Metallic Nanoparticles Obtained via “Green” Synthesis as a Platform for Biosensor Construction
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Enzymes
2.3. Green Synthesis of Nanoparticles
2.4. Characterization of the Synthesized NPs
2.5. Biosensor Evaluation
2.5.1. Apparatus
2.5.2. Immobilization of the Enzymes onto Electrodes
2.6. Statistical Analysis
3. Results and Discussion
3.1. Characterization of the Green-Synthesized Nanoparticles
3.2. Biosensor Evaluation
3.2.1. Laccase-Based Amperometric Biosensors
3.2.2. AO-Based ABSs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NPs | Nanoparticles |
| gNPs | Nanoparticles obtained via green synthesis |
| gPdNPs | Pd-based gNPs |
| gCr2O3NPs | Cr2O3-based gNPs |
| AO | Alcohol oxidase |
| HRP | Horseradish peroxidase |
| ABS | Amperometric biosensor |
| GE | Graphite electrode |
| ABTS | 2,2′-azinobis (3-ethylbenzothiazoline-6-sulfonate) diammonium salt |
| PB | Phosphate buffer |
| SEM | Scanning electron microscope |
| SEM-XRM | Scanning electron microscopy coupled with X-ray microanalysis |
| FM | Fluorescence microscope |
| RM | Reaction mixture |
| FTIR | Fourier transform infrared spectroscopy |
| BP | Bisphenol A |
| KMapp | Michaelis–Menten constant |
| Imax | Maximal current response on tested analyte at substrate saturation |
References
- Sastry, M.; Ahmad, A.; Khan, M.I.; Kumar, R. Microbial nanoparticle production. In Nanobiotechnology: Concepts, Applications and Perspectives; Niemeyer, C.M., Mirkin, C.A., Eds.; Wiley-VCH: Weinheim, Germany, 2004; pp. 126–135. [Google Scholar]
- Mandal, D.; Mishra, S.; Singh, R.K. Green synthesized nanoparticles as potential nanosensors. In Environmental, Chemical and Medical Sensors; Bhattacharya, S., Agarwal, A.K., Chanda, N., Pandey, A., Sen, A.K., Eds.; Springer: Singapore, 2018; pp. 137–164. ISBN 978-981-10-7750-0. [Google Scholar]
- Ali, J.; Zainab, S.; Ali, N. Green synthesis of metal nanoparticles by microorganisms; a current prospective. JNA 2015, 2, 32–38. [Google Scholar] [CrossRef]
- Soni, M.; Mehta, P.; Soni, A.; Goswami, G.K. Green nanoparticles: Synthesis and applications. IOSR J. Biotechnol. Biochem. 2018, 4, 78–83. [Google Scholar] [CrossRef]
- Naik, G.K.; Mishra, P.M.; Parida, K. Green synthesis of Au/TiO2 for effective dye degradation in aqueous system. Chem. Eng. J. 2013, 229, 492–497. [Google Scholar] [CrossRef]
- Yedurkar, S.M.; Maurya, C.B.; Mahanwar, P.A. Synthesis of nanoparticles by green chemistry process and their application in surface coatings: A review. Arch. Appl. Sci. Res. 2016, 8, 55–69. [Google Scholar] [CrossRef]
- Glatstein, D.A.; Bruna, N.; Gallardo-Benavente, C.; Bravo, D.; Pérez, M.E.; Francisca, C.F.M.; Pérez-Donoso, J.M. Arsenic and cadmium bioremediation by antarctic bacteria capable of biosynthesizing CdS fluorescent nanoparticles. J. Environ. Eng. 2018, 144, 04017107. [Google Scholar] [CrossRef]
- Mukherjee, P.; Ahmad, A.; Mandal, D.; Senapati, S.; Sainkar, S.R.; Khan, M.I.; Ramani, R.; Parischa, R.; Ajayakumar, P.V.; Alam, M.; et al. Bioreduction of AuCl4− ions by the fungus, Verticillium sp. and surface trapping of the gold nanoparticles formed. Angew. Chem. Int. Ed. 2001, 40, 3585–3588. [Google Scholar] [CrossRef]
- Anbazhagan, S.; Azeez, S.; Morukattu, G.; Rajan, R.; Venkatesan, K.; Thangavelu, K.P. Synthesis, characterization and biological applications of mycosynthesized silver nanoparticles. 3 Biotech 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Khatami, M.; Alijani, H.Q.; Sharifi, I. Biosynthesis of bimetallic and core–shell nanoparticles: Their biomedical applications. IET Nanobiotechnol. 2018, 12, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Amina, M.; Alarfaj, N.A.; El-Tohamy, M.F.; Al Musayeib, N.M.; Oraby, H.F. Sequential injection-chemiluminescence evaluation of stigmasterol glucoside and luteolin via green synthesis of silver nanoparticles using biomass of Plectranthus asirensis. Green Chem. Lett. Rev. 2018, 11, 523–533. [Google Scholar] [CrossRef]
- Aswathy, A.S.; Philip, D. Green synthesis of gold nanoparticles using Trigonella foenum-graecum and its size-dependent catalytic activity. Spectrochim. Acta A 2012, 97, 1–5. [Google Scholar] [CrossRef]
- Martinotti, M.G.; Allegrone, G.; Cavallo, M.; Fracchia, L. Biosurfactants. In Sustainable Development in Chemical Engineering Innovative Technologies; Piemonte, V., De Falco, M., Basile, A., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2013; pp. 199–240. ISBN 978-1-509-52363-4. [Google Scholar]
