Tolerance in Rice
Abstract
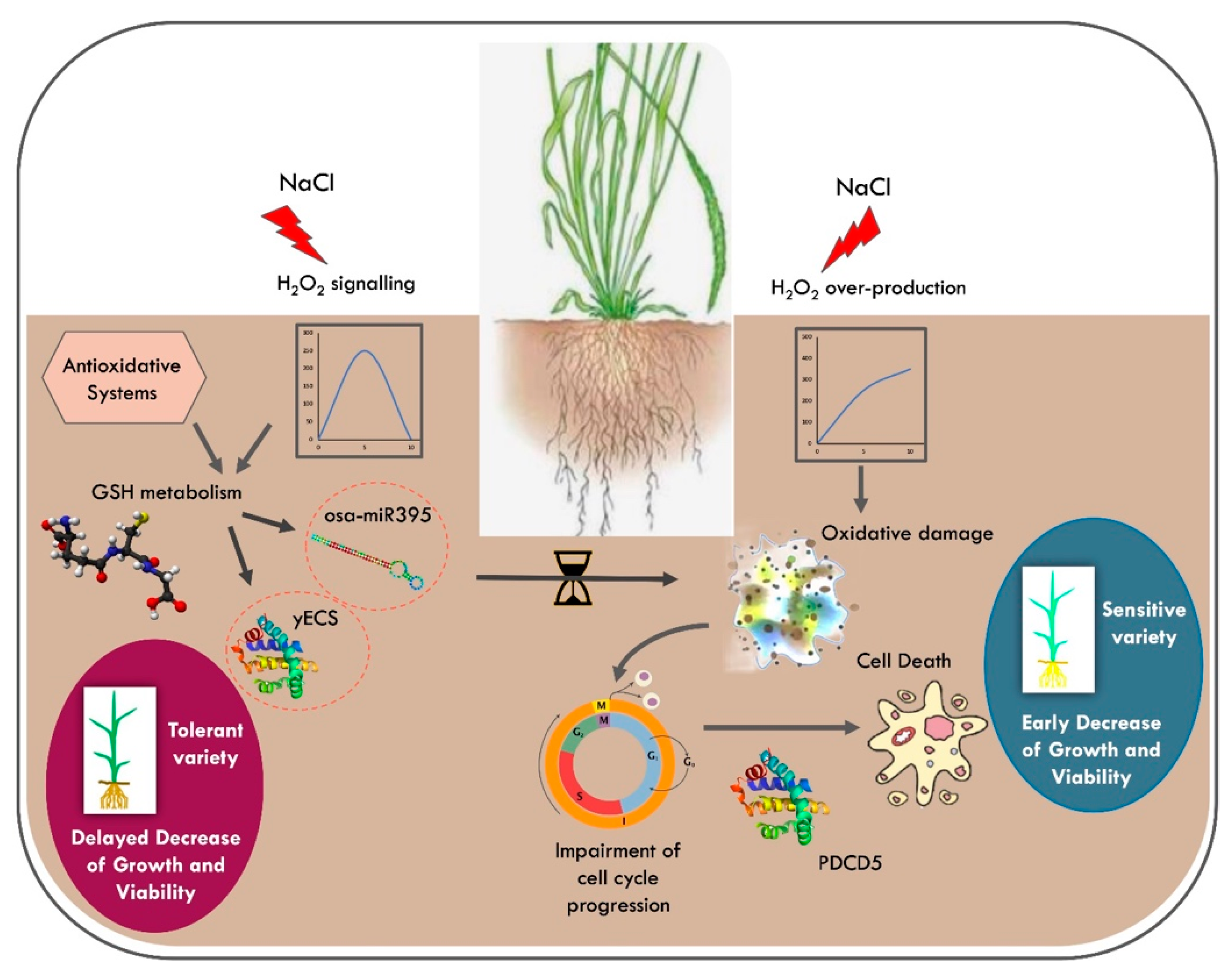
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Shoot and Root Phenotypic Characterization
2.2.1. Determination of Plant Growth
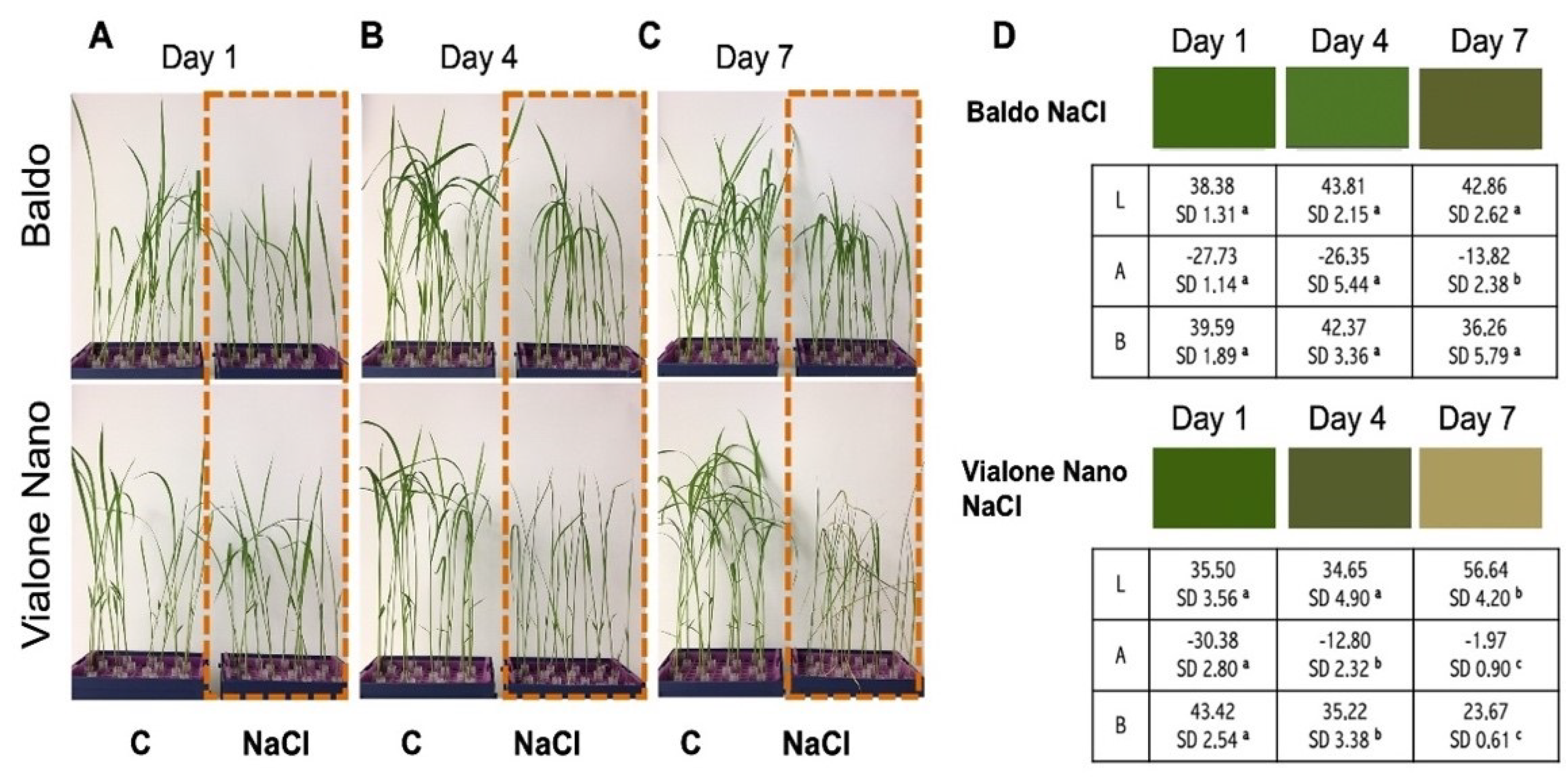
2.2.2. Leaf Color Quantification and Chlorophyll Content in Leaves
2.2.3. Cell Root Viability
2.3. Gene Expression Analysis
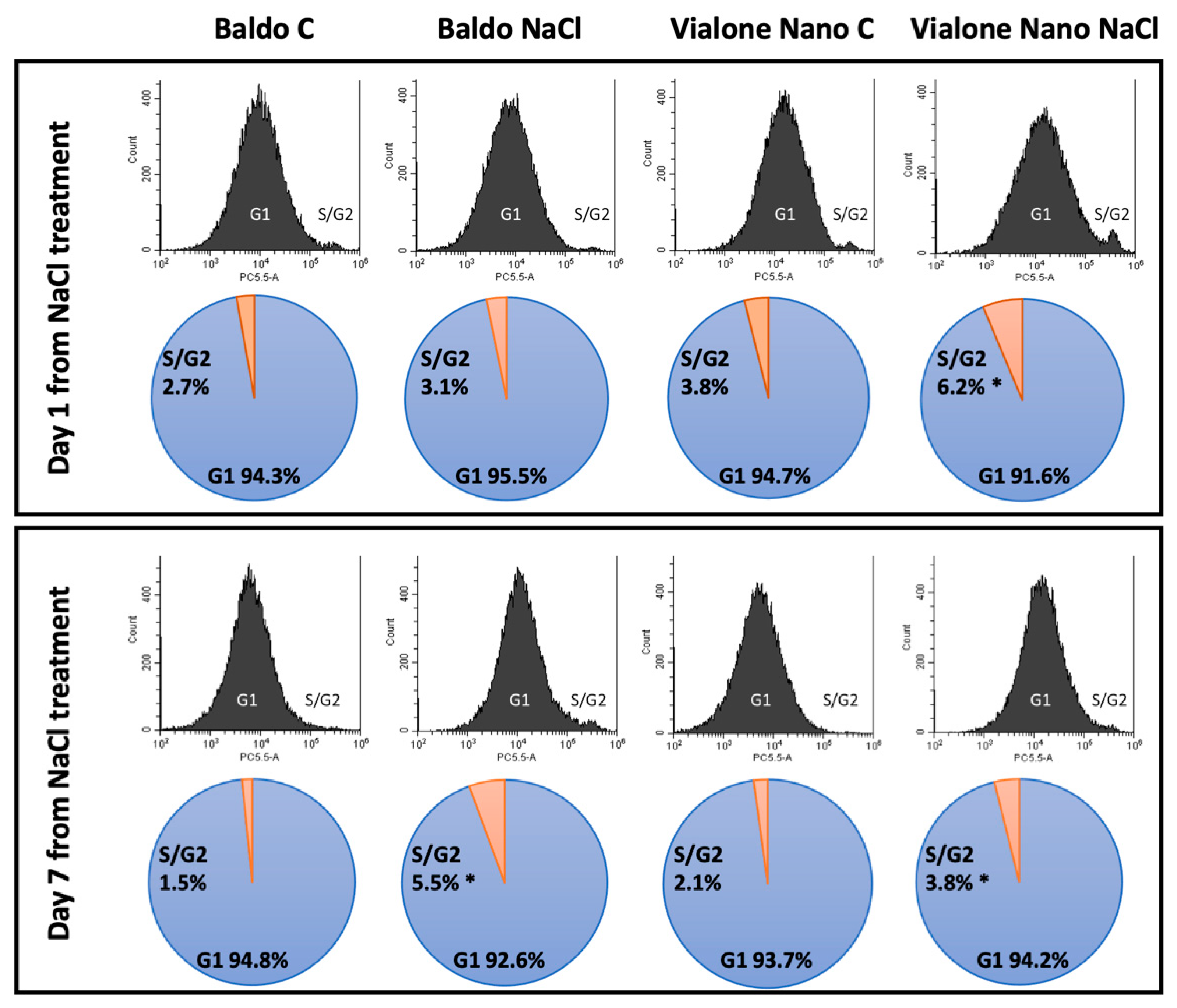
2.4. Flow Cytometry
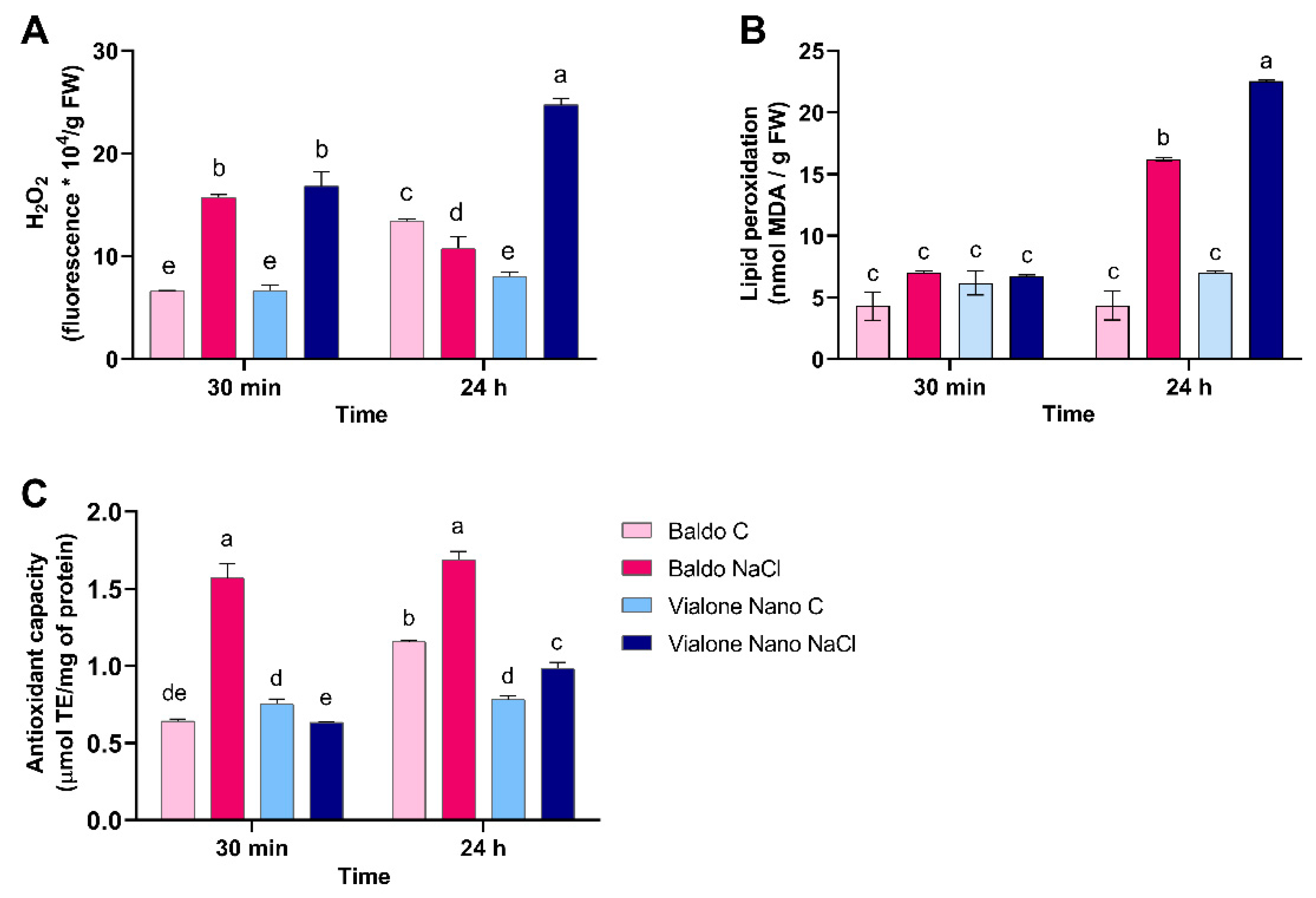
2.5. Analysis of Hydrogen Peroxide and Redox Systems
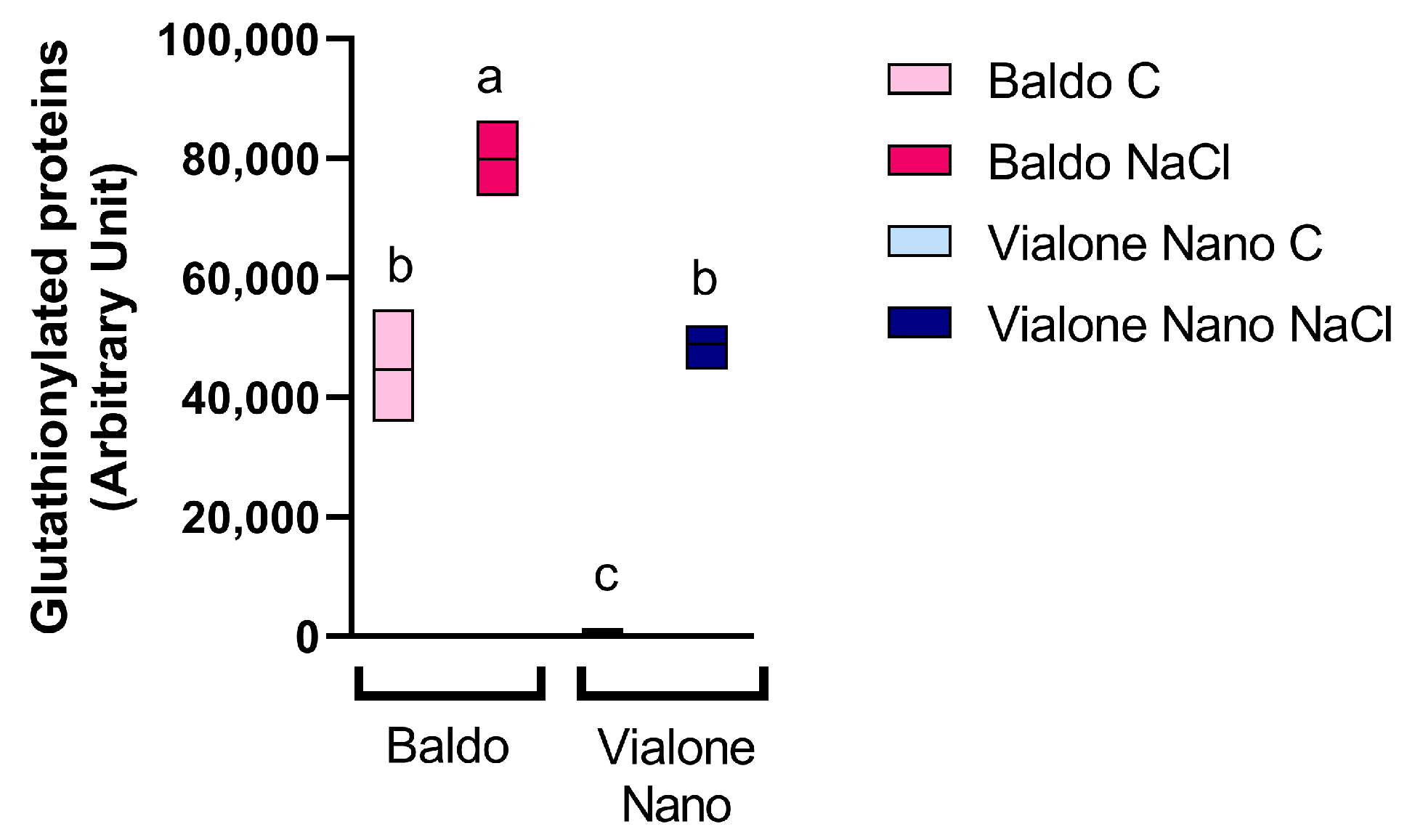
2.6. SDS/PAGE and Immunoblotting
2.7. Statistical Analysis
3. Results
3.1. Morpho-Phenotypical Characterization of Baldo and Vialone Nano Plants Showed Different Salt Susceptibility
3.2. Salt Stress Mainly Compromised the Root Meristematic Activity and Root Cell Viability in the Sensitive Variety
3.3. The Tolerant Variety Showed Enhanced Capabilities to Activate the ROS Scavenging Systems That Enabled H2O2 to Act as a Signal Molecule
3.4. Early Modulation of the Glutathione Metabolism and Sulfur Assimilation in the Tolerant Variety
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaki Mostafa Ali, F. The Determinants of Salinity Tolerance in Maize (Zea Mays L.); University of Groningen: Groningen, The Netherlands, 2011. [Google Scholar]
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R.; Sarwar, M.I. Abbas Shoukat, Maqsood Ul Hussan, Muhammad Ishtiaq Sarwar. A Review: Impact of Salinity on Plant Growth. Nat. Sci. 2019, 1, 34–40. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant Cellular and Molecular Responses to High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Guo, Y. Elucidating the Molecular Mechanisms Mediating Plant Salt-Stress Responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, T.C.; Xu, L. Sensitivity of Growth of Roots versus Leaves to Water Stress: Biophysical Analysis and Relation to Water Transport. J. Exp. Bot. 2000, 51, 1595–1616. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Dietz, K.