Prediction of Alzheimer’s Disease Based on Multi-Modal Domain Adaptation
Abstract
1. Introduction
1.1. Analysis of AD Based on Multi-Modal Data
1.2. Similarity Analysis Between Different Models
1.3. Domain Adaptation Development Status
1.4. The Present Study
- This work presents a MM-DDA model, which fully leverages the complementary information from two modalities of data to enhance the classification accuracy. Furthermore, MM-DDA operates without requiring class labels from the target domain, thereby reducing the cost and complexity of data annotation.
- The cross-entropy loss function combined with the Gaussian kernel was employed to calculate the correlation loss between modalities, quantify and optimize the semantic similarity between different modalities, and enhance the synergy between modalities.
- This paper employs the multi-head attention mechanism to dynamically adjust the weights among different modality features and capture richer semantic information.
2. Materials and Methods
2.1. Source and Preprocessing of the Dataset
2.2. Problem Formulation
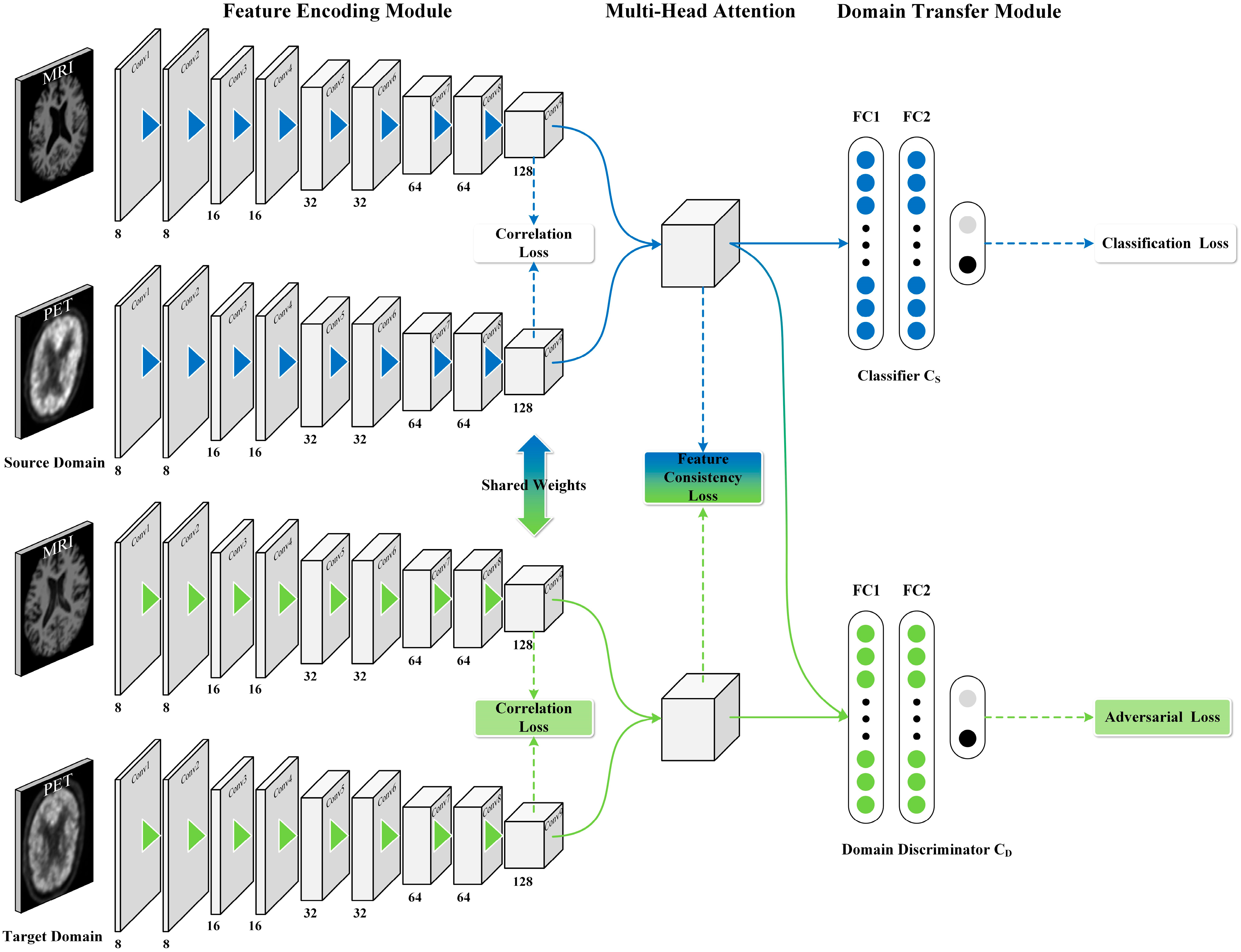
2.3. The Proposed Approach
2.3.1. Feature Coding Module
2.3.2. Multi-Head Attention Feature Fusion Module
2.3.3. Domain Transfer Module
3. Results
3.1. Experimental Setup
3.2. Comparison Methods
- (1)
- DAAN. Based on single-modal image data, DAAN achieves unsupervised domain adaptation through dynamic adversarial adaptation networks, which can obtain domain consistent features while dynamically evaluating the priority of global and local feature space layouts.
