Characterization of Biofilm Extracts from Two Marine Bacteria
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
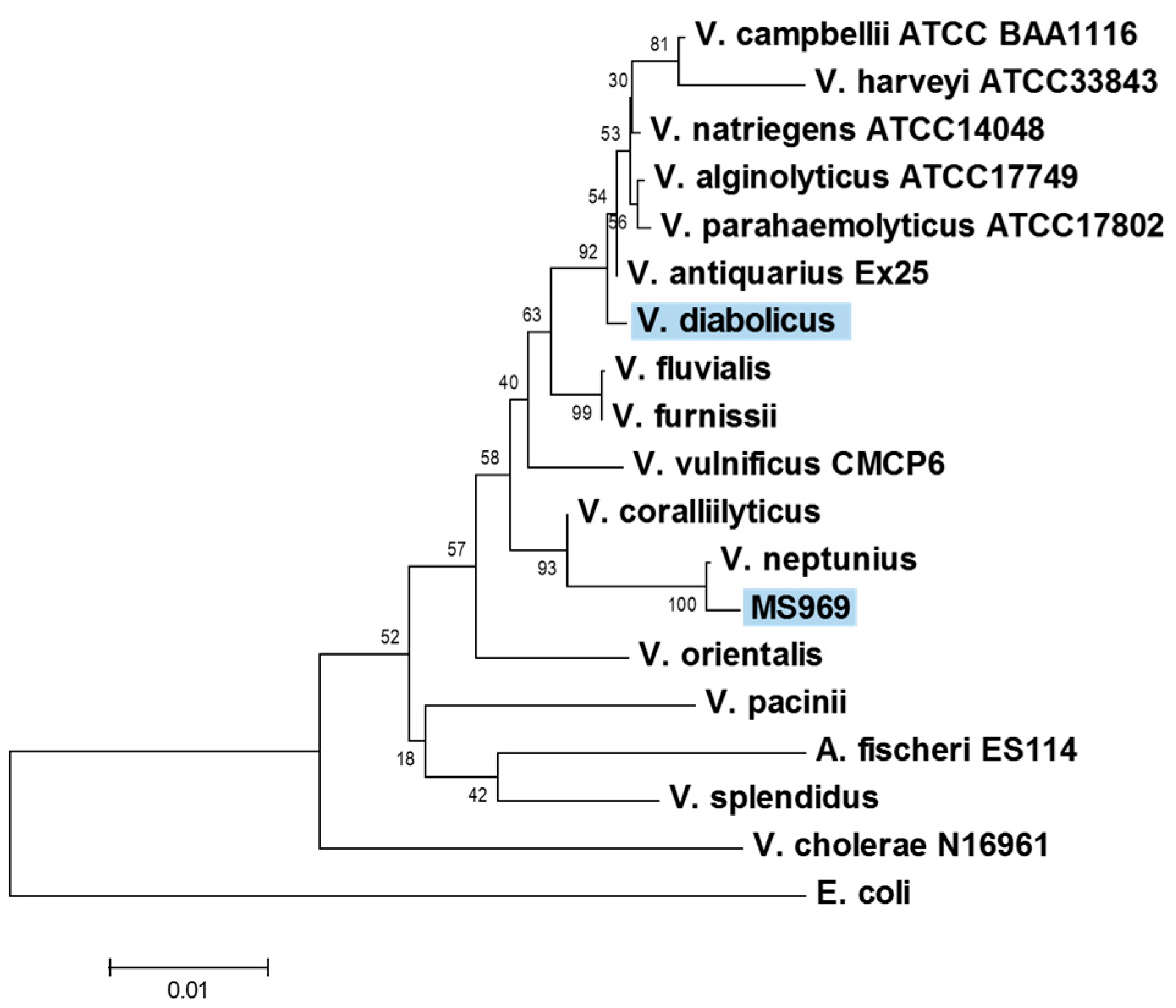
2.2. Identification of the MS969 Strain
2.3. Biofilm Assays
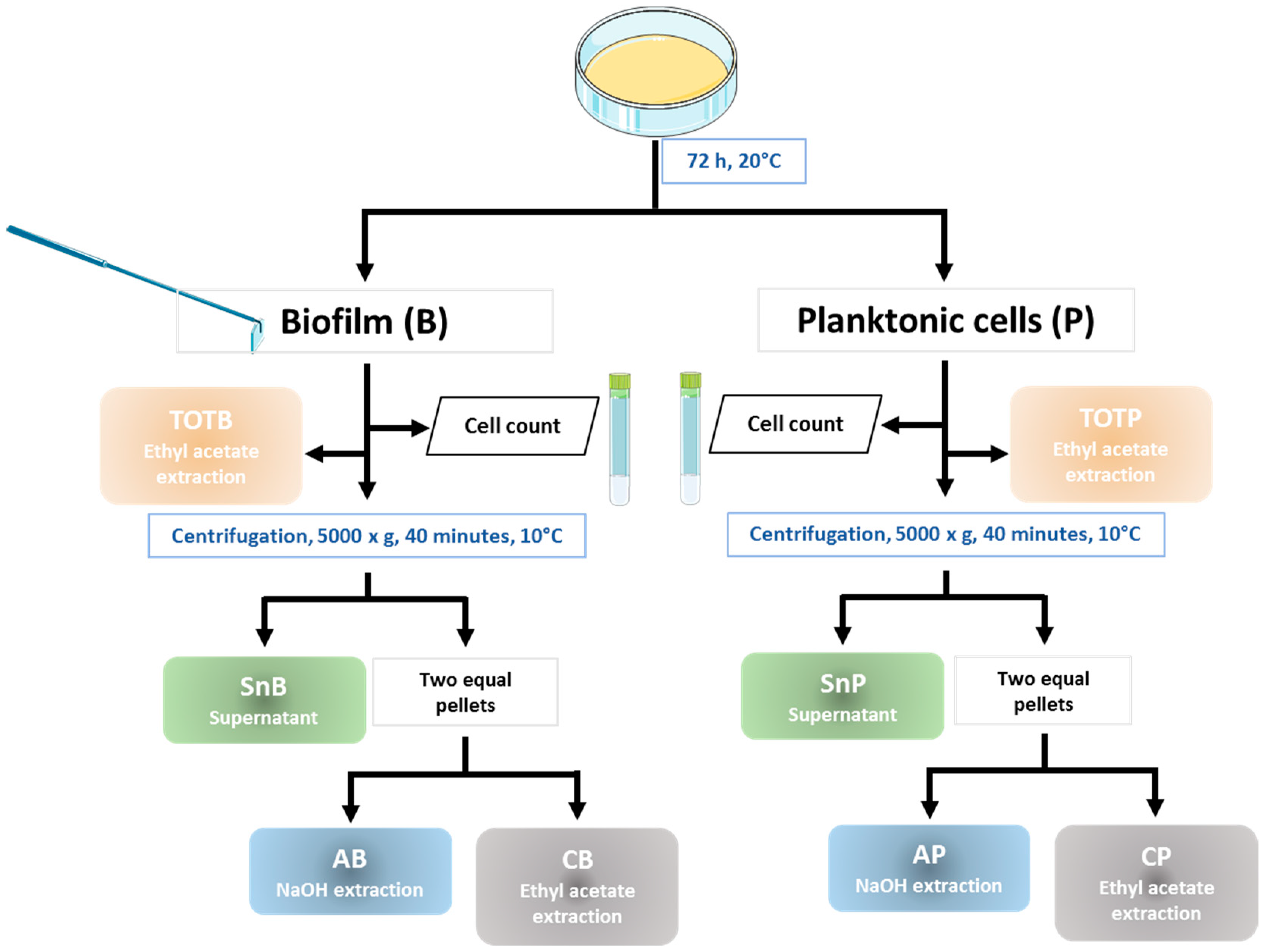
2.4. Biofilm Recovery
2.5. Preparation of Extracts
2.6. Chemistry of the Biofilm Matrix
2.6.1. Composition Analysis
2.6.2. Electrophoretic Analysis of Proteins, DNA, and Glycopolymers
2.6.3. Carbohydrate Characterization Using Gas Chromatography
2.6.4. Determination of Molecular Weight
2.7. Antimicrobial Activity Assay
2.8. Effect of Extracts on Quorum Sensing
2.9. Motility Assays
3. Results
3.1. Identification of the MS969 Strain
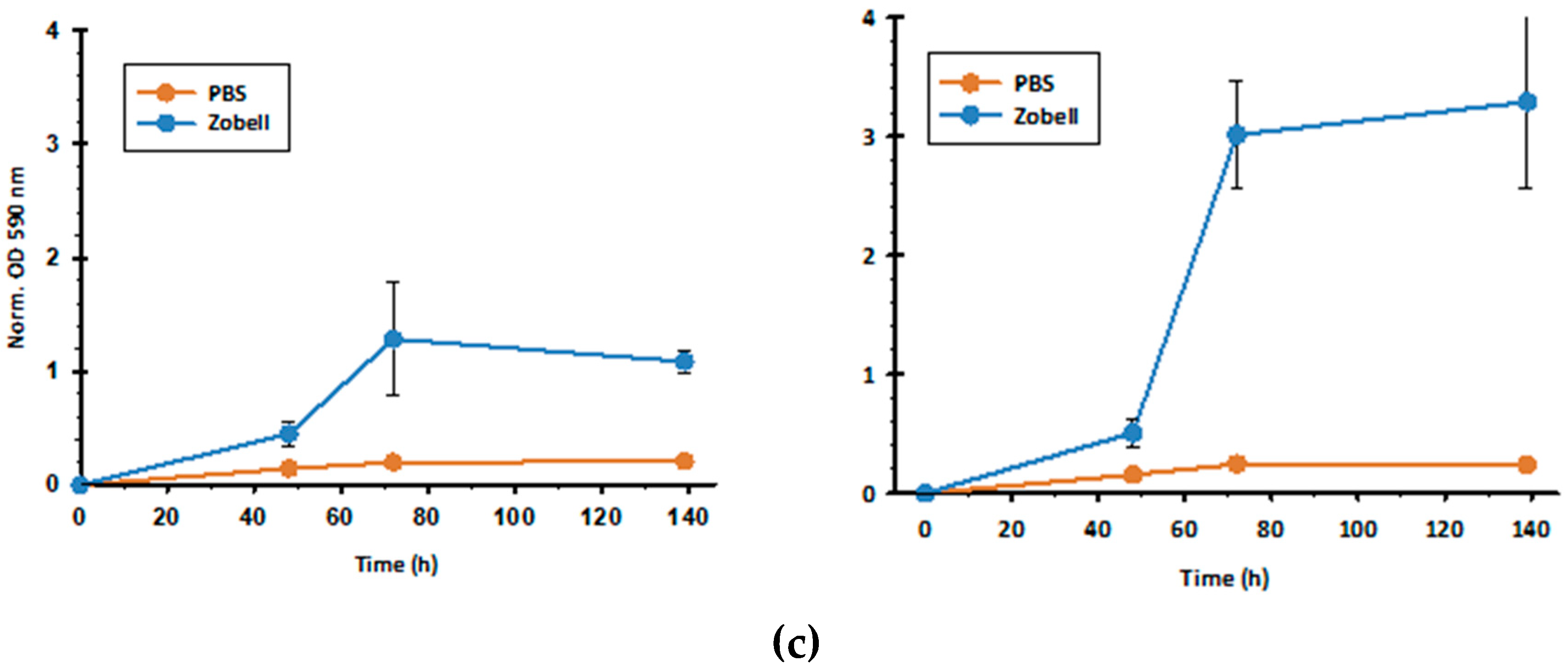
3.2. Biofilm Formation by V. diabolicus CNCM I-1629 and Vibrio sp. MS969
3.3. Preparation of Biofilm Extracts
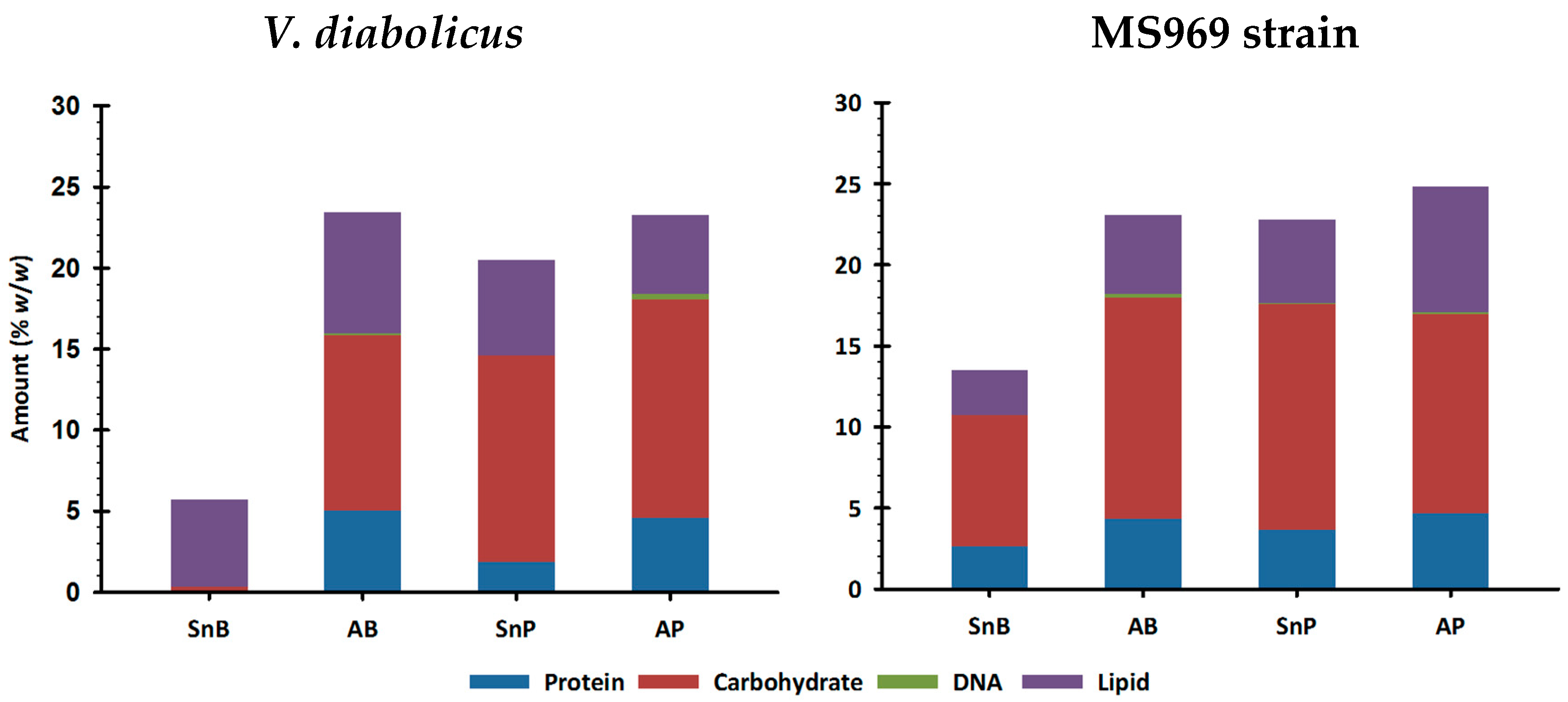
3.4. Biofilm Water-Soluble Components
3.4.1. Biochemical Composition Determination Using Colorimetric or Fluorimetric Assays
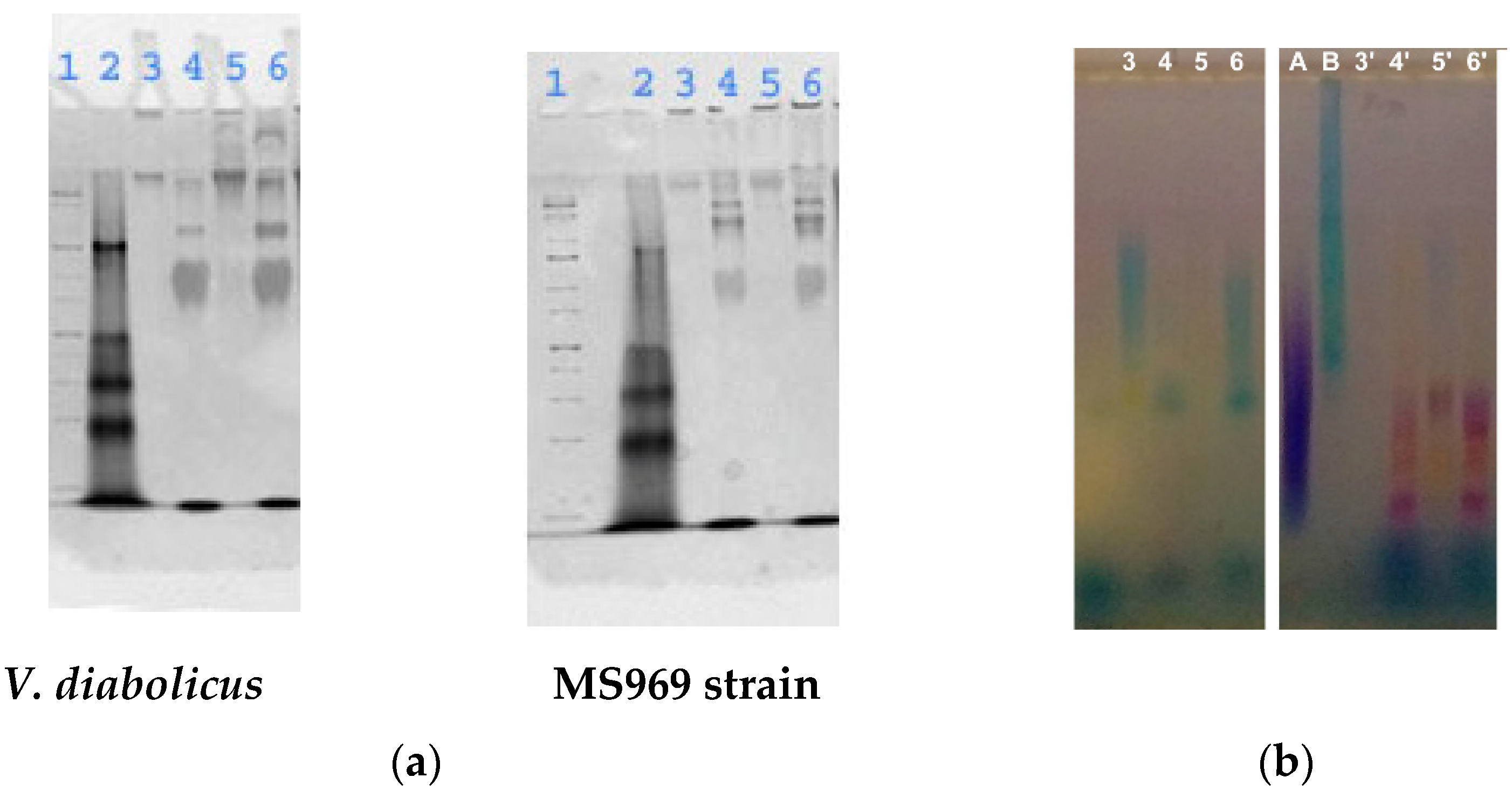
3.4.2. Electrophoretic Analysis
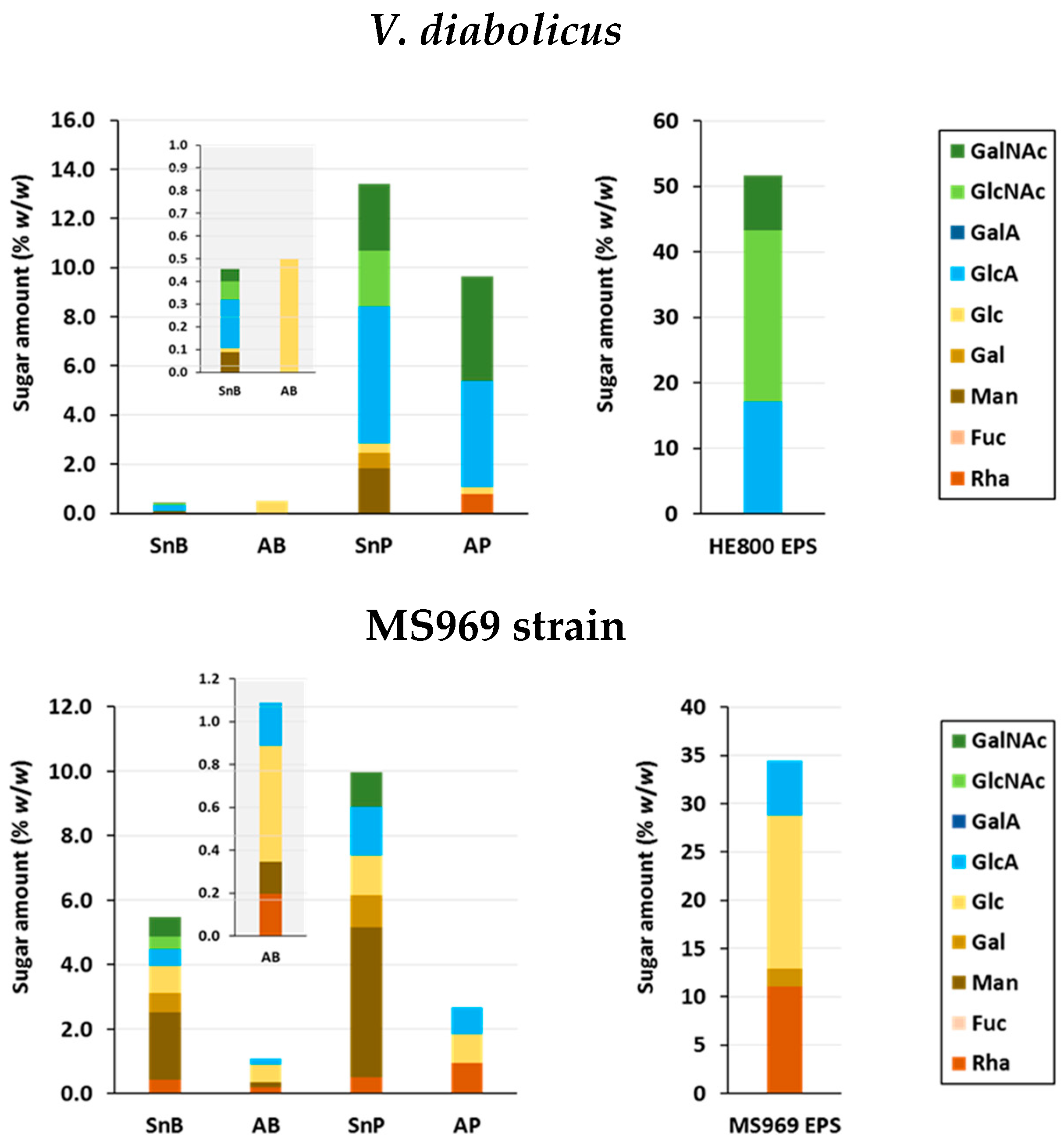
3.4.3. Osidic Composition
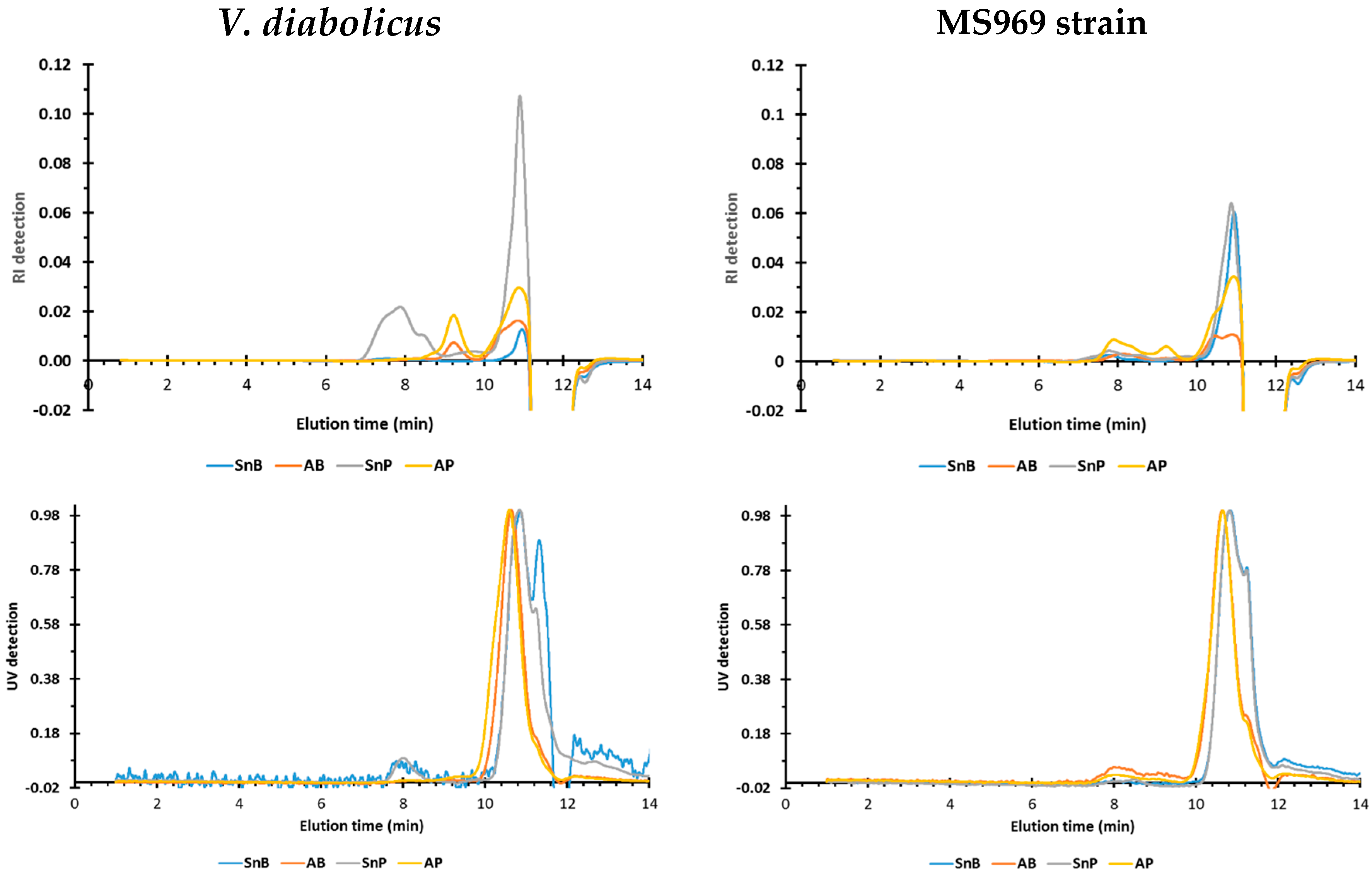
3.4.4. Size Exclusion Chromatographic Profiles
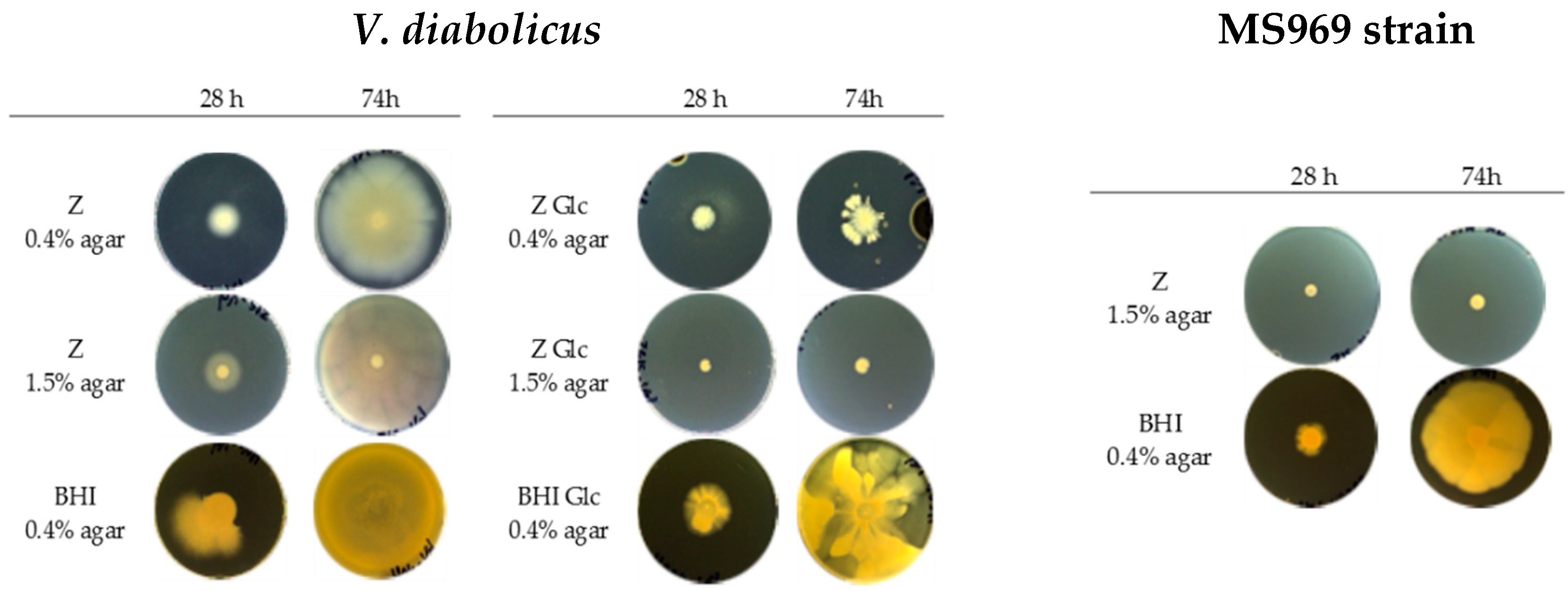
3.5. Motility Assay
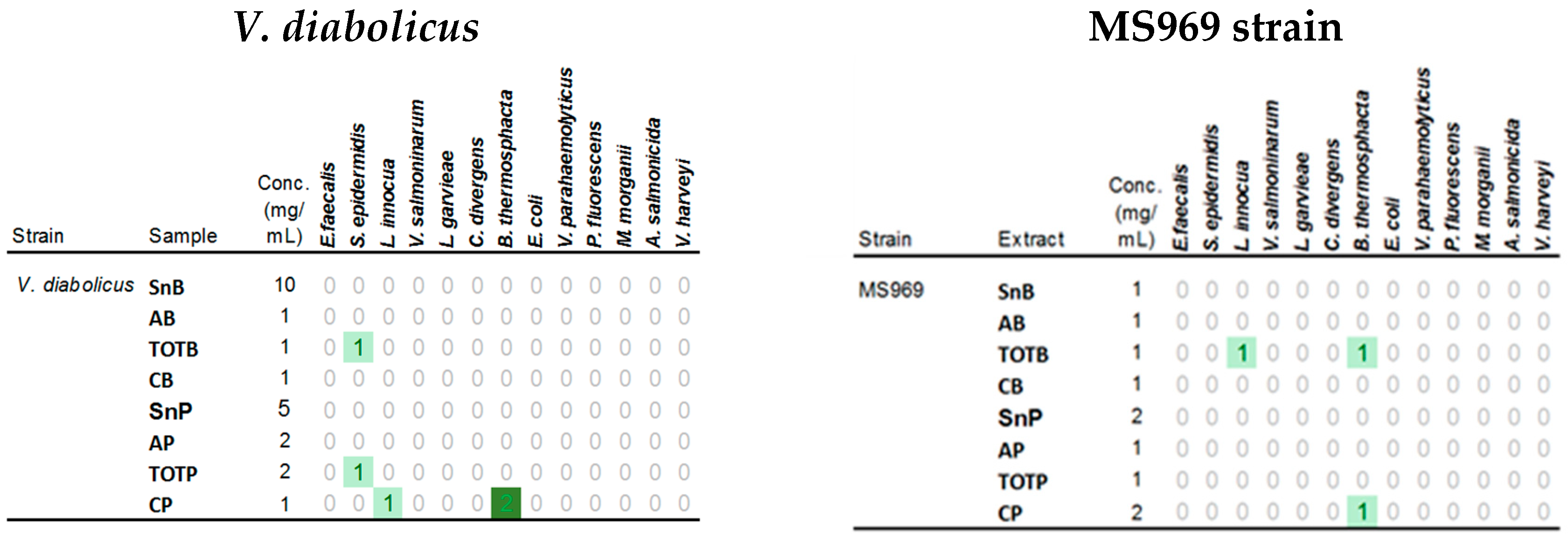
3.6. Antimicrobial Activities
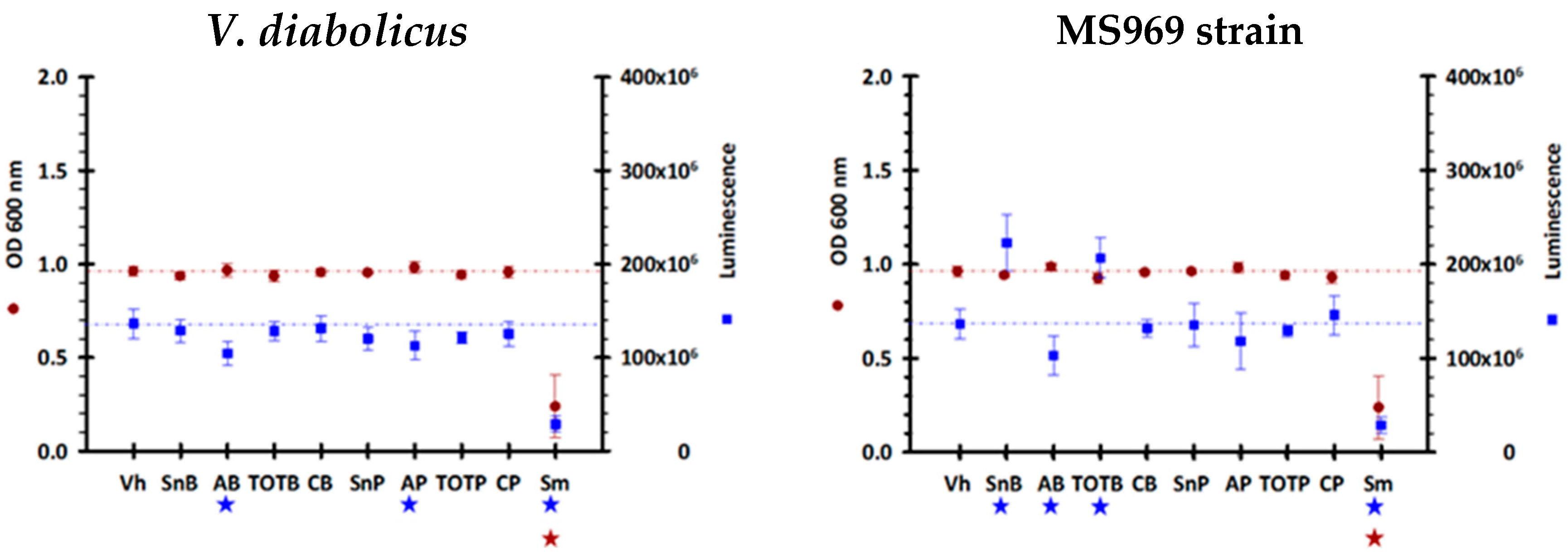
3.7. Quorum-Sensing Signaling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flemming, H.-C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Genet. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Neu, T.R.; Lawrence, J.R. Chapter 37—Extracellular polymeric substances in microbial biofilms. In Microbial Glycobiology; Academic Press: San Diego, CA, USA, 2010; pp. 733–758. [Google Scholar]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Genet. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Marine Biofilms: A successful microbial strategy with economic implications. Front. Mar. Sci. 2018, 5, 126. [Google Scholar] [CrossRef]
- Nakhamchik, A.; Wilde, C.; Rowe-Magnus, D.A. Cyclic-di-GMP Regulates Extracellular Polysaccharide Production, Biofilm Formation, and Rugose Colony Development by Vibrio vulnificus. Appl. Environ. Microbiol. 2008, 74, 4199–4209. [Google Scholar] [CrossRef] [PubMed]