-J. Tuning of Redox Regulatory Mechanisms, Reactive Oxygen Species and Redox Homeostasis under Salinity Stress. Front. Plant Sci. 2016, 7, 548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demidchik, V.; Cuin, T.; Svistunenko, D.; Smith, S.; Miller, A.; Shabala, S.; Sokolik, A.; Yurin, V. Arabidopsis Root K+-Efflux Conductance Activated by Hydroxyl Radicals: Single-Channel Properties, Genetic Basis and Involvement in Stress-Induced Cell Death. J. Cell Sci. 2010, 123, 1468–1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locato, V.; Gadaleta, C.; De Gara, L.; De Pinto, M.C. Production of Reactive Species and Modulation of Antioxidant Network in Response to Heat Shock: A Critical Balance for Cell Fate. Plant Cell Environ. 2008, 31, 1606–1619. [Google Scholar] [CrossRef]
- Locato, V.; Paradiso, A.; Sabetta, W.; De Gara, L.; de Pinto, M.C. Chapter Nine-Nitric Oxide and Reactive Oxygen Species in PCD Signaling. In Advances in Botanical Research; Nitric Oxide and Signaling in Plants; Wendehenne, D., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 77, pp. 165–192. [Google Scholar]
- Considine, M.J.; Foyer, C.H. Stress Effects on the Reactive Oxygen Species-Dependent Regulation of Plant Growth and Development. J. Exp. Bot. 2021, 72, 5795–5806. [Google Scholar] [CrossRef]
- Zagorchev, L.; Seal, C.E.; Kranner, I.; Odjakova, M. A Central Role for Thiols in Plant Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2013, 14, 7405–7432. [Google Scholar] [CrossRef] [Green Version]
- Fatma, M.; Asgher, M.; Masood, A.; Khan, N.A. Excess Sulfur Supplementation Improves Photosynthesis and Growth in Mustard under Salt Stress through Increased Production of Glutathione. 2014, 107, 55–63. Environ. Exp. Bot. 2014, 107, 55–63. [Google Scholar] [CrossRef]
- Fatma, M.; Masood, A.; Per, T.S.; Khan, N.A. Nitric Oxide Alleviates Salt Stress Inhibited Photosynthetic Performance by Interacting with Sulfur Assimilation in Mustard. Front. Plant Sci. 2016, 7, 521. [Google Scholar] [CrossRef] [Green Version]
- Kaur, N.; Dhawan, M.; Sharma, I.; Pati, P.K. Interdependency of Reactive Oxygen Species Generating and Scavenging System in Salt Sensitive and Salt Tolerant Cultivars of Rice. BMC Plant Biol. 2016, 16, 131. [Google Scholar] [CrossRef] [Green Version]
- Bertazzini, M.; Sacchi, G.A.; Forlani, G. A Differential Tolerance to Mild Salt Stress Conditions among Six Italian Rice Genotypes Does Not Rely on Na+ Exclusion from Shoots. J. Plant Physiol. 2018, 226, 145–153. [Google Scholar] [CrossRef]