- (2)
- AD2A. The AD2A framework combines adversarial training and attention-directed feature learning to realize automatic brain disease recognition from multi-site MRI data. The framework can automatically locate brain regions associated with disease through attention mechanisms and use adversarial learning for cross-domain knowledge transfer.
- (3)
- PMDA. Based on the MRI data, the PMDA framework realizes the automatic assistant diagnosis of MRI data under domain bias problems by integrating multi-scale feature extraction, prototype-constrained maximum density divergence (Pro-MDD), and adversarial domain alignment.
4. Discussion
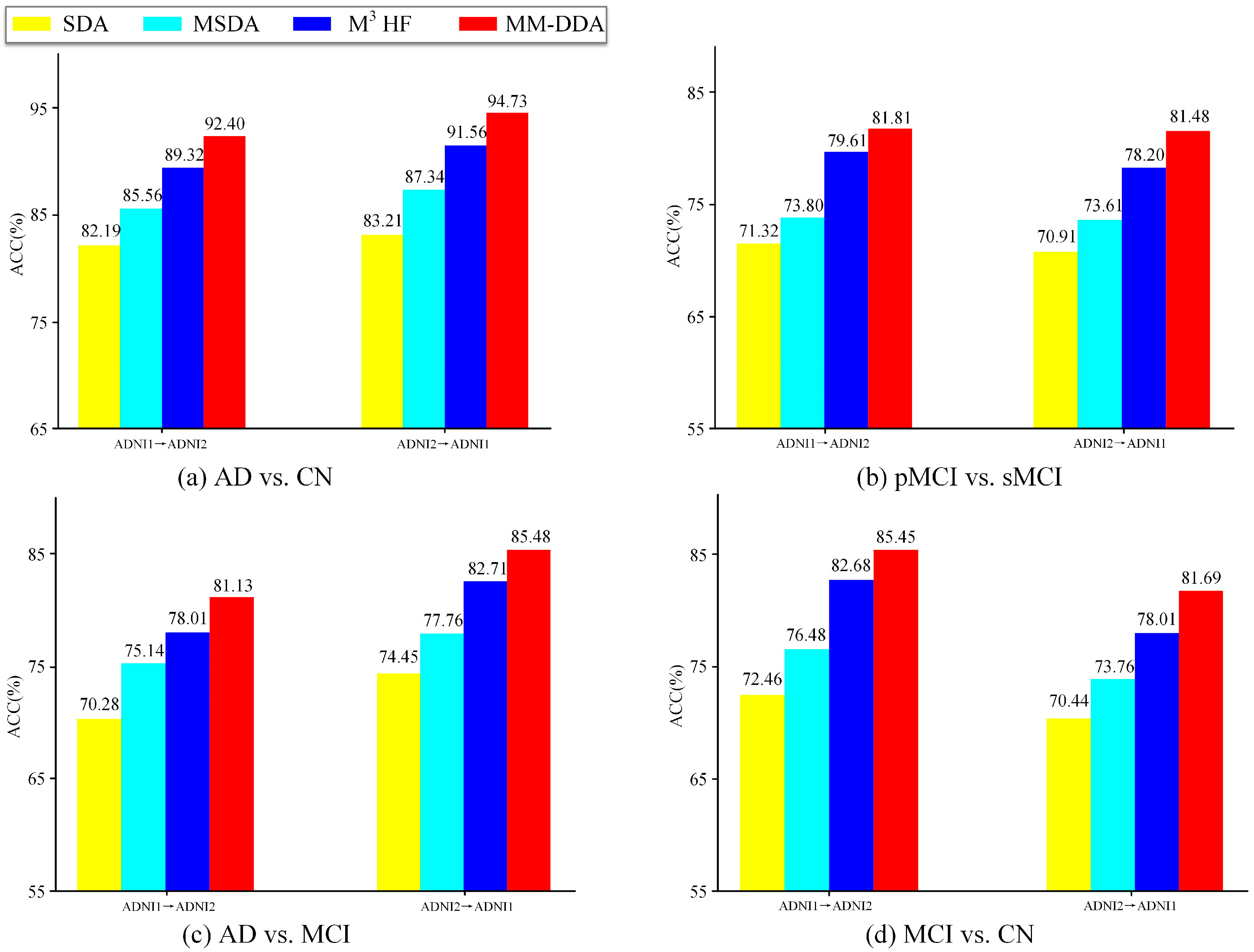
4.1. Ablation Experiment
4.2. Determination of Head Numbers
4.3. Limitations and Prospects
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jack, C.R.; Graf, A.; Burnham, S.C.; Doty, E.G.; Moebius, H.J.; Montenigro, P.; Siemers, E.; Sink, K.M.; Shaw, L.M.; Hansen, C.T.; et al. Application of the revised criteria for diagnosis and staging of Alzheimer’s disease: Drug development and clinical practice. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2024, 10, e70013. [Google Scholar] [CrossRef]
- Airlangga, G. Advancing Alzheimer’s Diagnosis: A Comparative Analysis of Deep Learning Architectures on Multidimensional Health Data. J. Inform. Ekon. Bisnis 2024, 6, 810–814. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan on the Public Health Response to Dementia 2017–2025. 2017. Available online: https://www.who.int/publications/i/item/9789241513487 (accessed on 2 March 2025).
- Chételat, G.; Arbizu, J.; Barthel, H.; Garibotto, V.; Law, I.; Morbelli, S.; van de Giessen, E.; Agosta, F.; Barkhof, F.; Brooks, D.J.; et al. Amyloid-PET and 18F-FDG-PET in the diagnostic investigation of Alzheimer’s disease and other dementias. Lancet Neurol. 2020, 19, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Ravienna, K.; Salobrar-Garcia, E. Early indicators of cognitive decline, Alzheimer’s disease, and related dementias captured by neurophysiological tools. Front. Psychol. 2024, 15, 1393724. [Google Scholar] [CrossRef]
- Leng, Y.; Cui, W.; Peng, Y.; Yan, C.; Cao, Y.; Yan, Z.; Chen, S.; Jiang, X.; Zheng, J. Multimodal cross enhanced fusion network for diagnosis ofAlzheimer’s disease and subjective memory complaints. Comput. Biol. Med. 2023, 157, 106788. [Google Scholar] [CrossRef] [PubMed]