- Dickschat, J.S. Quorum sensing and bacterial biofilms. Nat. Prod. Rep. 2010, 27, 343–369. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Cai, X.; Wang, P.; Guo, Y.; Liu, X.; Li, B.; Wang, X. Biofilm Formation and Heat Stress Induce Pyomelanin Production in Deep-Sea Pseudoalteromonas sp. SM9913. Front. Microbiol. 2017, 8, 1822. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Thawrani, D.; Banerjee, P.; Gachhui, R.; Mukherjee, J. Induced Biofilm Cultivation Enhances Riboflavin Production by an Intertidally Derived Candida famata. Appl. Biochem. Biotechnol. 2012, 166, 1991–2006. [Google Scholar] [CrossRef] [PubMed]
- Mnif, I.; Ghribi, D. Glycolipid biosurfactants: Potential related biomedical and biotechnological applications. Carbohydr. Res. 2015, 416, 59–69. [Google Scholar]
- Victor, I.U.; Kwiencien, M.; Tripathi, L.; Cobice, D.; McClean, S.; Marchant, R.; Banat, I.M. Quorum sensing as a potential target for increased production of rhamnolipid biosurfactant in Burkholderia thailandensis E264. Appl. Microbiol. Biotechnol. 2019, 103, 6505–6517. [Google Scholar] [CrossRef]
- Schmidt, S.; Blom, J.F.; Pernthaler, J.; Berg, G.; Baldwin, A.; Mahenthiralingam, E.; Eberl, L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of theBurkholderia cepaciacomplex. Environ. Microbiol. 2009, 11, 1422–1437. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. Relevance of microbial extracellular polymeric substances (EPSs)—Part I: Structural and ecological aspects. Water Sci. Technol. 2001, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Moppert, X.; Fardeau, M.; Carrero, J.; Valadéz-González, A.; Guezennec, J.; Ortega-Morales, B.; Santiago-García, J.; Chan-Bacab, M.; Miranda-Tello, E.; Bartolo-Pérez, P.; et al. Characterization of extracellular polymers synthesized by tropical intertidal biofilm bacteria. J. Appl. Microbiol. 2007, 102, 254–264. [Google Scholar]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I.W. Biofilm exopolysaccharides: A strong and sticky framework. Microbiol. 2001, 147, 3–9. [Google Scholar] [CrossRef]
- Ostapska, H.; Howell, P.L.; Sheppard, D.C. Deacetylated microbial biofilm exopolysaccharides: It pays to be positive. PLoS Pathog. 2018, 14, e1007411. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Tolker-Nielsen, T. The biofilm matrix—A sticky framework. In The Biofilm Mode of Life; Kjelleberg, S., Givskov, M., Eds.; Horizon Press: Wymondham, UK, 2007; pp. 37–69. [Google Scholar]
- Senni, K.; Pereira, J.; Gueniche, F.; Delbarre-Ladrat, C.; Sinquin, C.; Ratiskol, J.; Godeau, G.; Fischer, A.-M.; Helley, D.; Colliec-Jouault, S. Marine Polysaccharides: A Source of Bioactive Molecules for Cell Therapy and Tissue Engineering. Mar. Drugs 2011, 9, 1664–1681. [Google Scholar] [CrossRef]
