Analysis of Protein–Receptor Interactions on an Example of Leptin–Leptin Receptor Interaction Using the Resonant Recognition Model
Abstract
Featured Application
Abstract
1. Introduction
2. Methods and Materials
2.1. Methods—Resonant Recognition Model (RRM)
2.2. Materials—Protein Sequences Analyzed by RRM
2.2.1. Twenty Leptin Proteins:
2.2.2. Five Leptin Receptor Proteins:
2.2.3. Five Neuropeptide Y Proteins:
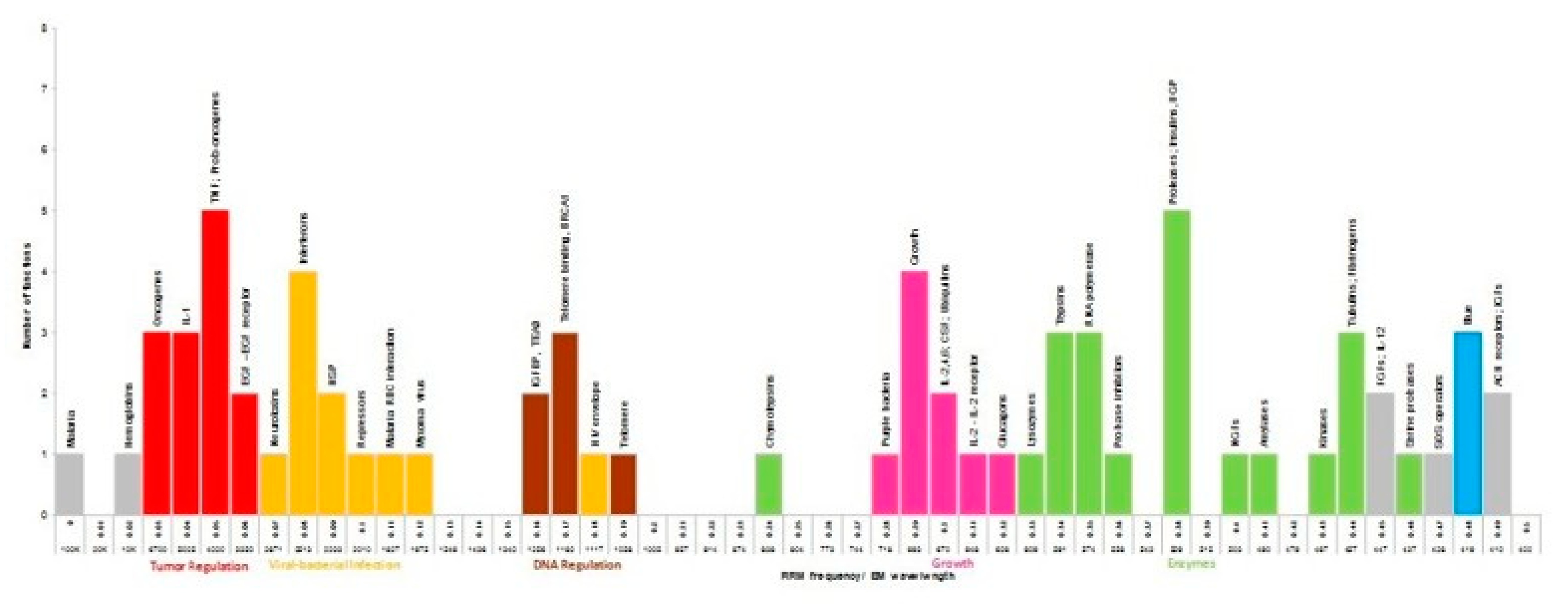
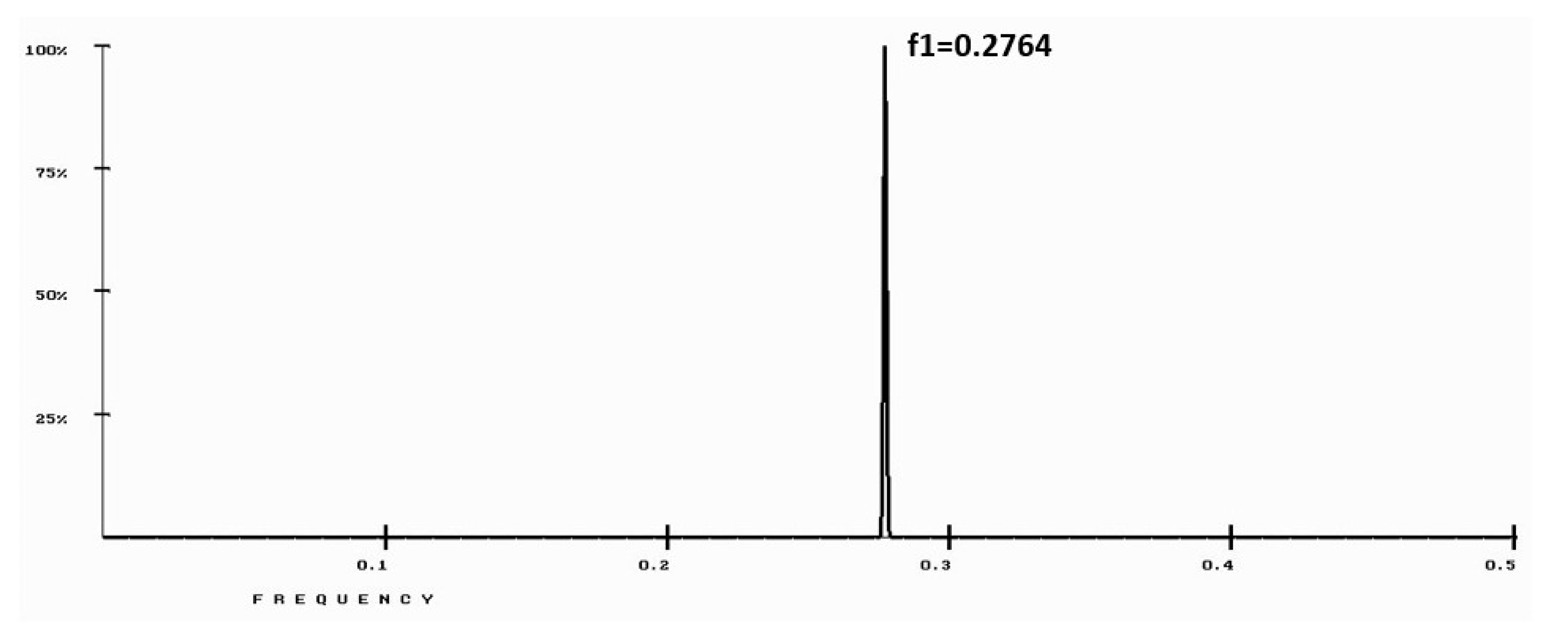
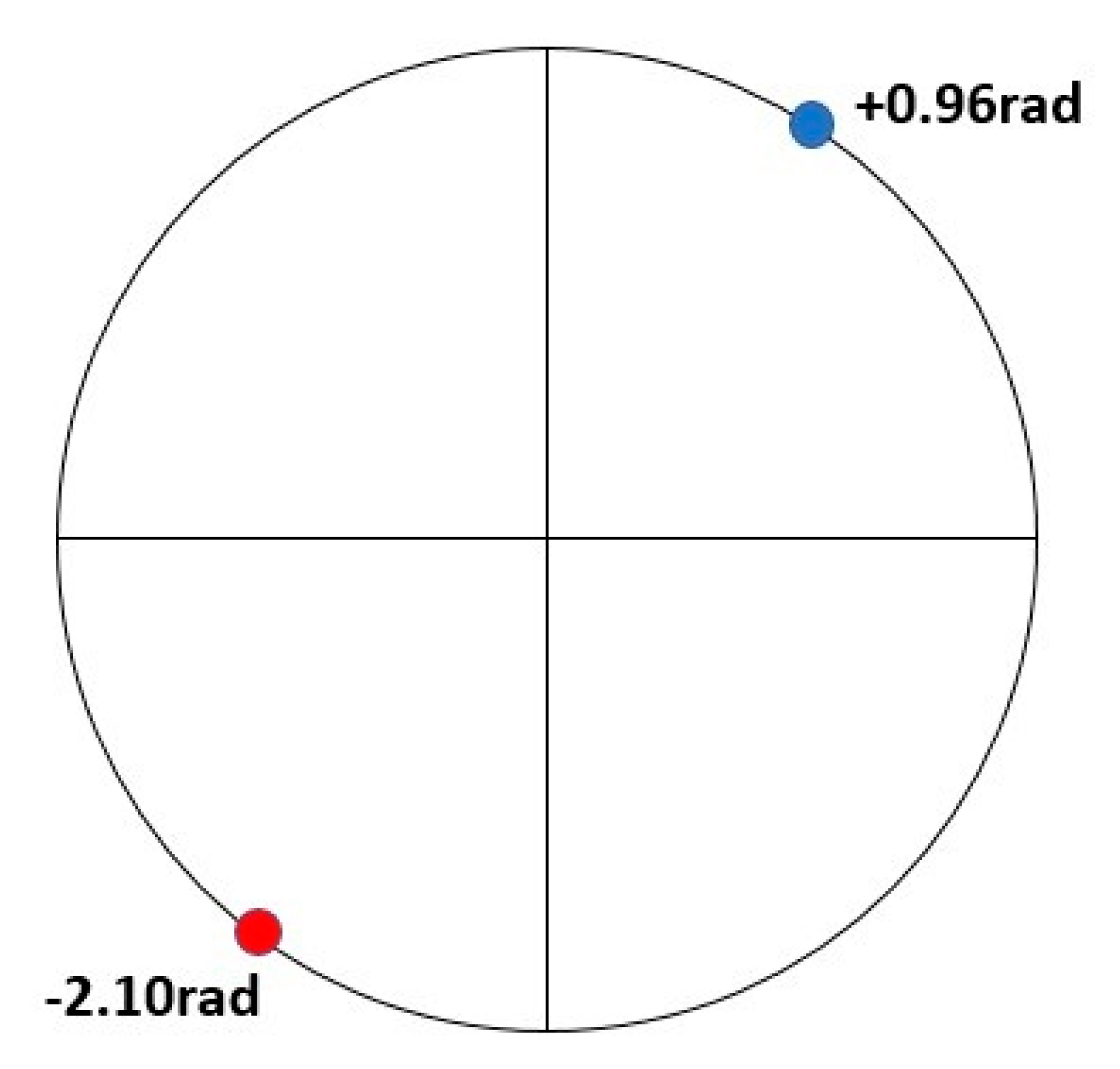
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cosic, I. Macromolecular Bioactivity: Is it Resonant Interaction between Macromolecules? Theory and Applications. IEEE Trans Biomed. Eng. 1994, 41, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I. Virtual spectroscopy for fun and profit. Biotechnology 1995, 13, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I. The Resonant Recognition Model of Macromolecular Bioactivity: Theory and Applications; Birkhauser Verlag: Basel, Switzerland, 1997. [Google Scholar]
- Cosic, I.; Drummond, A.E.; Underwood, J.R.; Hearn, M.T.W. In vitro inhibition of the actions of basic FGF by novel 16 amino acid peptides. Mol. Cell. Biochem. 1994, 130, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Cosic, D. Balancing insulin stability and activity—Rational approach. Med. Data 2014, 6, 7–10. [Google Scholar]
- Cosic, I.; Cosic, D.; Lazar, K. Analysis of Tumor Necrosis Factor Function Using the Resonant Recognition Model. Cell Biochem. Biophys. 2015. [Google Scholar] [CrossRef]
- Cosic, I.; Paspaliaris, V.; Cosic, D.; Kolios, G. Analysis of Interleukin-12 and Interleukin-23 Pathways to Distinguish between Immune Activation and Inflammation Functions. Med. Data 2019, 11, 7–14. [Google Scholar]
- Haslam, D.W.; James, W.P. Obesity. Lancet 2005, 366, 1197–1209. [Google Scholar] [CrossRef]