- Formentin, E.; Sudiro, C.; Ronci, M.B.; Locato, V.; Barizza, E.; Stevanato, P.; Ijaz, B.; Zottini, M.; De Gara, L.; Lo Schiavo, F. H2O2 Signature and Innate Antioxidative Profile Make the Difference Between Sensitivity and Tolerance to Salt in Rice Cells. Front. Plant Sci. 2018, 9, 1549. [Google Scholar] [CrossRef] [Green Version]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and Cell Physiological Analyses in Different Rice Cultivars Provide New Insights Into Adaptive and Salinity Stress Responses. Front. Plant Sci. 2018, 9, 204. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Counce, P.A.; Keisling, T.C.; Mitchell, A.J. A Uniform, Objective, and Adaptive System for Expressing Rice Development. Crop Sci. 2000, 40, 436–443. [Google Scholar] [CrossRef] [Green Version]
- Willis, O.O.; Mouti, M.E.; Sila, D.N.; Mwasaru, M.; Thiongo, G.; Murage, H.; Ojijo, N.O. Physico-Chemical Properties and Antioxidant Potential of Syrup Prepared from ‘Madhura’ Sweet Sorghum (Sorghum Bicolor L. Moench) Cultivar Grown at Different Locations in Kenya. Sugar Tech. 2013, 15, 263–270. [Google Scholar] [CrossRef]
- Ferrer, M.A.; Cimini, S.; López-Orenes, A.; Calderón, A.A.; De Gara, L. Differential Pb Tolerance in Metallicolous and Non-Metallicolous Zygophyllum Fabago Populations Involves the Strengthening of the Antioxidative Pathways. Environ. Exp. Bot. 2018, 150, 141–151. [Google Scholar] [CrossRef]
- Romero-Puertas, M.C.; Rodríguez-Serrano, M.; Corpas, F.J.; Gómez, M.; Del Río, L.A.; Sandalio, L.M. Cadmium-Induced Subcellular Accumulation of O2− and H2O2 in Pea Leaves. Plant Cell Environ. 2004, 27, 1122–1134. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A Highly Sensitive RT-PCR Method for Detection and Quantification of MicroRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, P.; Hou, H.; Zhang, H.; Wang, Y.; Yan, S.; Huang, Y.; Li, H.; Tan, J.; Hu, A.; et al. Transcriptional Regulation of Cell Cycle Genes in Response to Abiotic Stresses Correlates with Dynamic Changes in Histone Modifications in Maize. PLoS ONE 2014, 9, e106070. [Google Scholar] [CrossRef]
- Sabetta, W.; Vandelle, E.; Locato, V.; Costa, A.; Cimini, S.; Bittencourt Moura, A.; Luoni, L.; Graf, A.; Viggiano, L.; De Gara, L.; et al. Genetic Buffering of Cyclic AMP in Arabidopsis Thaliana Compromises the Plant Immune Response Triggered by an Avirulent Strain of Pseudomonas Syringae Pv. Tomato. Plant J. 2019, 98, 590–606. [Google Scholar] [CrossRef]