- Redcorn, R.M.; Hillman, E.T.; Solomon, K.V.; Engelberth, A.S. Xanthobacter-dominated biofilm as a novel source for high-value rhamnose. Appl. Microbiol. Biotechnol. 2019, 103, 4525–4538. [Google Scholar] [CrossRef]
- Caruso, C.; Rizzo, C.; Mangano, S.; Poli, A.; di Donato, P.; Finore, I.; Nicolaus, B.; di Marco, G.; Michaud, L.; Giudice, A.L. Production and biotechnological potential of extracellular polymeric substances from sponge-associated antarctic bacteria. Appl. Environ. Microbiol. 2018, 84, e01624-17. [Google Scholar] [CrossRef]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a Role for Bacterial Cellulose in Environmental Interactions: Lessons Learned from Diverse Biofilm-Producing Proteobacteria. Front. Microbiol. 2015, 6, 402. [Google Scholar] [CrossRef]
- Tielker, D.; Hacker, S.; Loris, R.; Strathmann, M.; Wingender, J.; Wilhelm, S.; Rosenau, F.; Jaeger, K.-E. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 2005, 151, 1313–1323. [Google Scholar] [CrossRef]
- Lasa, Í.; Penadés, J.R. Bap: A family of surface proteins involved in biofilm formation. Res. Microbiol. 2006, 157, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Sinsabaugh, R.L.; Repert, D.; Weiland, T.; Golladay, S.W.; Linkins, A.E. Exoenzyme accumulation in epilithic biofilms. Hydrobiologia 1991, 222, 29–37. [Google Scholar] [CrossRef]
- Seper, A.; Fengler, V.H.I.; Roier, S.; Wolinski, H.; Kohlwein, S.D.; Bishop, A.L.; Camilli, A.; Reidl, J.; Schild, S. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 2011, 82, 1015–1037. [Google Scholar] [CrossRef] [PubMed]
- Pedraza, M.C.C.; Novais, T.F.; Faustoferri, R.C.; Quivey, R.G.; Terekhov, A.; Hamaker, B.R.; Klein, M.I. Extracellular DNA and lipoteichoic acids interact with exopolysaccharides in the extracellular matrix of Streptococcus mutansbiofilms. Biofouling 2017, 33, 722–740. [Google Scholar] [CrossRef]
- Kavanaugh, J.S.; Flack, C.E.; Lister, J.; Ricker, E.B.; Ibberson, C.B.; Jenul, C.; Moormeier, D.E.; Delmain, E.A.; Bayles, K.W.; Horswill, A.R. Identification of Extracellular DNA-Binding Proteins in the Biofilm Matrix. mBio 2019, 10, e01137-19. [Google Scholar] [CrossRef]
- Molin, S.; Tolker-Nielsen, T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 2003, 14, 255–261. [Google Scholar] [CrossRef]
- Shibata, S.; Visick, K.L. Sensor kinase RscS induces the production of antigenically distinct outer membrane vesicles that depend on the symbiosis polysaccharide locus in Vibrio fischeri. J. Bacteriol. 2012, 194, 185–194. [Google Scholar] [CrossRef]
