Aging, Rather than Genotype, Is the Principal Contributor to Differential Gene Expression Within Targeted Replacement APOE2, APOE3, and APOE4 Mouse Brain
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA Sequencing
2.3. Sequence Analysis
2.4. Statistical Analysis and Bioinformatics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ADRDs | Alzheimer’s disease-related dementias |
| ApoE | Apolipoprotein E |
| CSF | Cerebrospinal fluid |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| GTC | Genome Technology Center |
| GWAS | Genome-wide association studies |
| hiPSC | Human-induced pluripotent stem cell |
| IPA | Ingenuity Pathway Analysis |
| iPSC | Induced pluripotent stem cell |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LBD | Lewy body disease |
| logFC | Log-fold change |
| MO | Months of age |
| NYUGSOM | New York University Grossman School of Medicine |
| PPIs | Protein–protein interactions |
| RNA-seq | RNA sequencing |
| RIN | RNA integrity number |
References
- Mahley, R.W. Apolipoprotein E: From cardiovascular disease to neurodegenerative disorders. J. Mol. Med. 2016, 94, 739–746. [Google Scholar] [CrossRef]
- Koutsodendris, N.; Nelson, M.R.; Rao, A.; Huang, Y. Apolipoprotein E and Alzheimer’s Disease: Findings, Hypotheses, and Potential Mechanisms. Annu. Rev. Pathol. 2022, 17, 73–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef]
- Bertram, L.; McQueen, M.B.; Mullin, K.; Blacker, D.; Tanzi, R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genet. 2007, 39, 17–23. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef]
- Andrews, S.J.; Fulton-Howard, B.; Goate, A. Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol. 2020, 19, 326–335. [Google Scholar] [CrossRef]
- Abrego-Guandique, D.M.; Saraceno, G.F.; Cannataro, R.; Manzzo de Burnside, M.; Caroleo, M.C.; Cione, E. Apolipoprotein E and Alzheimer’s Disease in Italian Population: Systematic Review and Meta-Analysis. Brain Sci. 2024, 14, 908. [Google Scholar] [CrossRef]
- Chartier-Harlin, M.C.; Parfitt, M.; Legrain, S.; Pérez-Tur, J.; Brousseau, T.; Evans, A.; Berr, C.; Vidal, O.; Roques, P.; Gourlet, V.; et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: Analysis of the 19q13.2 chromosomal region. Hum. Mol. Genet. 1994, 3, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Ren, Y.; Yamazaki, Y.; Qiao, W.; Li, F.; Felton, L.M.; Mahmoudiandehkordi, S.; Kueider-Paisley, A.; Sonoustoun, B.; Arnold, M.; et al. Alzheimer’s Risk Factors Age, APOE Genotype, and Sex Drive Distinct Molecular Pathways. Neuron 2020, 106, 727–742.e6. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Risch, N.J.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C., Jr.; Rimmler, J.B.; Locke, P.A.; Conneally, P.M.; Schmader, K.E.; et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 1994, 7, 180–184. [Google Scholar] [CrossRef]