- Mandal, D.; Bolander, M.E.; Mukhopadhyay, D.; Sarkar, G.; Mukherjee, P. The use of microorganisms for the formation of metal nanoparticles and their application. Appl. Environ. Microb. 2006, 69, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Filice, M. Biosynthesis of metal nanoparticles: Novel efficient heterogeneous nanocatalysts. Nanomaterials 2016, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Rajan, A.; Meena, K.M.; Philip, D. Shape tailored green synthesis and catalytic properties of gold nanocrystals. Spectrochim. Acta A 2014, 118, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Adelere, I.A.; Lateef, A. A novel approach to the green synthesis of metallic nanoparticles: The use of agro-wastes, enzymes, and pigments. Nanotechnol. Rev. 2016, 5, 567–587. [Google Scholar] [CrossRef]
- Schlüter, M.; Hentzel, T.; Suarez, C.; Koch, M.; Lorenz, W.G.; Böhm, L.; Düring, R.A.; Koinig, K.A.; Bunge, M. Synthesis of novel palladium(0) nanocatalysts by microorganisms from heavy-metal-influenced high-alpine sites for dehalogenation of polychlorinated dioxins. Chemosphere 2014, 117, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Sunkari, S.; Gangapuram, B.R.; Dadigala, R.; Bandi, R.; Alle, M.; Guttena, V. Microwave-irradiated green synthesis of gold nanoparticles for catalytic and anti-bacterial activity. J. Anal. Sci. Technol. 2017, 8, 9. [Google Scholar] [CrossRef]
- Kalpana, V.N.; Rajeswari, D. A Review on green synthesis, biomedical applications, and toxicity studies of ZnO NPs. Bioinorg. Chem. Appl. 2018, 2018, 3569758. [Google Scholar] [CrossRef]
- Ramesh, C.; Mohan Kumar, K.T.; Latha, N.; Ragunathan, V. Green synthesis of Cr2O3 nanoparticles using tridax procumbens leaf extract and its antibacterial activity on Escherichia coli. Curr. Nanosci. 2012, 8, 603–607. [Google Scholar] [CrossRef]
- Sonker, A.S.; Richa; Pathak, J.; Rajneesh; Kannaujiya, V.K.; Sinha, R.P. Characterization and in vitro antitumor, antibacterial and antifungal activities of green synthesized silver nanoparticles using cell extract of Nostoc sp. strain HKAR-2. Can. J. Biotech. 2017, 1, 26–37. [Google Scholar] [CrossRef]
- Ali, S.G.; Ansari, M.A.; Khan, H.M.; Jalal, M.; Mahdi, A.A.; Cameotra, S.S. Antibacterial and antibiofilm potential of green synthesized silver nanoparticles against imipenem resistant clinical isolates of P. aeruginosa. Bionanoscience 2018, 8, 544–553. [Google Scholar] [CrossRef]
- Owaid, M.N.; Zaidan, T.A.; Muslim, R.F.; Hammood, M.A. Biosynthesis, characterization and cytotoxicity of zinc nanoparticles using Panax ginseng roots, Araliaceae. Acta Pharm. Sci. 2019, 57. [Google Scholar] [CrossRef]
- Al-Bahrani, R.M.; Majeed, S.M.A.; Owaid, M.N.; Mohammed, A.B.; Rheem, D.A. Phyto-fabrication, characteristics and anticandidal effects of silver nanoparticles from leaves of Ziziphus mauritiana Lam. Acta Pharm. Sci. 2018, 56, 85–92. [Google Scholar] [CrossRef]
- Kharissova, O.V.; Dias, H.V.; Kharisov, B.I. The greener synthesis of nanoparticles. Trends Biotechnol. 2013, 31, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Hulkoti, N.I.; Taranath, T.C. Biosynthesis of nanoparticles using microbes—A review. Colloids Surf. B Biointerfaces 2014, 121, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Abbas, S.; Fariq, A.; Yasmin, A. Microbes: nature’s cell factories of nanoparticles synthesis. In Exploring the Realms of Nature for Nanosynthesis; Prasad, R., Jha, A., Prasad, K., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 25–50. ISBN 978-3-319-99570-0. [Google Scholar]
- Manivasagan, P.; Nam, S.Y.; Oh, J. Marine microorganisms as potential biofactories for synthesis of metallic nanoparticles. Crit. Rev. Microbiol. 2016, 42, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.R.; Yong, P.; Macaskie, L.E. Enzymatic recovery of elemental palladium by using sulfate-reducing bacteria. Appl. Environ. Microb. 1998, 64, 4607–4609. [Google Scholar]
- Rehman, F.U.; Jiang, H.; Selke, M.; Wang, X. Mammalian cells: A unique scaffold for in situ biosynthesis of metallic nanomaterials and biomedical applications. J. Mater. Chem. B 2018, 6, 6501–6514. [Google Scholar] [CrossRef]
- Wang, C.; Gao, X.; Chen, Z.; Chen, Y.H. Preparation, Characterization and application of polysaccharide-based metallic nanoparticles: A Review. Polymers 2017, 9, 689. [Google Scholar] [CrossRef]
- Płaza, G.A.; Chojniak, J.; Banat, I.M. Biosurfactant mediated biosynthesis of selected metallic nanoparticles. Int. J. Mol. Sci. 2014, 15, 13720–13737. [Google Scholar] [CrossRef]
- Kaliaraj, G.S.; Bharathiraja, S.; Manivasagan, P.; Kim, S.-K. Green synthesis of metal nanoparticles using seaweed polysaccharides. In Seaweed Polysaccharides: Isolation, Biological and Biomedical Applications; Venkatesan, J., Anil, S., Kim, S.-K., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 101–109. ISBN 978-0-12-809816-5. [Google Scholar]