- Decho, A.W.; Gutierrez, T. Microbial Extracellular Polymeric Substances (EPSs) in Ocean Systems. Front. Microbiol. 2017, 8, 922. [Google Scholar] [CrossRef]
- Gehrke, T.; Telegdi, J.; Thierry, D.; Sand, W. Importance of Extracellular Polymeric Substances from Thiobacillus ferrooxidans for Bioleaching. Appl. Environ. Microbiol. 1998, 64, 2743–2747. [Google Scholar]
- Ojha, A.K.; Baughn, A.D.; Sambandan, D.; Hsu, T.; Trivelli, X.; Guerardel, Y.; Alahari, A.; Kremer, L.; Jacobs, W.R., Jr.; Hatfull, G.F. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 2008, 69, 164–174. [Google Scholar] [CrossRef]
- Davey, M.E.; Caiazza, N.C.; O’Toole, G.A. Rhamnolipid Surfactant Production Affects Biofilm Architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Seviour, T.; Derlon, N.; Dueholm, M.S.; Flemming, H.-C.; Girbal-Neuhauser, E.; Horn, H.; Kjelleberg, S.; Van Loosdrecht, M.C.; Lotti, T.; Malpei, M.F.; et al. Extracellular polymeric substances of biofilms: Suffering from an identity crisis. Water Res. 2019, 151, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.M.A.; Nerland, A.H.; Al-Haroni, M.; Bakken, V. Characterization of extracellular polymeric matrix, and treatment of Fusobacterium nucleatum and Porphyromonas gingivalis biofilms with DNase I and proteinase K. J. Oral Microbiol. 2013, 5, 623. [Google Scholar]
- Wai, S.N.; Mizunoe, Y.; Takade, A.; Kawabata, S.-I.; Yoshida, S.-I. Vibrio cholerae O1 Strain TSI-4 Produces the Exopolysaccharide Materials That Determine Colony Morphology, Stress Resistance, and Biofilm Formation. Appl. Environ. Microbiol. 1998, 64, 3648–3655. [Google Scholar] [PubMed]
- Ramirez-Mora, T.; Retana-Lobo, C.; Valle-Bourrouet, G. Biochemical characterization of extracellular polymeric substances from endodontic biofilms. PLoS ONE 2018, 13, e0204081. [Google Scholar] [CrossRef]
- Chang, S.; Lee, Y. Comparison of two chemical extraction methods for proteins and polysaccharides of Spirogyra fluviatilis in extracellular polymeric substances. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2017; Volume 64, p. 012122. [Google Scholar]
- Brian-Jaisson, F.; Molmeret, M.; Fahs, A.; Guentas-Dombrowsky, L.; Culioli, G.; Blache, Y.; Cérantola, S.; Ortalo-Magné, A. Characterization and anti-biofilm activity of extracellular polymeric substances produced by the marine biofilm-forming bacteriumPseudoalteromonas ulvaestrain TC14. Biofouling 2016, 32, 547–560. [Google Scholar] [CrossRef]