- Panza, F.; Solfrizzi, V.; Torres, F.; Mastroianni, F.; Colacicco, A.M.; Basile, A.M.; Capurso, C.; D’Introno, A.; Del Parigi, A.; Capurso, A. Apolipoprotein E in Southern Italy: Protective effect of epsilon 2 allele in early- and late-onset sporadic Alzheimer’s disease. Neurosci. Lett. 2000, 292, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Belloy, M.E.; Napolioni, V.; Greicius, M.D. A Quarter Century of APOE and Alzheimer’s Disease: Progress to Date and the Path Forward. Neuron 2019, 101, 820–838. [Google Scholar] [CrossRef]
- Riedel, B.C.; Thompson, P.M.; Brinton, R.D. Age, APOE and sex: Triad of risk of Alzheimer’s disease. J. Steroid Biochem. Mol. Biol. 2016, 160, 134–147. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Qian, J.; Monsell, S.E.; Betensky, R.A.; Hyman, B.T. APOEε2 is associated with milder clinical and pathological Alzheimer disease. Ann. Neurol. 2015, 77, 917–929. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Das, S.; Hyman, B.T. APOE and Alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021, 20, 68–80. [Google Scholar] [CrossRef]
- Blanchard, J.W.; Bula, M.; Davila-Velderrain, J.; Akay, L.A.; Zhu, L.; Frank, A.; Victor, M.B.; Bonner, J.M.; Mathys, H.; Lin, Y.T.; et al. Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. Nat. Med. 2020, 26, 952–963. [Google Scholar] [CrossRef]
- Salvadó, G.; Grothe, M.J.; Groot, C.; Moscoso, A.; Schöll, M.; Gispert, J.D.; Ossenkoppele, R. Differential associations of APOE-ε2 and APOE-ε4 alleles with PET-measured amyloid-β and tau deposition in older individuals without dementia. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 2212–2224. [Google Scholar] [CrossRef] [PubMed]
- Tsuang, D.; Leverenz, J.B.; Lopez, O.L.; Hamilton, R.L.; Bennett, D.A.; Schneider, J.A.; Buchman, A.S.; Larson, E.B.; Crane, P.K.; Kaye, J.A.; et al. APOE ε4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 2013, 70, 223–228. [Google Scholar] [CrossRef]
- Dickson, D.W.; Heckman, M.G.; Murray, M.E.; Soto, A.I.; Walton, R.L.; Diehl, N.N.; van Gerpen, J.A.; Uitti, R.J.; Wszolek, Z.K.; Ertekin-Taner, N.; et al. APOE ε4 is associated with severity of Lewy body pathology independent of Alzheimer pathology. Neurology 2018, 91, e1182–e1195. [Google Scholar] [CrossRef]
- Reiman, E.M.; Arboleda-Velasquez, J.F.; Quiroz, Y.T.; Huentelman, M.J.; Beach, T.G.; Caselli, R.J.; Chen, Y.; Su, Y.; Myers, A.J.; Hardy, J.; et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5000-person neuropathological study. Nat. Commun. 2020, 11, 667. [Google Scholar] [CrossRef]