- Luppino, F.S.; de Wit, L.M.; Bouvy, P.F.; Stijnen, T.; Cuijpers, P.; Penninx, B.W.; Zitman, F.G. Overweight, obesity, and depression: A systematic review and meta-analysis of longitudinal studies. Arch. Gener. Psychiatry 2010, 67, 220–229. [Google Scholar] [CrossRef]
- Yazdi, F.T.; Clee, S.M.; Meyre, D. Obesity genetics in mouse and human: Back and forth, and back again. PeerJ 2015, 3, e856. [Google Scholar] [CrossRef]
- Brennan, A.M.; Mantzoros, C.S. Drug Insight: The Role of Leptin in Human Physiology and Pathophysiology—Emerging Clinical Applications. Nat. Clin. Pract. Endocrinol. Metab. 2006, 2, 318–327. [Google Scholar] [CrossRef]
- Baicy, K.; London, E.D.; Monterosso, J.; Wong, M.L.; Delibasi, T.; Sharma, A.; Licinio, J. Leptin Replacement Alters Brain Response to Food Cues in Genetically Leptin-Deficient Adults. Proc. Natl. Acad. Sci. USA 2007, 104, 18276–18279. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Lazar, K.; Cosic, D. Prediction of Tubulin resonant frequencies using the Resonant Recognition Model (RRM). IEEE Trans. Nanobiosci. 2015, 12, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Cosic, D.; Lazar, K. Is it possible to predict electromagnetic resonances in proteins, DNA and RNA? Nonlinear Biomed. Phys. 2015, 3. [Google Scholar] [CrossRef]
- Cosic, I.; Cosic, D.; Lazar, K. Environmental Light and Its Relationship with Electromagnetic Resonances of Biomolecular Interactions, as Predicted by the Resonant Recognition Model. Int. J. Environ. Res. Public Health 2016, 13, 647. [Google Scholar] [CrossRef] [PubMed]
- Cosic, I.; Cosic, D. The Treatment of Crigler-Najjar Syndrome by Blue Light as Explained by Resonant Recognition Model. EPJ Nonlinear Biomed. Phys. 2016, 4. [Google Scholar] [CrossRef]
- Vojisavljevic, V.; Pirogova, E.; Cosic, I. The Effect of Electromagnetic Radiation (550nm-850nm) on I-Lactate Dehydrogenase Kinetics. Int. J. Radiat. Biol. 2007, 83, 221–230. [Google Scholar] [CrossRef]