- Raguenes, G.; Christen, R.; Guezennec, J.; Pignet, P.; Barbier, G. Vibrio diabolicus sp. nov., a New Polysaccharide-Secreting Organism Isolated from a Deep-Sea Hydrothermal Vent Polychaete Annelid, Alvinella pompejana. Int. J. Syst. Bacteriol. 1997, 47, 989–995. [Google Scholar] [CrossRef]
- Guezennec, J. Deep-sea hydrothermal vents: A new source of innovative bacterial exopolysaccharides of biotechnological interest? J. Ind. Microbiol. Biotechnol. 2002, 29, 204–208. [Google Scholar] [CrossRef]
- Delbarre-Ladrat, C.; Salas, M.L.; Sinquin, C.; Zykwinska, A.; Colliec-Jouault, S. Bioprospecting for Exopolysaccharides from Deep-Sea Hydrothermal Vent Bacteria: Relationship between Bacterial Diversity and Chemical Diversity. Microorganisms 2017, 5, 63. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Boil. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Smith, P.; Krohn, R.; Hermanson, G.; Mallia, A.; Gartner, F.; Provenzano, M.; Fujimoto, E.; Goeke, N.; Olson, B.; Klenk, D. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Tillmans, J.; Philippi, K. Über den Gehalt der wichtigsten Proteine der Nahrungsmittel an Kohlenhydrat und über ein kolorimetrisches Verfahren zur quantitativen Bestimmung von stickstoff-freiem Zucker im Eiweiß. Biochem. Z 1929, 215, 36–60. [Google Scholar]
- Rimington, C. The carbohydrate complex of the serum proteins improved method for isolation and re-determination of structure. Isolation of glucosaminodimannose from proteins of ox blood. Biochem. J. 1931, 25, 1062–1071. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Axelsson, M.; Gentili, F. A Single-Step Method for Rapid Extraction of Total Lipids from Green Microalgae. PLoS ONE 2014, 9, e89643. [Google Scholar] [CrossRef]
- Frings, C.S.; Fendley, T.W.; Dunn, R.T.; A Queen, C. Improved determination of total serum lipids by the sulfo-phospho-vanillin reaction. Clin. Chem. 1972, 18, 673–674. [Google Scholar]
- Zykwinska, A.; Marchand, L.; Bonnetot, S.; Sinquin, C.; Colliec-Jouault, S.; Delbarre-Ladrat, C. Deep-sea Hydrothermal Vent Bacteria as a Source of Glycosaminoglycan-Mimetic Exopolysaccharides. Molecules 2019, 24, 1703. [Google Scholar] [CrossRef]