- Martens, Y.A.; Zhao, N.; Liu, C.C.; Kanekiyo, T.; Yang, A.J.; Goate, A.M.; Holtzman, D.M.; Bu, G. ApoE Cascade Hypothesis in the pathogenesis of Alzheimer’s disease and related dementias. Neuron 2022, 110, 1304–1317. [Google Scholar] [CrossRef]
- Chen, Y.; Strickland, M.R.; Soranno, A.; Holtzman, D.M. Apolipoprotein E: Structural Insights and Links to Alzheimer Disease Pathogenesis. Neuron 2021, 109, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Wennberg, A.M.; Tosakulwong, N.; Lesnick, T.G.; Murray, M.E.; Whitwell, J.L.; Liesinger, A.M.; Petrucelli, L.; Boeve, B.F.; Parisi, J.E.; Knopman, D.S.; et al. Association of Apolipoprotein E ε4 With Transactive Response DNA-Binding Protein 43. JAMA Neurol. 2018, 75, 1347–1354. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.S.; Yu, L.; White, C.C.; Chibnik, L.B.; Chhatwal, J.P.; Sperling, R.A.; Bennett, D.A.; Schneider, J.A.; De Jager, P.L. Evaluation of TDP-43 proteinopathy and hippocampal sclerosis in relation to APOE ε4 haplotype status: A community-based cohort study. Lancet Neurol. 2018, 17, 773–781. [Google Scholar] [CrossRef]
- Berge, G.; Sando, S.B.; Rongve, A.; Aarsland, D.; White, L.R. Apolipoprotein E ε2 genotype delays onset of dementia with Lewy bodies in a Norwegian cohort. J. Neurol. Neurosurg. Psychiatry 2014, 85, 1227–1231. [Google Scholar] [CrossRef] [PubMed]
- Sabir, M.S.; Blauwendraat, C.; Ahmed, S.; Serrano, G.E.; Beach, T.G.; Perkins, M.; Rice, A.C.; Masliah, E.; Morris, C.M.; Pihlstrom, L.; et al. Assessment of APOE in atypical parkinsonism syndromes. Neurobiol. Dis. 2019, 127, 142–146. [Google Scholar] [CrossRef]
- Therriault, J.; Pascoal, T.A.; Benedet, A.L.; Tissot, C.; Savard, M.; Chamoun, M.; Lussier, F.; Kang, M.S.; Berzgin, G.; Wang, T.; et al. Frequency of Biologically Defined Alzheimer Disease in Relation to Age, Sex, APOE ε4, and Cognitive Impairment. Neurology 2021, 96, e975–e985. [Google Scholar] [CrossRef]
- Torres-Perez, E.; Ledesma, M.; Garcia-Sobreviela, M.P.; Leon-Latre, M.; Arbones-Mainar, J.M. Apolipoprotein E4 association with metabolic syndrome depends on body fatness. Atherosclerosis 2016, 245, 35–42. [Google Scholar] [CrossRef]
- Nuriel, T.; Peng, K.Y.; Ashok, A.; Dillman, A.A.; Figueroa, H.Y.; Apuzzo, J.; Ambat, J.; Levy, E.; Cookson, M.R.; Mathews, P.M.; et al. The Endosomal-Lysosomal Pathway is Dysregulated by APOE4 Expression in Vivo. Front. Neurosci. 2017, 11, 702. [Google Scholar] [CrossRef]
- Peng, K.Y.; Liemisa, B.; Pasato, J.; D’Acunzo, P.; Pawlik, M.; Heguy, A.; Penikalapati, S.C.; Labuza, A.; Pidikiti, H.; Alldred, M.J.; et al. Apolipoprotein E2 Expression Alters Endosomal Pathways in a Mouse Model With Increased Brain Exosome Levels During Aging. Traffic 2024, 25, e12937. [Google Scholar] [CrossRef] [PubMed]