- Dotta, B.T.; Murugan, N.J.; Karbowski, L.M.; Lafrenie, R.M.; Persinger, M.A. Shifting wavelength of ultraweak photon emissions from dying melanoma cells: Their chemical enhancement and blocking are predicted by Cosic’s theory of resonant recognition model for macromolecules. Naturwissenschaften 2014, 101. [Google Scholar] [CrossRef]
- Murugan, N.J.; Karbowski, L.M.; Persinger, M.A. Cosic’s Resonance Recognition Model for Protein Sequences and Photon Emission Differentiates Lethal and Non-Lethal Ebola Strains: Implications for Treatment. Open J. Biophys. 2014, 5, 35. [Google Scholar] [CrossRef]
- Karbowski, L.M.; Murugan, N.J.; Persinger, M.A. Novel Cosic resonance (standing wave) solutions for components of the JAK-STAT cellular signalling pathway: A convergence of spectral density profiles. FEBS Open Bio 2015, 5, 245–250. [Google Scholar] [CrossRef]
- Cosic, I.; Paspaliaris, V.; Cosic, D. Explanation of Osteoblastic Differentiation of Stem Cells by Photo Biomodulation Using the Resonant Recognition Model. Appl. Sci. 2019, 9, 1979. [Google Scholar] [CrossRef]
- Krsmanovic, V.; Biquard, J.M.; Sikorska-Walker, M.; Cosic, I.; Desgranges, C.; Trabaud, M.A.; Whitfield, J.F.; Durkin, J.P.; Achour, A.; Hearn, M.T. Investigation Into the Cross-reactivity of Rabbit Antibodies Raised against Nonhomologous Pairs of Synthetic Peptides Derived from HIV-1 gp120 proteins. J. Peptide Res. 1998, 52, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Hearn, M.T.W.; Biquard, J.M.; Cosic, I.; Krsmanovic, V. Peptides Immunologically related to proteins expressed by a viral agent, having a sequence of amino acids ordered by means of protein informational method. US Patent 6,294,174, 2001. [Google Scholar]