- Rigouin, C.; Ladrat, C.D.; Sinquin, C.; Colliec-Jouault, S.; Dion, M. Assessment of biochemical methods to detect enzymatic depolymerization of polysaccharides. Carbohydr. Polym. 2009, 76, 279–284. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Kamerling, J.P.; Gerwig, G.J.; Vliegenthart, J.F.G.; Clamp, J.R. Characterization by gas-liquid chromatography mass spectrometry of permethylsilylglycosides obtained in the methanolysis of glycoproteins and glycolipids. Biochem. J. 1975, 151, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Montreuil, J.; Bouquelet, S.; Debray, H.; Fournet, B.; Spik, G.; Strecker, G. Glycoproteins. In Carbohydrate Analysis: A Practical Approach, 2nd ed.; Chaplin, M.F., Kennedy, J.K., Eds.; IRL Press: Oxford, UK, 1986; pp. 143–204. [Google Scholar]
- Gratia, A. Techniques sélectives pour la recherche systématique des germes antibiotiques. Comptes Rendus Seances Societe Biologie Ses Filiales 1946, 140, 1053–1055. [Google Scholar]
- Sawabe, T.; Ogura, Y.; Matsumura, Y.; Feng, G.; Amin, A.R.; Mino, S.; Nakagawa, S.; Sawabe, T.; Kumar, R.; Fukui, Y.; et al. Updating the Vibrio clades defined by multilocus sequence phylogeny: Proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front. Microbiol. 2013, 4, 414. [Google Scholar] [CrossRef] [PubMed]
- Romalde, J.L.; Dieguez, A.L.; Lasa, A.; Balboa, S. New Vibrio species associated to molluscan microbiota: A review. Front. Microbiol. 2013, 4, 413. [Google Scholar] [CrossRef]
- Senni, K.; Guéniche, F.; Changotade, S.; Septier, D.; Sinquin, C.; Ratiskol, J.; Lutomski, D.; Godeau, G.; Guezennec, J.; Colliec-Jouault, S. Unusual Glycosaminoglycans from a Deep Sea Hydrothermal Bacterium Improve Fibrillar Collagen Structuring and Fibroblast Activities in Engineered Connective Tissues. Mar. Drugs 2013, 11, 1351–1369. [Google Scholar] [CrossRef]
- Goudenège, D.; Boursicot, V.; Versigny, T.; Bonnetot, S.; Ratiskol, J.; Sinquin, C.; Lapointe, G.; Le Roux, F.; Delbarre-Ladrat, C. Genome sequence of Vibrio diabolicus and identification of the exopolysaccharide HE800 biosynthesis locus. Appl. Microbiol. Biotechnol. 2014, 98, 10165–10176. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef]
- Hsieh, M.-L.; Hinton, D.M.; Waters, C.M. VpsR and cyclic di-GMP together drive transcription initiation to activate biofilm formation in Vibrio cholerae. Nucleic Acids Res. 2018, 46, 8876–8887. [Google Scholar] [CrossRef]
- Yildiz, F.; Fong, J.; Sadovskaya, I.; Grard, T.; Vinogradov, E. Structural Characterization of the Extracellular Polysaccharide from Vibrio cholerae O1 El-Tor. PLoS ONE 2014, 9, e86751. [Google Scholar] [CrossRef] [PubMed]