- De Pinto, M.C.; Francis, D.; De Gara, L. The Redox State of the Ascorbate-Dehydroascorbate Pair as a Specific Sensor of Cell Division in Tobacco BY-2 Cells. Protoplasma 1999, 209, 90–97. [Google Scholar] [CrossRef]
- García-Ulloa, A.; Sanjurjo, L.; Cimini, S.; Encina, A.; Martínez-Rubio, R.; Bouza, R.; Barral, L.; Estévez-Pérez, G.; Novo-Uzal, E.; De Gara, L.; et al. Overexpression of ZePrx in Nicotiana Tabacum Affects Lignin Biosynthesis without Altering Redox Homeostasis. Front. Plant Sci. 2020, 11, 900. [Google Scholar] [CrossRef]
- Hodges, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the Thiobarbituric Acid-Reactive-Substances Assay for Estimating Lipid Peroxidation in Plant Tissues Containing Anthocyanin and Other Interfering Compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Pasqualetti, V.; Locato, V.; Fanali, C.; Mulinacci, N.; Cimini, S.; Morgia, A.M.; Pasqua, G.; De Gara, L. Comparison between In Vitro Chemical and Ex Vivo Biological Assays to Evaluate Antioxidant Capacity of Botanical Extracts. Antioxidants 2021, 10, 1136. [Google Scholar] [CrossRef]
- Locato, V.; Uzal, E.; Cimini, S.; Zonno, M.; Evidente, A.; Micera, A.; Foyer, C.; De Gara, L. Low Concentrations of the Toxin Ophiobolin A Lead to an Arrest of the Cell Cycle and Alter the Intracellular Partitioning of Glutathione between the Nuclei and Cytoplasm. J. Exp. Bot. 2015, 66, 2991–3000. [Google Scholar] [CrossRef] [Green Version]
- Pushpam, R.; Manonmani, S.; Varthini, N.V.; Robin, S. Studies on Yield, Root Characters Related to Drought Tolerance and Their Association in Upland Rice Genotypes. Electron. J. Plant Breed. 2018, 9, 856. [Google Scholar] [CrossRef]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J. Rice Transcription Factor OsMADS25 Modulates Root Growth and Confers Salinity Tolerance via the ABA–Mediated Regulatory Pathway and ROS Scavenging. PLOS Genet. 2018, 14, e1007662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, D.H.; Seomun, S.; Choi, Y.D.; Jang, G. Root Development and Stress Tolerance in Rice: The Key to Improving Stress Tolerance without Yield Penalties. Int. J. Mol. Sci. 2020, 21, 1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, W.; Wu, J.; Wei, C.; Li, K.; He, G.; Attla, K.; Qian, X.; Yang, J. Interaction between Programmed Cell Death 5 and Calcineurin B-like Interacting Protein Kinase 23 in Oryza Sativa. Plant Sci. 2006, 6, 1150–1155. [Google Scholar] [CrossRef]