- Hebert, L.E.; Bienias, J.L.; Aggarwal, N.T.; Wilson, R.S.; Bennett, D.A.; Shah, R.C.; Evans, D.A. Change in risk of Alzheimer disease over time. Neurology 2010, 75, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Huentelman, M.; Ryan, L. More than just risk for Alzheimer’s disease: APOE ε4’s impact on the aging brain. Trends Neurosci. 2023, 46, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Verduzco Espinoza, A.P.; Na, N.; Campanati, L.; Ngo, P.; Baldwin, K.K.; Cline, H.T. Microglia-to-neuron signaling links APOE4 and inflammation to enhanced neuronal lipid metabolism and network activity. Proc. Natl. Acad. Sci. USA 2025, 122, e2516103122. [Google Scholar] [CrossRef]
- Feng, J.; Xiang, L.; Wan, G.; Qi, K.; Sun, L.; Huang, Z.; Zheng, C.; Lv, Z.; Hu, C.; Yang, Z. Is APOEε3 a favourable factor for the longevity: An association study in Chinese population. J. Genet. 2011, 90, 343–347. [Google Scholar] [CrossRef]
- Shinohara, M.; Kanekiyo, T.; Tachibana, M.; Kurti, A.; Shinohara, M.; Fu, Y.; Zhao, J.; Han, X.; Sullivan, P.M.; Rebeck, G.W.; et al. APOE2 is associated with longevity independent of Alzheimer’s disease. Elife 2020, 9, e62199. [Google Scholar] [CrossRef]
- Raulin, A.C.; Doss, S.V.; Trottier, Z.A.; Ikezu, T.C.; Bu, G.; Liu, C.C. ApoE in Alzheimer’s disease: Pathophysiology and therapeutic strategies. Mol. Neurodegener. 2022, 17, 72. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Devanney, N.A.; Golden, L.R.; Smith, C.T.; Schwartz, J.L.; Walsh, A.E.; Clarke, H.A.; Goulding, D.S.; Allenger, E.J.; Morillo-Segovia, G.; et al. APOE modulates microglial immunometabolism in response to age, amyloid pathology, and inflammatory challenge. Cell Rep. 2023, 42, 112196. [Google Scholar] [CrossRef]
- Lin, Y.T.; Seo, J.; Gao, F.; Feldman, H.M.; Wen, H.L.; Penney, J.; Cam, H.P.; Gjoneska, E.; Raja, W.K.; Cheng, J.; et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated With Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 2018, 98, 1141–1154.e7. [Google Scholar] [CrossRef]
- He, L.; Davila-Velderrain, J.; Sumida, T.S.; Hafler, D.A.; Kellis, M.; Kulminski, A.M. NEBULA is a fast negative binomial mixed model for differential or co-expression analysis of large-scale multi-subject single-cell data. Commun. Biol. 2021, 4, 629. [Google Scholar] [CrossRef]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. [Google Scholar] [CrossRef]
- Konijnenberg, E.; Tijms, B.M.; Gobom, J.; Dobricic, V.; Bos, I.; Vos, S.; Tsolaki, M.; Verhey, F.; Popp, J.; Martinez-Lage, P.; et al. APOE ε4 genotype-dependent cerebrospinal fluid proteomic signatures in Alzheimer’s disease. Alzheimers Res. Ther. 2020, 12, 65. [Google Scholar] [CrossRef]