- Minoshima, S.; Cross, D.; Thientunyakit, T.; Foster, N.L.; Drzezga, A. 18F-FDG PET imaging in neurodegenerative dementing disorders: Insights into subtype classification, emerging disease categories, and mixed dementia with copathologies. J. Nucl. Med. 2022, 63, 2S–12S. [Google Scholar] [CrossRef]
- Liu, L.; Sun, S.; Kang, W.; Wu, S.; Lin, L. A review of neuroimaging-based data-driven approach for Alzheimer’s disease heterogeneity analysis. Rev. Neurosci. 2024, 35, 121–139. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, X.; Shi, G.; Xiao, N.; Song, K.; Zhao, J.; Hao, R.; Li, K. Semantic consistency generative adversarial network for cross-modality domain adaptation in ultrasound thyroid nodule classification. Appl. Intell. 2022, 52, 10369–10383. [Google Scholar] [CrossRef]
- Gonneaud, J.; Baria, A.T.; Pichet Binette, A.; Gordon, B.A.; Chhatwal, J.P.; Cruchaga, C.; Jucker, M.; Levin, J.; Salloway, S.; Farlow, M.; et al. Accelerated functional brain aging in pre-clinical familial Alzheimer’s disease. Nat. Commun. 2021, 12, 5346. [Google Scholar] [CrossRef]
- Zheng, B.; He, J.; Zhu, J.; Xie, Y.; Zaki, N.; Qin, W. Style Enhanced Domain Adaptation Neural Network for Cross-Modality Cervical Tumor Segmentation. In Proceedings of the International Workshop on Computational Mathematics Modeling in Cancer Analysis, Cham, Switzerland, 8 October 2023. [Google Scholar]
- Ponisio, M.R.; Zempel, J.M.; Willie, J.T.; Tomko, S.R.; McEvoy, S.D.; Roland, J.L.; Williams, J.P. FDG-PET/MRI in the presurgical evaluation of pediatric epilepsy. Pediatr. Radiol. 2024, 54, 1589–1602. [Google Scholar] [CrossRef]
- Vega, F.; Addeh, A.; Ganesh, A.; Smith, E.E. Image translation for estimating two-dimensional axial amyloid-beta PET from structural MRI. J. Magn. Reson. Imaging 2024, 59, 1021–1031. [Google Scholar] [CrossRef]
- Yuan, Z.; Qi, N.; Zhou, Z.; Ding, J.; Chen, X.; Wu, J.; Wang, J.; Zhao, J. Diagnosis of Alzheimer’s disease using transfer learning with multi-modal 3DInception-v4. Quant. Imaging Med. Surg. 2025, 15, 1455–1467. [Google Scholar] [CrossRef]
- Yuan, P.; Xin, M.; Li, X.; Zhang, C.; Liu, J. Multi-modal Multi-probe PET/MR Imaging in evaluation of the effect of CSUL surgery in AD patients. Eur. J. Nucl. Med. Mol. Imaging 2024, 51, S265–S266. [Google Scholar]