- Roger, O.; Kervarec, N.; Ratiskol, J.; Colliec-Jouault, S.; Chevolot, L. Structural studies of the main exopolysaccharide produced by the deep-sea bacterium Alteromonas infernus. Carbohydr. Res. 2004, 339, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Rougeaux, H.; Kervarec, N.; Pichon, R.; Guezennec, J. Structure of the exopolysaccharide of Vibriodiabolicus isolated from a deep-sea hydrothermal vent. Carbohydr. Res. 1999, 322, 40–45. [Google Scholar] [CrossRef]
- Andrade, J.P.S.; Oliveira, C.P.; Tovar, A.M.F.; Mourão, P.A.D.S.; Vilanova, E. A color-code for glycosaminoglycans identification by means of polyacrylamide gel electrophoresis stained with the cationic carbocyanine dye Stains-all. Electrophoresis 2018, 39, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Yip, E.S.; Quirke, K.P.; Ondrey, J.M.; Visick, K.L. Roles of the Structural Symbiosis Polysaccharide (syp) Genes in Host Colonization, Biofilm Formation, and Polysaccharide Biosynthesis in Vibrio fischeri. J. Bacteriol. 2012, 194, 6736–6747. [Google Scholar] [CrossRef] [PubMed]
- Lebellenger, L.; Verrez-Bagnis, V.; Passerini, D.; Delbarre-Ladrat, C. Comparative genomics reveals a widespread distribution of an exopolysaccharide biosynthesis gene cluster among Vibrionaceae. BMC Res. Notes 2018, 11, 102. [Google Scholar] [CrossRef]
- Kavita, K.; Mishra, A.; Jha, B. Isolation and physico-chemical characterisation of extracellular polymeric substances produced by the marine bacterium Vibrio parahaemolyticus. Biofouling 2011, 27, 309–317. [Google Scholar] [CrossRef]
- Guo, Y.; Rowe-Magnus, D.A. Identification of a c-di-GMP-Regulated Polysaccharide Locus Governing Stress Resistance and Biofilm and Rugose Colony Formation in Vibrio vulnificus. Infect. Immun. 2010, 78, 1390–1402. [Google Scholar] [CrossRef]
- Jarrell, K.F.; McBride, M.J. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Genet. 2008, 6, 466–476. [Google Scholar] [CrossRef]
- Seper, A.; Pressler, K.; Kariisa, A.; Haid, A.G.; Roier, S.; Leitner, D.R.; Reidl, J.; Tamayo, R.; Schild, S. Identification of genes induced in Vibrio cholerae in a dynamic biofilm system. Int. J. Med Microbiol. 2014, 304, 749–763. [Google Scholar] [CrossRef]
- Tercero-Alburo, J.J.; González-Márquez, H.; Bonilla-González, E.; Quiñones-Ramírez, E.I.; Vázquez-Salinas, C. Identification of capsule, biofilm, lateral flagellum, and type IV pili in Vibrio mimicus strains. Microb. Pathog. 2014, 76, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Mansson, M.; Gram, L.; Larsen, T.O. Production of Bioactive Secondary Metabolites by Marine Vibrionaceae. Mar. Drugs 2011, 9, 1440–1468. [Google Scholar] [CrossRef] [PubMed]