- Sullivan, P.M.; Mezdour, H.; Aratani, Y.; Knouff, C.; Najib, J.; Reddick, R.L.; Quarfordt, S.H.; Maeda, N. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J. Biol. Chem. 1997, 272, 17972–17980. [Google Scholar] [CrossRef] [PubMed]
- Hixson, J.E.; Vernier, D.T. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J. Lipid Res. 1990, 31, 545–548. [Google Scholar] [CrossRef]
- Peng, K.Y.; Pérez-González, R.; Alldred, M.J.; Goulbourne, C.N.; Morales-Corraliza, J.; Saito, M.; Saito, M.; Ginsberg, S.D.; Mathews, P.M.; Levy, E. Apolipoprotein E4 genotype compromises brain exosome production. Brain 2019, 142, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Alldred, M.J.; Ibrahim, K.W.; Pidikiti, H.; Chiosis, G.; Mufson, E.J.; Stutzmann, G.E.; Ginsberg, S.D. Down syndrome frontal cortex layer III and layer V pyramidal neurons exhibit lamina specific degeneration in aged individuals. Acta Neuropathol. Commun. 2024, 12, 182. [Google Scholar] [CrossRef]
- Alldred, M.J.; Ibrahim, K.W.; Pidikiti, H.; Lee, S.H.; Heguy, A.; Chiosis, G.; Mufson, E.J.; Stutzmann, G.E.; Ginsberg, S.D. Profiling hippocampal neuronal populations reveals unique gene expression mosaics reflective of connectivity-based degeneration in the Ts65Dn mouse model of Down syndrome and Alzheimer’s disease. Front. Mol. Neurosci. 2025, 18, 1546375. [Google Scholar] [CrossRef]
- Labuza, A.; Alldred, M.J.; Pidikiti, H.; Malek-Ahmadi, M.H.; Lee, S.H.; Heguy, A.; Coleman, P.D.; Chakrabarty, S.; Chiosis, G.; Mufson, E.J.; et al. Frontal cortex pyramidal neuron expression profiles differentiate the prodromal stage from progressive degeneration across the Alzheimer’s disease spectrum. Alzheimers Dement. 2025, 21, e70395. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 October 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Picard Toolkit. Available online: https://broadinstitute.github.io/picard/ (accessed on 10 October 2025).
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Zehetmayer, S.; Posch, M.; Graf, A. Impact of adaptive filtering on power and false discovery rate in RNA-seq experiments. BMC Bioinform. 2022, 23, 388. [Google Scholar] [CrossRef] [PubMed]
- Rau, A.; Gallopin, M.; Celeux, G.; Jaffrézic, F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics 2013, 29, 2146–2152. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Roussos, P. Dream: Powerful differential expression analysis for repeated measures designs. Bioinformatics 2021, 37, 192–201. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Schadt, E.E. variancePartition: Interpreting drivers of variation in complex gene expression studies. BMC Bioinform. 2016, 17, 483. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Li, G.; Bien-Ly, N.; Andrews-Zwilling, Y.; Xu, Q.; Bernardo, A.; Ring, K.; Halabisky, B.; Deng, C.; Mahley, R.W.; Huang, Y. GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell 2009, 5, 634–645. [Google Scholar] [CrossRef]