- Liu, X.; Li, W.; Miao, S.; Liu, F.; Han, K.; Bezabih, T.T. HAMMF: Hierarchical attention-based multi-task and multi-modal fusion model for computer-aided diagnosis of Alzheimer’s disease. Comput. Biol. Med. 2024, 176, 108564. [Google Scholar] [CrossRef]
- Wang, J.; Liao, N.; Du, X.; Chen, Q.; Wei, B. A semi-supervised approach for the integration of multi-omics data based on transformer multi-head self-attention mechanism and graph convolutional networks. BMC Genom. 2024, 25, 86. [Google Scholar] [CrossRef]
- Zhang, C.; Fan, W.; Li, H.; Chen, C. Multi-level graph regularized robust multi-modal feature selection for Alzheimer’s disease classification. Knowl.-Based Syst. 2024, 293, 111676. [Google Scholar] [CrossRef]
- Lei, B.; Li, Y.; Fu, W.; Yang, P.; Chen, S.; Wang, T.; Xiao, X.; Niu, T.; Fu, Y.; Wang, S.; et al. Alzheimer’s disease diagnosis from multi-modal data via feature inductive learning and dual multilevel graph neural network. Med. Image Anal. 2024, 97, 103213. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, H.; Wei, S.; Mei, J.; Liu, F.; Zhang, G. Alzheimer’s disease prediction algorithm based on de-correlation constraint and multi-modal feature interaction. Comput. Biol. Med. 2024, 170, 108000. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, L.; Zhang, C.; Zhou, T.; Xiao, Z.; Geng, L.; Shen, D. Incomplete multi-modal representation learning for Alzheimer’s disease diagnosis. Med. Image Anal. 2021, 69, 101953. [Google Scholar] [CrossRef]
- Zhang, Q.; Cai, H.; Long, Y.; Yu, S. Multicentric prototype and pseudo-labeling based source-free domain adaptation for Alzheimer’s disease classification. Biomed. Signal Process. Control 2025, 103, 107483. [Google Scholar] [CrossRef]
- Guan, H.; Liu, Y.; Yang, E.; Yap, P.T.; Shen, D.; Liu, M. Multi-site MRI harmonization via attention-guided deep domain adaptation for brain disorder identification. Med. Image Anal. 2021, 71, 102076. [Google Scholar] [CrossRef]
- Turrisi, R.; Pati, S.; Pioggia, G.; Tartarisco, G. Adapting to evolving MRI data: A transfer learning approach for Alzheimer’s disease prediction. Neuroimage 2025, 307, 121016. [Google Scholar] [CrossRef]
- Dadsetan, S. Enhancing Alzheimer’s Prognostic Models with Cross-Domain Self-Supervised Learning and MRI Data Harmonization. Ph.D. Thesis, University of Pittsburgh, Pittsburgh, PA, USA, 2024. [Google Scholar]
- Alzheimer’s Disease Neuroimaging Initiative (ADNI). Available online: https://ida.loni.usc.edu (accessed on 9 July 2024).