- Al-Kalifawi, E.J.; Al-Obodi, E.E.; Al-Saadi, T.M. Characterization of Cr2O3 nanoparticles prepared by using different plant extracts. Acad. J. Agric. Res. 2018, 6, 026–032. [Google Scholar] [CrossRef]
- Sone, B.T.; Manikandan, E.; Gurib-Fakim, A.; Maaza, M. Single-phase α-Cr2O3 nanoparticles’ green synthesis using Callistemon viminalis’ red flower extract. Green Chem. Lett. Rev. 2016, 9, 85–90. [Google Scholar] [CrossRef]
- Kunoh, T.; Shimura, T.; Kasai, T.; Matsumoto, S.; Mahmud, H.; Khayrani, A.C.; Seno, M.; Kunoh, H.; Takada, J. Use of DNA-generated gold nanoparticles to radiosensitize and eradicate radioresistant glioma stem cells. Nanotechnology 2019, 30, 055101. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Narayan, S.; Chadha, A. Whole resting cells vs. cell free extracts of Candida parapsilosis ATCC 7330 for the synthesis of gold nanoparticles. AMB Express 2016, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.K.; Chisti, Y.; Banerjee, U.C. Synthesis of metallic nanoparticles using plant extracts. Biotechnol. Adv. 2013, 31, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Deplanche, K.; Caldelari, I.; Mikheenko, I.P.; Sargent, F.; Macaskie, L.E. Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia coli mutant strains. Microbiology 2010, 156, 2630–2640. [Google Scholar] [CrossRef] [PubMed]
- Pantidos, N.; Horsfall, L.E. Biological synthesis of metallic nanoparticles by bacteria, fungi and plants. J. Nanomed. Nanotechnol. 2014, 5, 1000233. [Google Scholar] [CrossRef]
- Chinnadayyala, S.R.; Santhosh, M.; Singh, N.K.; Goswami, P. Alcohol oxidase protein mediated in-situ synthesized and stabilized gold nanoparticles for developing amperometric alcohol biosensor. Biosens. Bioelectron. 2015, 69, 155–161. [Google Scholar] [CrossRef]
- Vetchinkina, E.P.; Loshchinina, E.A.; Vodolazov, I.R.; Kursky, V.F.; Dykman, L.A.; Nikitina, V.E. Biosynthesis of nanoparticles of metals and metalloids by basidiomycetes. Preparation of gold nanoparticles by using purified fungal phenol oxidases. Appl. Microbiol. Biotechnol. 2017, 101, 1047–1062. [Google Scholar] [CrossRef]
- Lin, P.C.; Lin, S.; Wang, P.C.; Sridhar, R. Techniques for physicochemical characterization of nanomaterials. Biotechnol. Adv. 2014, 32, 711–726. [Google Scholar] [CrossRef]
- Ramesh, A.; Sundari, M.T.; Thirugnanam, P.E. Microbial molecular mechanisms in biosynthesis of nanoparticles. In Bio-Nanoparticles: Biosynthesis and Sustainable Biotechnological Implications; Singh, O.V., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 53–81. [Google Scholar]
- Chokkareddy, R.; Redhi, G.G. Green synthesis of metal nanoparticles and its reaction mechanisms. In Green Metal Nanoparticles: Synthesis, Characterization and Their Application; Kanchi, S., Ahmed, S., Eds.; Scrivener Publishing LLC: Beverly, MA, USA, 2018; pp. 113–139. ISBN 978-1-119-41887-0. [Google Scholar]
- Marslin, G.; Siram, K.; Maqbool, Q.; Selvakesavan, R.K.; Kruszka, D.; Kachlicki, P.; Franklin, G. Secondary metabolites in the green synthesis of metallic nanoparticles. Materials 2018, 11, 940. [Google Scholar] [CrossRef]
- Pandey, S.; Oza, G.; Gupta, A.; Shah, R.; Sharon, M. The possible involvement of nitrate reductase from Asparagus racemosus in biosynthesis of gold nanoparticles. Eur. J. Exp. Biol. 2012, 2, 475–483. [Google Scholar]
- Lipke, P.N.; Ramsook, C.; Garcia-Sherman, M.C.; Jackson, D.N.; Chan, C.X.J.; Bois, M.; Klotz, S.A. Between amyloids and aggregation lies a connection with strength and adhesion. New J. Sci. 2014, 2014, 815102. [Google Scholar] [CrossRef] [PubMed]
- Raveendran, P.; Fu, J.; Wallen, S.L. Completely “green” synthesis and stabilization of metal nanoparticles. J. Am. Chem. Soc. 2003, 125, 13940–13941. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.S.; Naraian, R. Biosynthesis of nanoparticles by penicillium and their medical applications. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.G., Rodriguez-Couto, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 235–246. [Google Scholar]
- Ksheminska, H.P.; Honchar, T.M.; Gayda, G.Z.; Gonchar, M.V. Extra cellular chromate reducing activity of the yeast cultures. Cent. Eur. J. Biol. 2006, 1, 137–149. [Google Scholar] [CrossRef]
- Ksheminska, H.; Honchar, T.; Usatenko, Y.; Gonchar, M.V. The chromate resistance phenotype of some yeast mutants correlates with a lower level of Cr(V)-species generated in the extra-cellular medium. Biometals 2010, 23, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Ksheminska, H.P.; Gayda, G.Z.; Ivash, M.F.; Gonchar, M.V. Chromate-resistant mutants of the yeast Pichia guilliermondii: Selection and properties. Microbiology 2011, 80, 314–325. [Google Scholar] [CrossRef]