- Li, Z.; Shue, F.; Zhao, N.; Shinohara, M.; Bu, G. APOE2: Protective mechanism and therapeutic implications for Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 63. [Google Scholar] [CrossRef]
- Belaidi, A.A.; Bush, A.I.; Ayton, S. Apolipoprotein E in Alzheimer’s disease: Molecular insights and therapeutic opportunities. Mol. Neurodegener. 2025, 20, 47. [Google Scholar] [CrossRef]
- Liu, S.; Liu, J.; Weng, R.; Gu, X.; Zhong, Z. Apolipoprotein E gene polymorphism and the risk of cardiovascular disease and type 2 diabetes. BMC Cardiovasc. Disord. 2019, 19, 213. [Google Scholar] [CrossRef]
- Federoff, M.; Jimenez-Rolando, B.; Nalls, M.A.; Singleton, A.B. A large study reveals no association between APOE and Parkinson’s disease. Neurobiol. Dis. 2012, 46, 389–392. [Google Scholar] [CrossRef]
- Harhangi, B.S.; de Rijk, M.C.; van Duijn, C.M.; Van Broeckhoven, C.; Hofman, A.; Breteler, M.M. APOE and the risk of PD with or without dementia in a population-based study. Neurology 2000, 54, 1272–1276. [Google Scholar] [CrossRef]
- Arboleda-Velasquez, J.F.; Lopera, F.; O’Hare, M.; Delgado-Tirado, S.; Marino, C.; Chmielewska, N.; Saez-Torres, K.L.; Amarnani, D.; Schultz, A.P.; Sperling, R.A.; et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: A case report. Nat. Med. 2019, 25, 1680–1683. [Google Scholar] [CrossRef]
- Slooter, A.J.; Cruts, M.; Kalmijn, S.; Hofman, A.; Breteler, M.M.; Van Broeckhoven, C.; van Duijn, C.M. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: The Rotterdam Study. Arch. Neurol. 1998, 55, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Alldred, M.J.; Lee, S.H.; Stutzmann, G.E.; Ginsberg, S.D. Oxidative Phosphorylation Is Dysregulated Within the Basocortical Circuit in a 6-month old Mouse Model of Down Syndrome and Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 707950. [Google Scholar] [CrossRef] [PubMed]
- Alldred, M.J.; Penikalapati, S.C.; Lee, S.H.; Heguy, A.; Roussos, P.; Ginsberg, S.D. Profiling Basal Forebrain Cholinergic Neurons Reveals a Molecular Basis for Vulnerability Within the Ts65Dn Model of Down Syndrome and Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 5141–5162. [Google Scholar] [CrossRef] [PubMed]
- Koffie, R.M.; Hashimoto, T.; Tai, H.C.; Kay, K.R.; Serrano-Pozo, A.; Joyner, D.; Hou, S.; Kopeikina, K.J.; Frosch, M.P.; Lee, V.M.; et al. Apolipoprotein E4 effects in Alzheimer’s disease are mediated by synaptotoxic oligomeric amyloid-beta. Brain 2012, 135, 2155–2168. [Google Scholar] [CrossRef]