- Wietz, M.; Mansson, M.; Gotfredsen, C.H.; Larsen, T.O.; Gram, L. Antibacterial Compounds from Marine Vibrionaceae Isolated on a Global Expedition. Mar. Drugs 2010, 8, 2946–2960. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Miyashiro, T. Quorum Sensing in the Squid-Vibrio Symbiosis. Int. J. Mol. Sci. 2013, 14, 16386–16401. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, N.; Shin, J.Y.; Jung, K. Activity, Abundance, and Localization of Quorum Sensing Receptors in Vibrio harveyi. Front. Microbiol. 2017, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Henke, J.M.; Bassler, B.L. Three Parallel Quorum-Sensing Systems Regulate Gene Expression in Vibrio harveyi. J. Bacteriol. 2004, 186, 6902–6914. [Google Scholar] [CrossRef]
- Casillo, A.; Lanzetta, R.; Parrilli, M.; Corsaro, M.M. Exopolysaccharides from Marine and Marine Extremophilic Bacteria: Structures, Properties, Ecological Roles and Applications. Mar. Drugs 2018, 16, 69. [Google Scholar] [CrossRef]
- Zarins-Tutt, J.S.; Barberi, T.T.; Gao, H.; Mearns-Spragg, A.; Zhang, L.; Newman, D.J.; Goss, R.J.M. Prospecting for new bacterial metabolites: A glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 2016, 33, 54–72. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Passerini, D.; Fécamp, F.; Marchand, L.; Kolypczuk, L.; Bonnetot, S.; Sinquin, C.; Verrez-Bagnis, V.; Hervio-Heath, D.; Colliec-Jouault, S.; Delbarre-Ladrat, C. Characterization of Biofilm Extracts from Two Marine Bacteria. Appl. Sci. 2019, 9, 4971. https://doi.org/10.3390/app9224971
Passerini D, Fécamp F, Marchand L, Kolypczuk L, Bonnetot S, Sinquin C, Verrez-Bagnis V, Hervio-Heath D, Colliec-Jouault S, Delbarre-Ladrat C. Characterization of Biofilm Extracts from Two Marine Bacteria. Applied Sciences. 2019; 9(22):4971. https://doi.org/10.3390/app9224971
Chicago/Turabian StylePasserini, Delphine, Florian Fécamp, Laetitia Marchand, Laetitia Kolypczuk, Sandrine Bonnetot, Corinne Sinquin, Véronique Verrez-Bagnis, Dominique Hervio-Heath, Sylvia Colliec-Jouault, and Christine Delbarre-Ladrat. 2019. "Characterization of Biofilm Extracts from Two Marine Bacteria" Applied Sciences 9, no. 22: 4971. https://doi.org/10.3390/app9224971
APA StylePasserini, D., Fécamp, F., Marchand, L., Kolypczuk, L., Bonnetot, S., Sinquin, C., Verrez-Bagnis, V., Hervio-Heath, D., Colliec-Jouault, S., & Delbarre-Ladrat, C. (2019). Characterization of Biofilm Extracts from Two Marine Bacteria. Applied Sciences, 9(22), 4971. https://doi.org/10.3390/app9224971