- Ksheminska, H.P.; Gayda, G.Z.; Ivash, M.F.; Gonchar, M.V. The details of extra-cellular chromate reduction by the yeasts Pichia guilliermondii. First Indep. Sci. J. 2015, 1–2, 18–23. [Google Scholar]
- Stasyuk, N.; Gayda, G.; Serkiz, R.; Gonchar, M. The “green” synthesis of gold nanoparticles by the yeast Hansenula polymorpha. Visnyk Lviv. Univ. (Ser. Biol.) 2016, 73, 96–102. [Google Scholar]
- Lootsik, M.D.; Bilyy, R.A.; Lutsyk, M.M.; Stoika, R.S. Preparation of chitosan with high blood clotting activity and its hemostatic potential assessment. Biotechnol. Acta 2015, 8, 32–40. [Google Scholar] [CrossRef]
- Shleev, S.V.; Shumakovich, G.P.; Nikitina, O.V.; Morozova, O.V.; Pavlishko, H.M.; Gayda, G.Z.; Gonchar, M.V. Purification and characterization of alcohol oxidase from the genetically constructed over-producing strain of the methylotrophic yeast Hansenula polymorpha. Biochemistry (Moscow) 2006, 71, 245–250. [Google Scholar] [CrossRef]
- Stasyk, O.V.; Boretsky, Y.R.; Gonchar, M.V.; Sibirny, A.A. Recombinant arginine-degrading enzymes in metabolic anticancer therapy and bioanalytics. Cell Biol. Int. 2015, 39, 246–252. [Google Scholar] [CrossRef]
- Gonchar, M.V.; Maidan, M.M.; Pavlishko, H.M.; Sibirny, A.A. A new oxidase-peroxidase kit for ethanol assays in alcoholic beverages. Food Technol. Biotech. 2001, 39, 37–42. [Google Scholar]
- Zdansky, K.; Kacerovsky, P.; Zavadil, J.; Lorincik, J.; Fojtik, A. Layers of metal nanoparticles on semiconductors deposited by electrophoresis from solutions with reverse micelles. Nanoscale Res. Lett. 2007, 2, 450–454. [Google Scholar] [CrossRef]
- Stasyuk, N.Y.; Gayda, G.Z.; Serkiz, R.J.; Gonchar, M.V. Cell imaging with fluorescent bi-metallic nanoparticles. J. Adv. Chem. 2015, 11, 3499–3511. [Google Scholar] [CrossRef]
- Tortolini, C.; Rea, S.; Carota, E.; Cannistraro, S.; Mazzei, F. Influence of the immobilization procedures on the electroanalytical performances of Trametes versicolor laccase based bioelectrode. Microchem. J. 2012, 100, 8–13. [Google Scholar] [CrossRef]
- Favero, G.; Fusco, G.; Mazzei, F.; Tasca, F.; Antiochia, R. Electrochemical characterization of graphene and MWCNT screen-printed electrodes modified with AuNPs for laccase biosensor development. Nanomaterials 2015, 5, 1995–2006. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qu, X.; Guo, H.; Chen, H.; Liu, B.; Dong, S. Facile preparation of amperometric laccase biosensor with multifunction based on the matrix of carbon nanotubes–chitosan composite. Biosens. Bioelectron. 2006, 21, 2195–2201. [Google Scholar] [CrossRef]
- Gonzalez-Rivera, J.C.; Hindawi, J.F.O. Fabrication of an amperometric flow-injection microfluidic biosensor based on laccase for in situ determination of phenolic compounds. BioMed Res. Int. 2015, 2015, 845261. [Google Scholar] [CrossRef]
- Pelle, F.D.; Compagnone, D. Nanomaterial-based sensing and biosensing of phenolic compounds and related antioxidant capacity in food. Sensors 2018, 18, 462. [Google Scholar] [CrossRef]
- Jarosz-Wilkołazka, A.; Ruzgas, T.; Gorton, L. Amperometric detection of mono- and diphenols at Cerrena unicolor laccase-modified graphite electrode: Correlation between sensitivity and substrate structure. Talanta 2005, 66, 1219–1224. [Google Scholar] [CrossRef]
- Rodríguez-Delgado, M.M.; Alemán-Nava, G.S.; Rodríguez-Delgado, J.-M.; Dieck-Assad, G.; Martínez-Chapa, S.O.; Barceló, D.; Parra, R. Laccase-based biosensors for detection of phenolic compounds. TrAC Trend Anal. Chem. 2015, 74, 21–45. [Google Scholar] [CrossRef]
- Mousty, C.; Vieille, L.; Cosnier, S. Laccase immobilization in redox active layered double hydroxides: A reagentless amperometric biosensor. Biosens. Bioelectron. 2007, 22, 1733–1738. [Google Scholar] [CrossRef] [PubMed]
- Smutok, O.; Ngounou, B.; Pavlishko, H.; Gayda, G.; Gonchar, M.; Schuhmann, W. A reagentless bienzyme amperometric biosensor based on alcohol oxidase/peroxidase and an Os-complex modified electrodeposition paint. Sens. Actuators B Chem. 2006, 113, 590–598. [Google Scholar] [CrossRef]
- Smutok, O.; Gayda, G.; Dmytruk, K.; Klepach, H.; Nisnevitch, M.; Sibirny, A.; Puchalski, C.; Broda, D.; Schuhmann, W.; Gonchar, M.; et al. Amperometric biosensors for lactate, alcohols, and glycerol assays in clinical diagnostics, In Biosensors—Emerging Materials and Applications; Serra, P.A., Ed.; InTech: Rijeka, Croatia, 2011; pp. 401–446. Available online: https://www.intechopen.com/books/biosensors-emerging-materials-and-applications/amperometric-biosensors-for-lactate-alcohols-and-glycerol-assays-in-clinical-diagnostics (accessed on 20 December 2018).