- Mao, A.Q.; Mohri, M.; Zhong, Y.T. Cross-entropy loss functions: Theoretical analysis and applications. In Proceedings of the International Conference on Machine Learning, Baltimore, MD, USA, 17–23 July 2023. [Google Scholar]
- Huang, F.; Jiang, M.; Liu, F.; Xu, D.; Fan, Z.; Wang, Y. Classification of heads in multi-head attention mechanisms. In Proceedings of the International Conference on Knowledge Science, Engineering and Management, Singapore, 6–8 August 2022. [Google Scholar]
- Zhuang, Z.; Si, L.; Wang, S.; Xuan, K.; Ouyang, X.; Zhan, Y.; Xue, Z.; Zhang, L.; Shen, D.; Yao, W.; et al. Knee Cartilage Defect Assessment by Graph Representation and Surface Convolution. IEEE Trans. Med. Imaging 2023, 42, 368–379. [Google Scholar] [CrossRef]
- Lei, B.; Zhu, Y.; Liang, E.; Yang, P.; Chen, S.; Hu, H.; Xie, H.; Wei, Z.; Hao, F.; Song, X.; et al. Federated domain adaptation via transformer for multi-site Alzheimer’s disease diagnosis. IEEE Trans. Med. Imaging 2023, 42, 3651–3664. [Google Scholar] [CrossRef]
- Cao, T.; Lin, R.; Zheng, Y.; Shen, D.; Xu, L. A novel approach analysing the dynamic brain functional connectivity for improved MCI detection. IEEE Trans. Biomed. Eng. 2023, 71, 207–216. [Google Scholar] [CrossRef]
- Yu, C.; Wang, J.; Chen, Y.; Huang, M. Transfer learning with dynamic adversarial adaptation network. In Proceedings of the 2019 IEEE International Conference on Data Mining (ICDM), Beijing, China, 8–11 November 2019. [Google Scholar]
- Cai, H.S.; Zhang, Q.M.; Long, Y. Prototype-guided multi-scale domain adaptation for Alzheimer’s disease detection. Comput. Biol. Med. 2023, 154, 106570. [Google Scholar] [CrossRef]

| Category | Gender (F/M) | Age | MMSE |
|---|---|---|---|
| AD | 70/110 | 75.7 ± 4.9 | 3.2 |
| CN | 137/142 | 5.2 | 1.3 |
| pMCI | 31/37 | 5.6 | 3.9 |
| sMCI | 34/78 | 6.8 | 2.2 |
| S → T | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) | F1 (%) |
|---|---|---|---|---|---|---|
| ADNI1→ADNI2 | DAAN | 84.98 | 82.45 | 88.16 | 83.61 | 82.21 |
| AD2A | 88.51 | 90.41 | 90.46 | 90.17 | 88.54 | |
| PMDA | 90.12 | 94.17 | 88.14 | 92.89 | 89.87 | |
| MM-DDA | 92.40 | 96.87 | 89.36 | 95.26 | 91.17 | |
| ADNI2→ADNI1 | DAAN | 86.23 | 87.51 | 85.46 | 86.55 | 84.71 |
| AD2A | 89.16 | 90.66 | 89.14 | 91.61 | 91.44 | |
| PMDA | 91.46 | 87.58 | 92.78 | 91.34 | 89.46 | |
| MM-DDA | 94.73 | 92.59 | 96.66 | 95.01 | 94.33 |
| S → T | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) | F1 (%) |