- Konings, S.C.; Torres-Garcia, L.; Martinsson, I.; Gouras, G.K. Astrocytic and Neuronal Apolipoprotein E Isoforms Differentially Affect Neuronal Excitability. Front. Neurosci. 2021, 15, 734001. [Google Scholar] [CrossRef]
- Chen, Y.; Durakoglugil, M.S.; Xian, X.; Herz, J. ApoE4 reduces glutamate receptor function and synaptic plasticity by selectively impairing ApoE receptor recycling. Proc. Natl. Acad. Sci. USA 2010, 107, 12011–12016. [Google Scholar] [CrossRef]
- Dumanis, S.B.; Tesoriero, J.A.; Babus, L.W.; Nguyen, M.T.; Trotter, J.H.; Ladu, M.J.; Weeber, E.J.; Turner, R.S.; Xu, B.; Rebeck, G.W.; et al. ApoE4 decreases spine density and dendritic complexity in cortical neurons in vivo. J. Neurosci. 2009, 29, 15317–15322. [Google Scholar] [CrossRef]
- Schmukler, E.; Solomon, S.; Simonovitch, S.; Goldshmit, Y.; Wolfson, E.; Michaelson, D.M.; Pinkas-Kramarski, R. Altered mitochondrial dynamics and function in APOE4-expressing astrocytes. Cell Death Dis. 2020, 11, 578. [Google Scholar] [CrossRef]
- Andrews-Zwilling, Y.; Bien-Ly, N.; Xu, Q.; Li, G.; Bernardo, A.; Yoon, S.Y.; Zwilling, D.; Yan, T.X.; Chen, L.; Huang, Y. Apolipoprotein E4 causes age- and Tau-dependent impairment of GABAergic interneurons, leading to learning and memory deficits in mice. J. Neurosci. 2010, 30, 13707–13717. [Google Scholar] [CrossRef] [PubMed]
- Knoferle, J.; Yoon, S.Y.; Walker, D.; Leung, L.; Gillespie, A.K.; Tong, L.M.; Bien-Ly, N.; Huang, Y. Apolipoprotein E4 produced in GABAergic interneurons causes learning and memory deficits in mice. J. Neurosci. 2014, 34, 14069–14078. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Fu, Y.; Yamazaki, Y.; Ren, Y.; Davis, M.D.; Liu, C.C.; Lu, W.; Wang, X.; Chen, K.; Cherukuri, Y.; et al. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat. Commun. 2020, 11, 5540. [Google Scholar] [CrossRef]
- Blumenfeld, J.; Yip, O.; Kim, M.J.; Huang, Y. Cell type-specific roles of APOE4 in Alzheimer disease. Nat. Rev. Neurosci. 2024, 25, 91–110. [Google Scholar] [CrossRef] [PubMed]




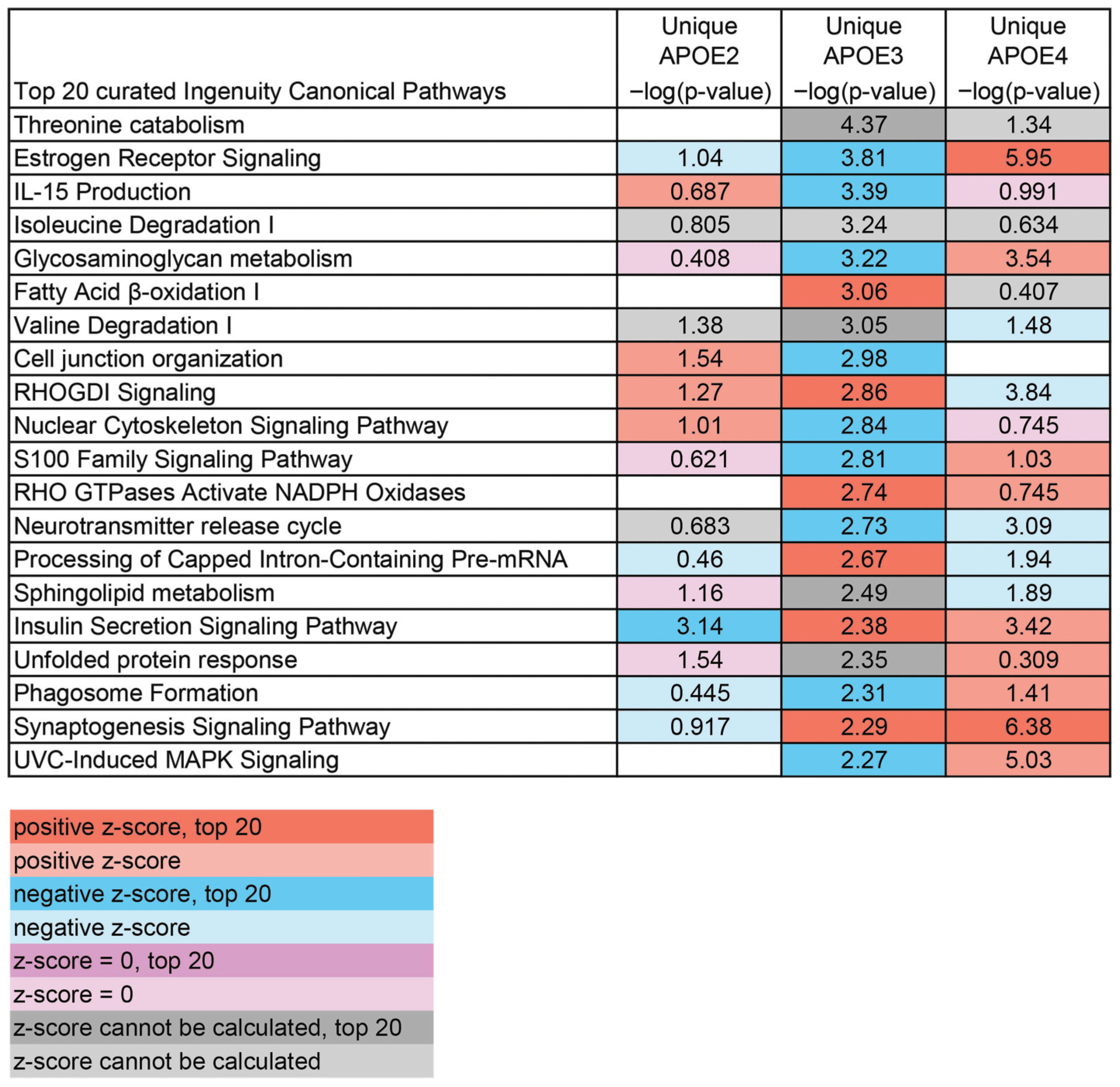

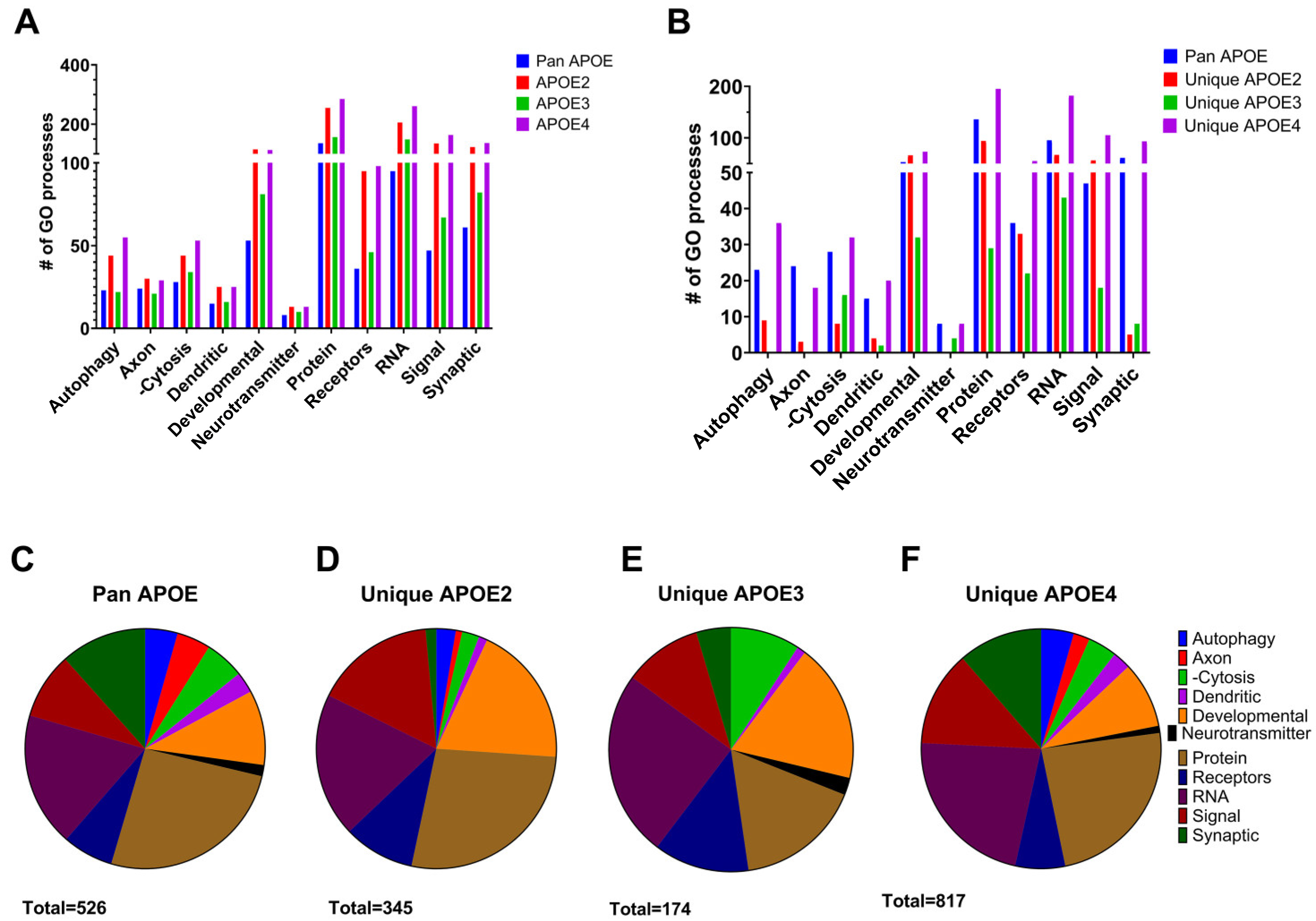

| Description | APOEZ −log(p-Value) | APOE3 −log(p-Value) | APOE4 −log(p-Value) | APOE4 Unique −log(p-Value) |
|---|---|---|---|---|
| Parkinson’s disease | 13.89 | 17.67 | 23.63 | |
| Huntington’s disease | 18.45 | 17.95 | 23.08 | |
| Alzheimer’s disease | 12.97 | 16.04 | 21.58 | |
| Amyotrophic lateral sclerosis | 15.74 | 16.42 | 18.28 | |
| Prion disease | 10.36 | 16.19 | 17.68 | |
| Retrograde endocannabinoid signaling | 8.50 | 5.03 | 14.14 | |
| Spinocerebellar ataxia | 8.92 | 7.39 | 13.63 | |
| Thermogenesis | 11.07 | 11.93 | 13.44 | |
| Autophagy–animal | 8.30 | 3.42 | 11.16 | |
| Oxidative phosphorylation | 11.97 | 15.45 | 10.93 | |