- Dmytruk, K.V.; Smutok, O.V.; Ryabova, O.B.; Gayda, G.Z.; Sibirny, V.A.; Schuhmann, W.; Gonchar, M.V.; Sibirny, A.A. Isolation and characterization of mutated alcohol oxidases from the yeast Hansenula polymorpha with decreased affinity toward substrates and their use as selective elements of an amperometric biosensor. BMC Biotechnol. 2007, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Karkovska, M.I.; Stasyuk, N.Y.; Gayda, G.Z.; Smutok, O.V.; Gonchar, M.V. Nanomaterials in construction of biosensors of biomedical purposes. In Multifunctional Nanomaterials for Biology and Medicine: Molecular Design, Synthesis and Application; Stoika, R., Ed.; Naukova Dumka: Kyiv, Ukraine, 2017; pp. 165–177. (In Ukrainian) [Google Scholar]
- Gonchar, M.V.; Ksheminska, G.P.; Hladarevska, N.M.; Sibirny, A.A. Catalase-minus mutants of methylotrophic yeast Hansenula polymorpha impaired in regulation of alcohol oxidase synthesis. In Genetics of Respiratory Enzymes in Yeasts; Lachowicz, T.M., Ed.; Wroclaw University Press: Wroclaw, Poland, 1990; pp. 222–228. [Google Scholar]
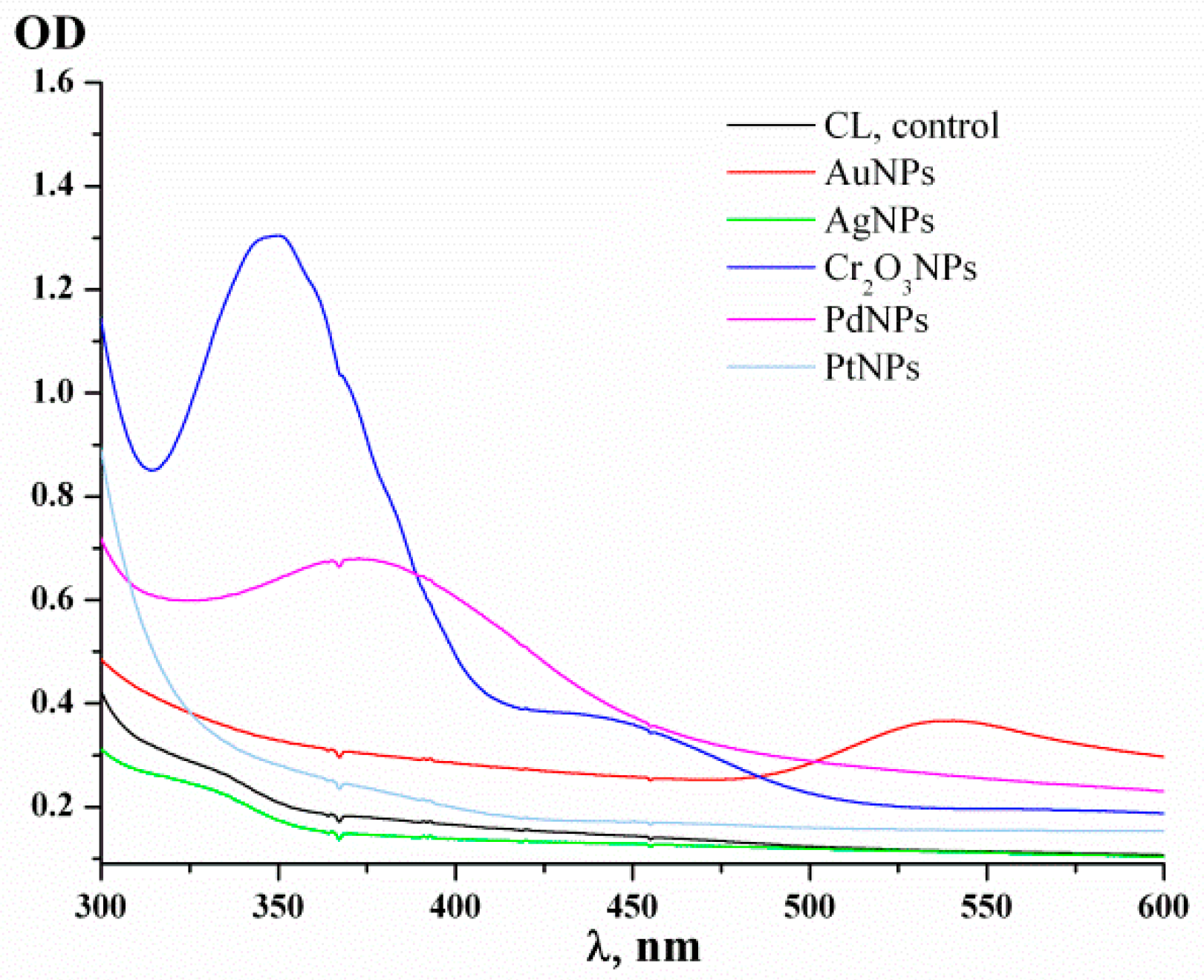
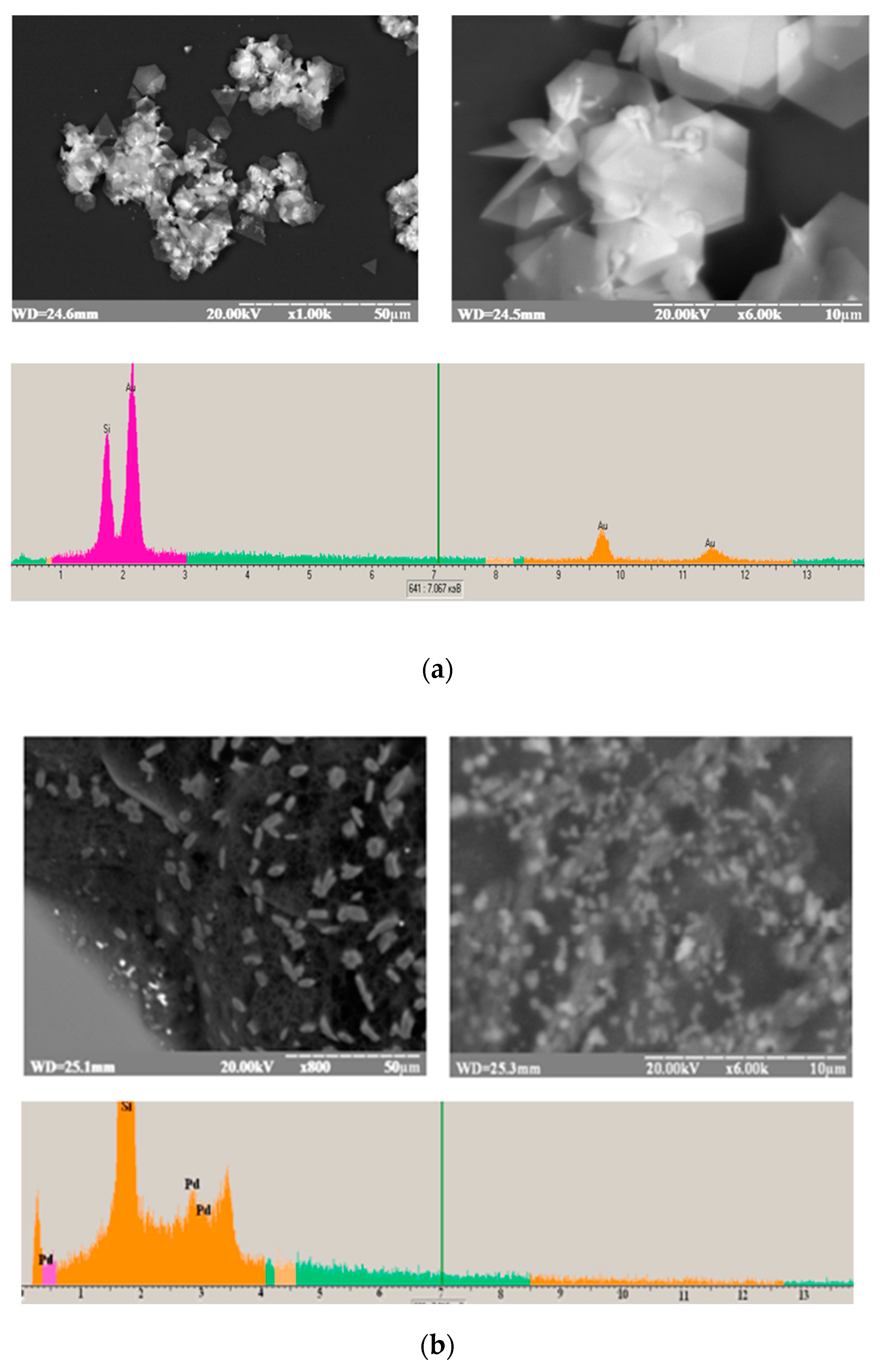
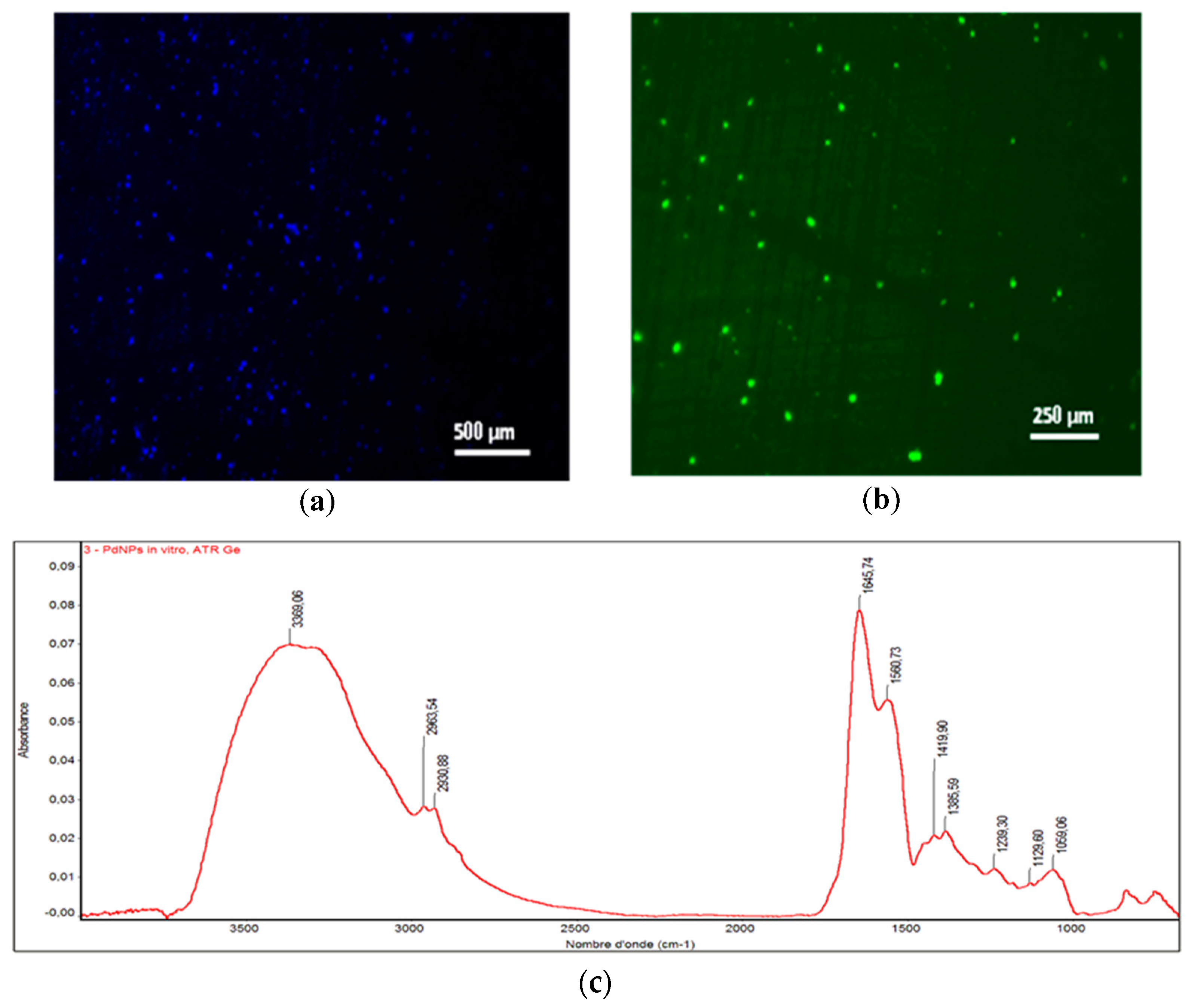

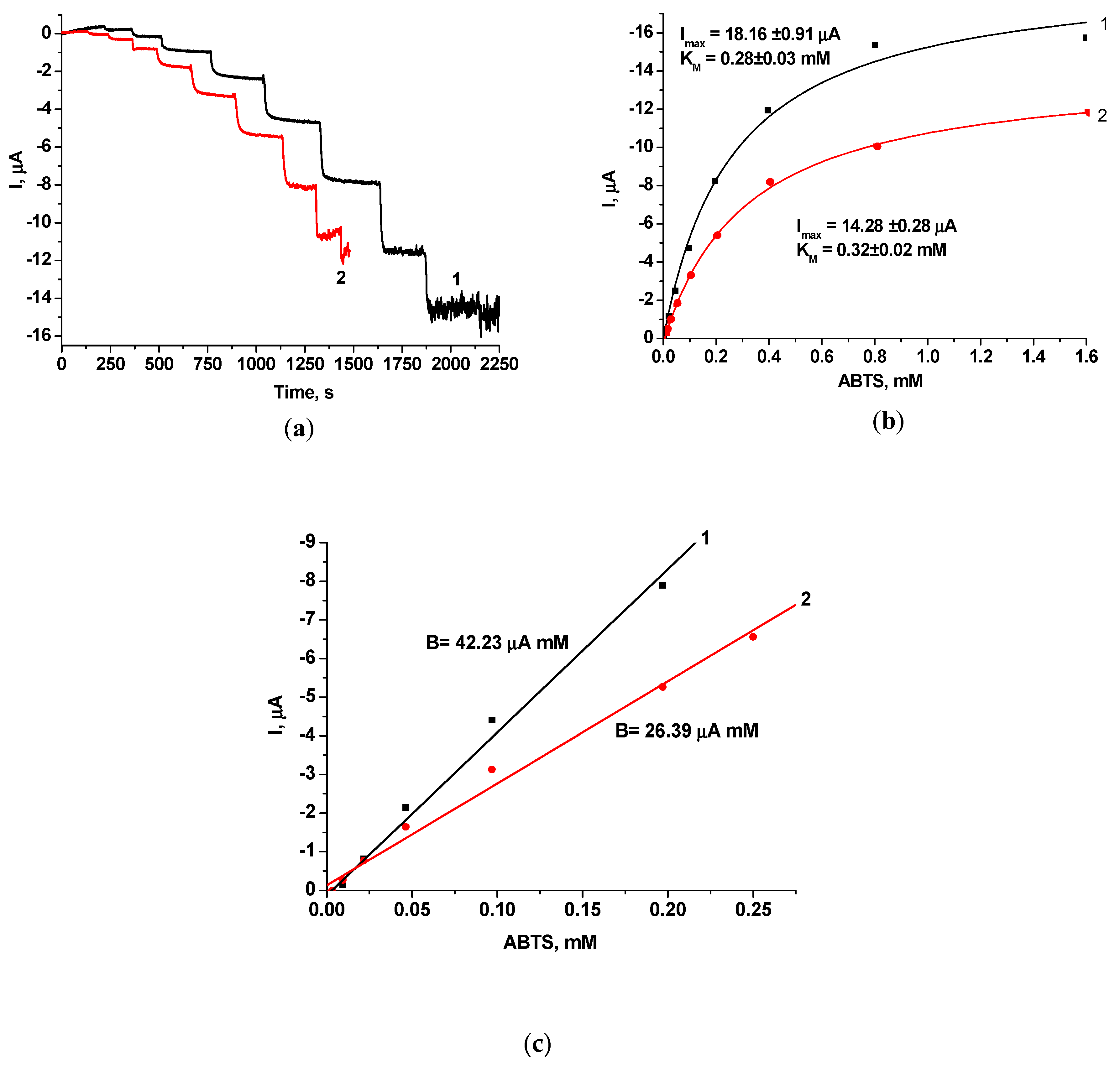
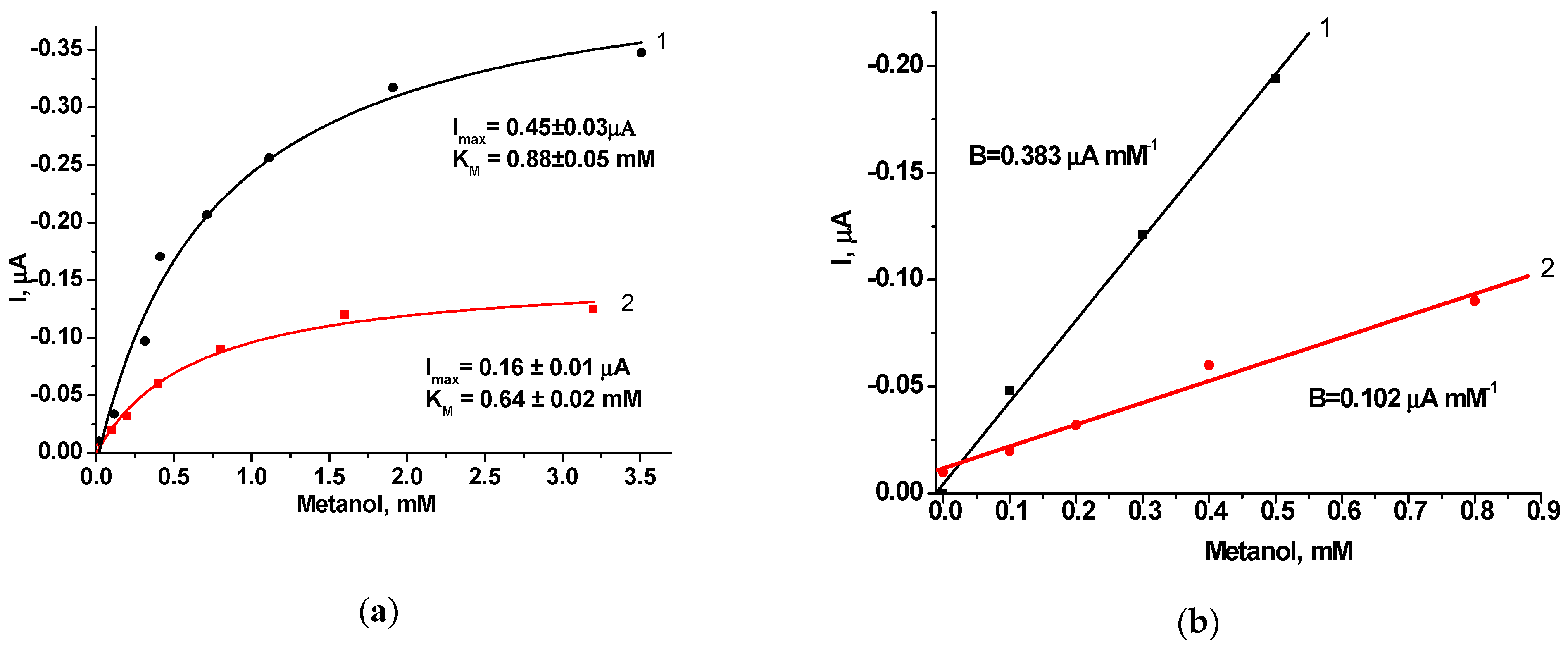
| Bioelectrode | Substrate | KMapp, mM | Imax, µA | Linear Range, up to mM | Sensitivity, A·M−1·m−2 | LOD 1, mM |
|---|---|---|---|---|---|---|
| Laccase/GE | BP | 0.73 ± 0.05 | 1.85 ± 0.04 | 0.35 | 232 | 0.050 |
| Laccase-gPdNPs/GE | BP | 1.80 ± 0.16 | 1.86 ± 0.05 | 0.50 | 137 | 0.145 |
| Laccase/GE | ABTS | 0.28 ± 0.03 | 19.16 ± 0.91 | 0.20 | 5780 | 0.008 |