- Achour, A.; Biquard, J.M.; Krsmanovic, V.; M’Bika, J.P.; Ficheux, D.; Sikorska, M.; Cozzone, A.J. Induction of Human Immunodeficiency Virus (HIV-1) Envelope Specific Cell-Mediated Immunity by a Non-Homologus Synthetic Peptide. PLoS ONE 2007, 11, 1–12. [Google Scholar] [CrossRef]
- Pirogova, E.; Istivan, T.; Gan, E.; Cosic, I. Advances in Methods for Therapeutic Peptide Discovery, Design and Development. Curr. Pharm. Biotechnol. 2011, 12, 1117–1127. [Google Scholar] [CrossRef]
- Almansour, N.; Pirogova, E.; Coloe, P.; Cosic, I.; Istivan, T. Investigation of cytotoxicity of negative control peptides versus bioactive peptides on skin cancer and normal cells: A comparative study. Future Med. Chem. 2012, 4, 1553–1565. [Google Scholar] [CrossRef]
- Istivan, T.; Pirogova, E.; Gan, E.; Almansour, N.; Coloe, P.; Cosic, I. Biological effects of a De Novo designed myxoma virus peptide analogue: Evaluation of cytotoxicity on tumor cells. PLoS ONE 2011, 6, 1–10. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cosic, I.; Paspaliaris, V.; Cosic, D. Analysis of Protein–Receptor Interactions on an Example of Leptin–Leptin Receptor Interaction Using the Resonant Recognition Model. Appl. Sci. 2019, 9, 5169. https://doi.org/10.3390/app9235169
Cosic I, Paspaliaris V, Cosic D. Analysis of Protein–Receptor Interactions on an Example of Leptin–Leptin Receptor Interaction Using the Resonant Recognition Model. Applied Sciences. 2019; 9(23):5169. https://doi.org/10.3390/app9235169
Chicago/Turabian StyleCosic, Irena, Vasilis Paspaliaris, and Drasko Cosic. 2019. "Analysis of Protein–Receptor Interactions on an Example of Leptin–Leptin Receptor Interaction Using the Resonant Recognition Model" Applied Sciences 9, no. 23: 5169. https://doi.org/10.3390/app9235169
APA StyleCosic, I., Paspaliaris, V., & Cosic, D. (2019). Analysis of Protein–Receptor Interactions on an Example of Leptin–Leptin Receptor Interaction Using the Resonant Recognition Model. Applied Sciences, 9(23), 5169. https://doi.org/10.3390/app9235169