- Sun, F.; Qi, W.; Qian, X.; Wang, Q.; Yang, M.; Dong, X.; Yang, J. Investigating the Role of OsPDCD5, a Homolog of the Mammalian PDCD5, in Programmed Cell Death by Inducible Expression in Rice. Plant Mol. Biol. Report. 2012, 30, 87–98. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Casadevall, R.; D’Andrea, L.; AbdElgawad, H.; Beemster, G.T.S.; Casati, P. AtPDCD5 Plays a Role in Programmed Cell Death after UV-B Exposure in Arabidopsis. Plant Physiol. 2016, 170, 2444–2460. [Google Scholar] [CrossRef]
- De Pinto, M.C.; Locato, V.; Paradiso, A.; De Gara, L. Role of Redox Homeostasis in Thermo-Tolerance under a Climate Change Scenario. Ann. Bot. 2015, 116, 487–496. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Farnese, F.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When Bad Guys Become Good Ones: The Key Role of Reactive Oxygen Species and Nitric Oxide in the Plant Responses to Abiotic Stress. Front. Plant Sci. 2016, 7, 471. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in Plants: An Integrated Overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in Plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced MiRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Jagadeeswaran, G.; Li, Y.-F.; Sunkar, R. Redox Signaling Mediates the Expression of a Sulfate-Deprivation-Inducible MicroRNA395 in Arabidopsis. Plant J. 2014, 77, 85–96. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Gulyás, Z.; Kalapos, B.; Boldizsár, Á.; Liu, X.; Pál, M.; Yao, Y.; Galiba, G.; Kocsy, G. Identification of a Redox-Dependent Regulatory Network of MiRNAs and Their Targets in Wheat. J. Exp. Bot. 2019, 70, 85–99. [Google Scholar] [CrossRef]
- Liang, G.; Yang, F.; Yu, D. MicroRNA395 Mediates Regulation of Sulfate Accumulation and Allocation in Arabidopsis Thaliana. Plant J. 2010, 62, 1046–1057. [Google Scholar] [CrossRef]
- Dixon, D.P.; Skipsey, M.; Grundy, N.M.; Edwards, R. Stress-Induced Protein S-Glutathionylation in Arabidopsis. Plant Physiol. 2005, 138, 2233–2244. [Google Scholar] [CrossRef] [Green Version]
- Zaffagnini, M.; Bedhomme, M.; Lemaire, S.D.; Trost, P. The Emerging Roles of Protein Glutathionylation in Chloroplasts. Plant Sci. 2012, 185–186, 86–96. [Google Scholar] [CrossRef]