| Laccase-gPdNPs/GE | ABTS | 0.32 ± 0.02 | 14.29 ± 0.28 | 0.25 | 3620 | 0.014 |
| Bioelectrode | KMapp, mM | Imax, µA | Linear Range, up to mM | Sensitivity, A·M−1·m−2 | LOD 2, mM |
|---|---|---|---|---|---|
| AO/HRP/GE | 0.88 ± 0.05 | 0.45 ± 0.03 | 0.5 | 52 | 0.005 |
| AO/HRP-gPdNPs/GE | 0.64 ± 0.02 | 0.16 ± 0.01 | 0.8 | 14 | 0.011 |
| AO/HRP-EDP 1/GE [71] | 0.48 | 4 | 1 | 66 | ND 3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gayda, G.Z.; Demkiv, O.M.; Stasyuk, N.Y.; Serkiz, R.Y.; Lootsik, M.D.; Errachid, A.; Gonchar, M.V.; Nisnevitch, M. Metallic Nanoparticles Obtained via “Green” Synthesis as a Platform for Biosensor Construction. Appl. Sci. 2019, 9, 720. https://doi.org/10.3390/app9040720
Gayda GZ, Demkiv OM, Stasyuk NY, Serkiz RY, Lootsik MD, Errachid A, Gonchar MV, Nisnevitch M. Metallic Nanoparticles Obtained via “Green” Synthesis as a Platform for Biosensor Construction. Applied Sciences. 2019; 9(4):720. https://doi.org/10.3390/app9040720
Chicago/Turabian StyleGayda, Galina Z., Olha M. Demkiv, Nataliya Ye. Stasyuk, Roman Ya. Serkiz, Maksym D. Lootsik, Abdelhamid Errachid, Mykhailo V. Gonchar, and Marina Nisnevitch. 2019. "Metallic Nanoparticles Obtained via “Green” Synthesis as a Platform for Biosensor Construction" Applied Sciences 9, no. 4: 720. https://doi.org/10.3390/app9040720
APA StyleGayda, G. Z., Demkiv, O. M., Stasyuk, N. Y., Serkiz, R. Y., Lootsik, M. D., Errachid, A., Gonchar, M. V., & Nisnevitch, M. (2019). Metallic Nanoparticles Obtained via “Green” Synthesis as a Platform for Biosensor Construction. Applied Sciences, 9(4), 720. https://doi.org/10.3390/app9040720