|---|---|---|---|---|---|---|
| ADNI1→ADNI2 | DAAN | 75.43 | 70.17 | 73.14 | 78.51 | 73.55 |
| AD2A | 79.24 | 75.36 | 81.46 | 86.01 | 77.23 | |
| PMDA | 80.76 | 78.96 | 81.41 | 85.03 | 74.34 | |
| MM-DDA | 81.81 | 85.71 | 80.00 | 88.57 | 75.00 | |
| ADNI2→ADNI1 | DAAN | 73.89 | 68.96 | 71.68 | 79.56 | 71.13 |
| AD2A | 75.30 | 82.17 | 81.34 | 81.21 | 75.23 | |
| PMDA | 78.21 | 80.98 | 82.17 | 84.51 | 76.14 | |
| MM-DDA | 81.48 | 81.81 | 81.25 | 87.77 | 78.24 |
| S → T | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) | F1 (%) |
|---|---|---|---|---|---|---|
| ADNI1→ADNI2 | DAAN | 70.56 | 74.38 | 70.01 | 76.18 | 71.21 |
| AD2A | 74.77 | 73.87 | 79.88 | 80.11 | 74.51 | |
| PMDA | 77.11 | 83.86 | 76.26 | 84.27 | 81.18 | |
| MM-DDA | 81.13 | 84.84 | 75.00 | 88.96 | 84.84 | |
| ADNI2→ADNI1 | DAAN | 73.69 | 68.66 | 71.83 | 75.18 | 72.51 |
| AD2A | 79.11 | 71.03 | 84.81 | 80.44 | 78.22 | |
| PMDA | 80.51 | 74.56 | 92.33 | 86.45 | 80.32 | |
| MM-DDA | 85.48 | 74.07 | 94.28 | 88.27 | 81.63 |
| S → T | Method | ACC (%) | SEN (%) | SPE (%) | AUC (%) | F1 (%) |
|---|---|---|---|---|---|---|
| ADNI1→ADNI2 | DAAN | 76.31 | 78.21 | 70.45 | 81.07 | 72.78 |
| AD2A | 79.21 | 77.48 | 74.44 | 85.11 | 79.01 | |
| PMDA | 81.56 | 84.11 | 82.61 | 86.21 | 81.21 | |
| MM-DDA | 85.45 | 87.50 | 83.87 | 87.10 | 84.00 | |
| ADNI2→ADNI1 | DAAN | 72.43 | 81.08 | 69.76 | 75.77 | 70.11 |
| AD2A | 77.02 | 83.44 | 75.88 | 80.19 | 74.37 | |
| PMDA | 78.57 | 84.61 | 77.37 | 82.73 | 77.07 | |
| MM-DDA | 81.69 | 86.66 | 78.05 | 85.36 | 79.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, B.; Shen, C.; Liao, S.; Wu, F.; Liao, B. Prediction of Alzheimer’s Disease Based on Multi-Modal Domain Adaptation. Brain Sci. 2025, 15, 618. https://doi.org/10.3390/brainsci15060618
Fu B, Shen C, Liao S, Wu F, Liao B. Prediction of Alzheimer’s Disease Based on Multi-Modal Domain Adaptation. Brain Sciences. 2025; 15(6):618. https://doi.org/10.3390/brainsci15060618
Chicago/Turabian StyleFu, Binbin, Changsong Shen, Shuzu Liao, Fangxiang Wu, and Bo Liao. 2025. "Prediction of Alzheimer’s Disease Based on Multi-Modal Domain Adaptation" Brain Sciences 15, no. 6: 618. https://doi.org/10.3390/brainsci15060618
APA StyleFu, B., Shen, C., Liao, S., Wu, F., & Liao, B. (2025). Prediction of Alzheimer’s Disease Based on Multi-Modal Domain Adaptation. Brain Sciences, 15(6), 618. https://doi.org/10.3390/brainsci15060618