- Voesenek, L.; Bailey-Serres, J. Plant Biology: Genetics of High-Rise Rice. Nature 2009, 460, 959–960. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhou, J.; Li, Y.; Quan, R.; Wang, J.; Huang, R.; Qin, H. Salt Stress Promotes Abscisic Acid Accumulation to Affect Cell Proliferation and Expansion of Primary Roots in Rice. Int. J. Mol. Sci. 2021, 22, 10892. [Google Scholar] [CrossRef]
- Zhou, B.-B.S.; Elledge, S.J. The DNA Damage Response: Putting Checkpoints in Perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef]
- Kadota, Y.; Watanabe, T.; Fujii, S.; Higashi, K.; Sano, T.; Nagata, T.; Hasezawa, S.; Kuchitsu, K. Crosstalk between Elicitor-Induced Cell Death and Cell Cycle Regulation in Tobacco BY-2 Cells. Plant J. 2004, 40, 131–142. [Google Scholar] [CrossRef]
- Zebell, S.G.; Dong, X. Cell Cycle Regulators and Cell Death in Immunity. Cell Host Microbe 2015, 18, 402–407. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Fei, H.; Wang, L.; Xu, H.; Zhang, H.; Zheng, L. PDCD5 Regulates Cell Proliferation, Cell Cycle Progression and Apoptosis. Oncol. Lett. 2018, 15, 1177–1183. [Google Scholar] [CrossRef] [Green Version]
- Attia, K.; Li, K.-G.; Wei, C.; He, G.-M.; Su, W.; Yang, J.-S. Overexpression of the OsPDCD5 Gene Induces Programmed Cell Death in Rice. J. Integr. Plant Biol. 2005, 47, 1115–1122. [Google Scholar] [CrossRef]
- Wang, Y.; Attia, K.; Zha, X.; Abdelkhalik, A.; Zhang, S.; Qian, X.; Dong, X.; Sun, F.; Yang, J.; Lightfoot, D.; et al. Down-Regulation of the OsPDCD5 Gene Induced Photoperiod-Sensitive Male Sterility in Rice. Nat. Preced. 2010, 178, 221–228. [Google Scholar] [CrossRef]
- Yang, M.; Sun, F.; Wang, S.; Qi, W.; Wang, Q.; Dong, X.; Yang, J.; Luo, X. Down-Regulation of OsPDCD5, a Homolog of the Mammalian PDCD5, Increases Rice Tolerance to Salt Stress. Mol. Breed. 2013, 31, 333–346. [Google Scholar] [CrossRef]
- Dong, S.; Dong, X.; Han, X.; Zhang, F.; Zhu, Y.; Xin, X.; Wang, Y.; Hu, Y.; Yuan, D.; Wang, J.; et al. OsPDCD5 Negatively Regulates Plant Architecture and Grain Yield in Rice. Proc. Natl. Acad. Sci. USA 2021, 118, e2018799118. [Google Scholar] [CrossRef]
- De Pinto, M.C.; Locato, V.; De Gara, L. Redox Regulation in Plant Programmed Cell Death. Plant Cell Environ. 2012, 35, 234–244. [Google Scholar] [CrossRef] [Green Version]