| Ubiquitin-mediated proteolysis | 7.50 | 4.58 | 8.08 | |
| Ribosome | 8.69 | 22.91 | 5.69 | |
| Proteasome | 3.22 | 3.54 | 4.05 | |
| Synaptic vesicle cycle | 2.74 | 2.58 | 3.75 | |
| RNA polymerase | 3.23 | 2.64 | 3.08 | |
| Pathways of neurodegeneration–multiple diseases | 14.32 | 16.36 | 21.97 | 2.87 |
| Mitophagy–animal | 5.30 | 2.76 | 12.08 | 3.05 |
| Protein processing in endoplasmic reticulum | 6.54 | 2.45 | 6.24 | 2.87 |
| Axon guidance | 7.35 | 12.31 | 6.10 | |
| Endocytosis | 5.42 | 10.93 | 4.57 | |
| Gap junction | 4.77 | 10.61 | 2.84 | |
| Long-term potentiation | 3.93 | 8.36 | 2.51 | |
| Glutamatergic synapse | 3.37 | 8.72 | 3.71 | |
| Inositol phosphate metabolism | 2.55 | 2.95 | 2.57 | |
| Choline metabolism in cancer | 2.42 | 5.80 | 2.49 | |
| Rap1 signaling pathway | 2.17 | 5.62 | 4.13 | |
| Adherens junction | 2.05 | 5.09 | 3.67 | |
| Dopaminergic synapse | 8.07 | 3.91 | ||
| Phospholipase D signaling pathway | 5.71 | 3.51 | ||
| EGFR tyrosine kinase inhibitor resistance | 5.38 | 2.45 | ||
| Amphetamine addiction | 5.09 | 4.75 | ||
| Cocaine addiction | 3.37 | 4.29 | ||
| MAPK signaling pathway | 3.12 | 3.90 | ||
| Nucleotide excision repair | 3.88 | 3.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labuza, A.; Pidikiti, H.; Alldred, M.J.; Ibrahim, K.W.; Peng, K.Y.; Pasato, J.; Heguy, A.; Mathews, P.M.; Ginsberg, S.D. Aging, Rather than Genotype, Is the Principal Contributor to Differential Gene Expression Within Targeted Replacement APOE2, APOE3, and APOE4 Mouse Brain. Brain Sci. 2025, 15, 1117. https://doi.org/10.3390/brainsci15101117
Labuza A, Pidikiti H, Alldred MJ, Ibrahim KW, Peng KY, Pasato J, Heguy A, Mathews PM, Ginsberg SD. Aging, Rather than Genotype, Is the Principal Contributor to Differential Gene Expression Within Targeted Replacement APOE2, APOE3, and APOE4 Mouse Brain. Brain Sciences. 2025; 15(10):1117. https://doi.org/10.3390/brainsci15101117
Chicago/Turabian StyleLabuza, Amanda, Harshitha Pidikiti, Melissa J. Alldred, Kyrillos W. Ibrahim, Katherine Y. Peng, Jonathan Pasato, Adriana Heguy, Paul M. Mathews, and Stephen D. Ginsberg. 2025. "Aging, Rather than Genotype, Is the Principal Contributor to Differential Gene Expression Within Targeted Replacement APOE2, APOE3, and APOE4 Mouse Brain" Brain Sciences 15, no. 10: 1117. https://doi.org/10.3390/brainsci15101117
APA StyleLabuza, A., Pidikiti, H., Alldred, M. J., Ibrahim, K. W., Peng, K. Y., Pasato, J., Heguy, A., Mathews, P. M., & Ginsberg, S. D. (2025). Aging, Rather than Genotype, Is the Principal Contributor to Differential Gene Expression Within Targeted Replacement APOE2, APOE3, and APOE4 Mouse Brain. Brain Sciences, 15(10), 1117. https://doi.org/10.3390/brainsci15101117