- Locato, V.; Cimini, S.; De Gara, L. ROS and Redox Balance as Multifaceted Players of Cross-Tolerance: Epigenetic and Retrograde Control of Gene Expression. J. Exp. Bot. 2018, 69, 3373–3391. [Google Scholar] [CrossRef] [PubMed]
- Pilarska, M.; Wiciarz, M.; Jajić, I.; Kozieradzka-Kiszkurno, M.; Dobrev, P.; Vanková, R.; Niewiadomska, E. A Different Pattern of Production and Scavenging of Reactive Oxygen Species in Halophytic Eutrema Salsugineum (Thellungiella Salsuginea) Plants in Comparison to Arabidopsis Thaliana and Its Relation to Salt Stress Signaling. Front. Plant Sci. 2016, 7, 1179. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as Key Players in Plant Stress Signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, B.; Formentin, E.; Ronci, B.; Locato, V.; Barizza, E.; Hyder, M.Z.; Schiavo, F.L.; Yasmin, T. Salt Tolerance in Indica Rice Cell Cultures Depends on a Fine Tuning of ROS Signalling and Homeostasis. PLoS ONE 2019, 14, e0213986. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in Plants: Biosynthesis and Physiological Role in Environmental Stress Tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Dorion, S.; Ouellet, J.C.; Rivoal, J. Glutathione Metabolism in Plants under Stress: Beyond Reactive Oxygen Species Detoxification. Metabolites 2021, 11, 641. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential Expression of MiRNAs in Response to Salt Stress in Maize Roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Cimini, S.; Gualtieri, C.; Macovei, A.; Balestrazzi, A.; De Gara, L.; Locato, V. Redox Balance-DDR-MiRNA Triangle: Relevance in Genome Stability and Stress Responses in Plants. Front. Plant Sci. 2019, 10, 989. [Google Scholar] [CrossRef]
- Xing, L.; Zhu, M.; Luan, M.; Zhang, M.; Jin, L.; Liu, Y.; Zou, J.; Wang, L.; Xu, M. MiR169q and NUCLEAR FACTOR YA8 Enhance Salt Tolerance by Activating PEROXIDASE1 Expression in Response to ROS. Plant Physiol. 2022, 188, 608–623. [Google Scholar] [CrossRef]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q.; et al. The MiR172/IDS1 Signaling Module Confers Salt Tolerance through Maintaining ROS Homeostasis in Cereal Crops. New Phytol. 2021, 230, 1017–1033. [Google Scholar] [CrossRef]
- Wang, M.; Guo, W.; Li, J.; Pan, X.; Pan, L.; Zhao, J.; Zhang, Y.; Cai, S.; Huang, X.; Wang, A.; et al. The MiR528-AO Module Confers Enhanced Salt Tolerance in Rice by Modulating the Ascorbic Acid and Abscisic Acid Metabolism and ROS Scavenging. J. Agric. Food Chem. 2021, 69, 8634–8648. [Google Scholar] [CrossRef]
- Takahashi, H.; Yamazaki, M.; Sasakura, N.; Watanabe, A.; Leustek, T.; Engler, J.d.A.; Engler, G.; Van Montagu, M.; Saito, K. Regulation of Sulfur Assimilation in Higher Plants: A Sulfate Transporter Induced in Sulfate-Starved Roots Plays a Central Role in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 1997, 94, 11102–11107. [Google Scholar] [CrossRef] [Green Version]
- Matthewman, C.A.; Kawashima, C.G.; Húska, D.; Csorba, T.; Dalmay, T.; Kopriva, S. MiR395 Is a General Component of the Sulfate Assimilation Regulatory Network in Arabidopsis. FEBS Lett. 2012, 586, 3242–3248. [Google Scholar] [CrossRef]
- Yang, Z.; Hui, S.; Lv, Y.; Zhang, M.; Chen, D.; Tian, J.; Zhang, H.; Liu, H.; Cao, J.; Xie, W.; et al. MiR395-Regulated Sulfate Metabolism Exploits Pathogen Sensitivity to Sulfate to Boost Immunity in Rice. Mol. Plant 2022, 15, 671–688. [Google Scholar] [CrossRef]
- Panda, S.K.; Sunkar, R. Nutrient- and Other Stress-Responsive MicroRNAs in Plants: Role for Thiol-Based Redox Signaling. Plant Signal. Behav. 2015, 10, e1010916. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cimini, S.; Locato, V.; Giacinti, V.; Molinari, M.; De Gara, L. A Multifactorial Regulation of Glutathione Metabolism behind Salt Tolerance in Rice. Antioxidants 2022, 11, 1114. https://doi.org/10.3390/antiox11061114
Cimini S, Locato V, Giacinti V, Molinari M, De Gara L. A Multifactorial Regulation of Glutathione Metabolism behind Salt Tolerance in Rice. Antioxidants. 2022; 11(6):1114. https://doi.org/10.3390/antiox11061114
Chicago/Turabian StyleCimini, Sara, Vittoria Locato, Valentina Giacinti, Michela Molinari, and Laura De Gara. 2022. "A Multifactorial Regulation of Glutathione Metabolism behind Salt Tolerance in Rice" Antioxidants 11, no. 6: 1114. https://doi.org/10.3390/antiox11061114
APA StyleCimini, S., Locato, V., Giacinti, V., Molinari, M., & De Gara, L. (2022). A Multifactorial Regulation of Glutathione Metabolism behind Salt Tolerance in Rice. Antioxidants, 11(6), 1114. https://doi.org/10.3390/antiox11061114







