Potential Marker Genes for Predicting Adipogenic Differentiation of Mesenchymal Stromal Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Adipogenic Differentiation of MSCs
2.3. Osteogenic Differentiation of MSCs
2.4. Quantitative RT-PCR
2.5. 3D Scatter Plot
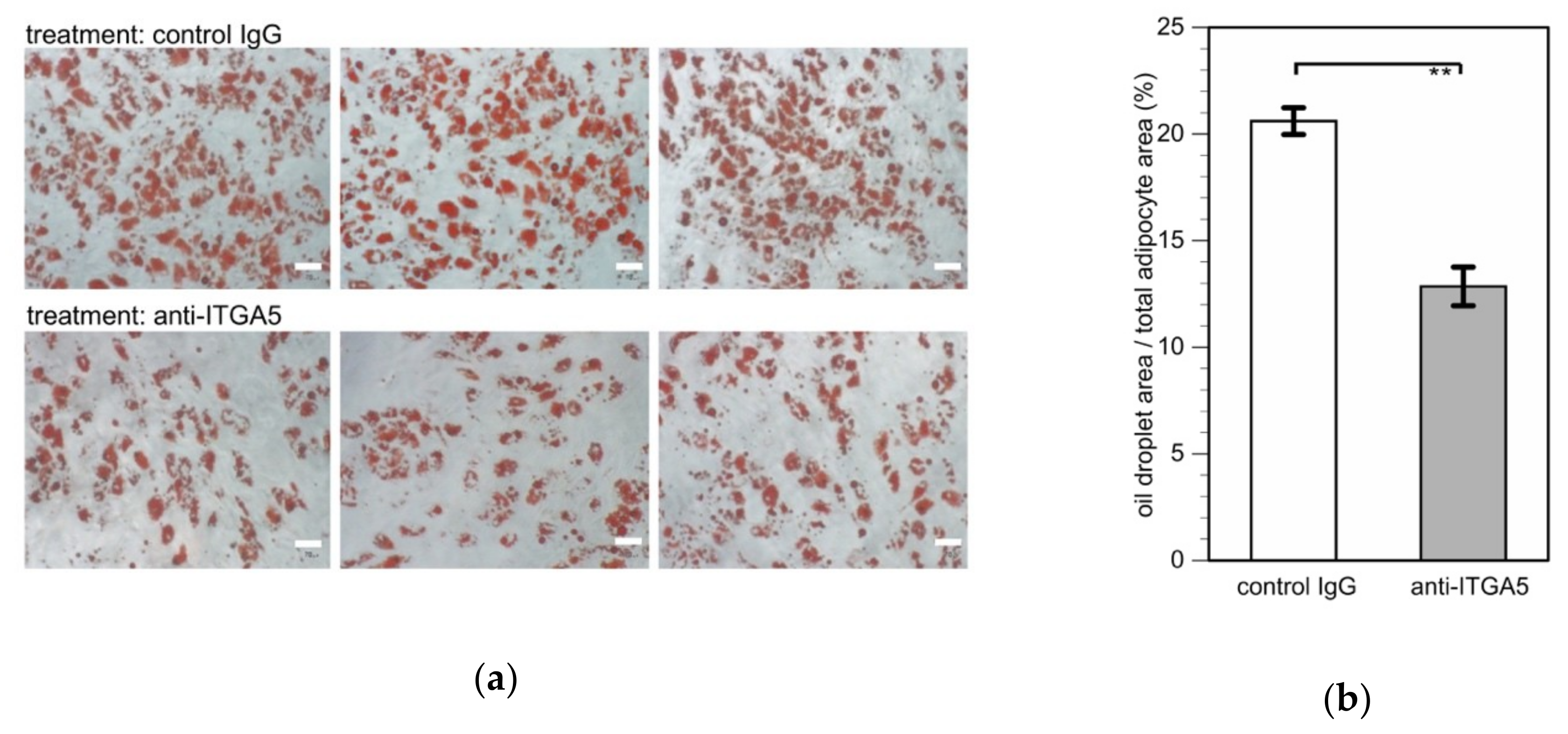
2.6. Effects of Anti-ITGA5 Antibody on Adipogenesis
2.7. Statistical Analysis
3. Results
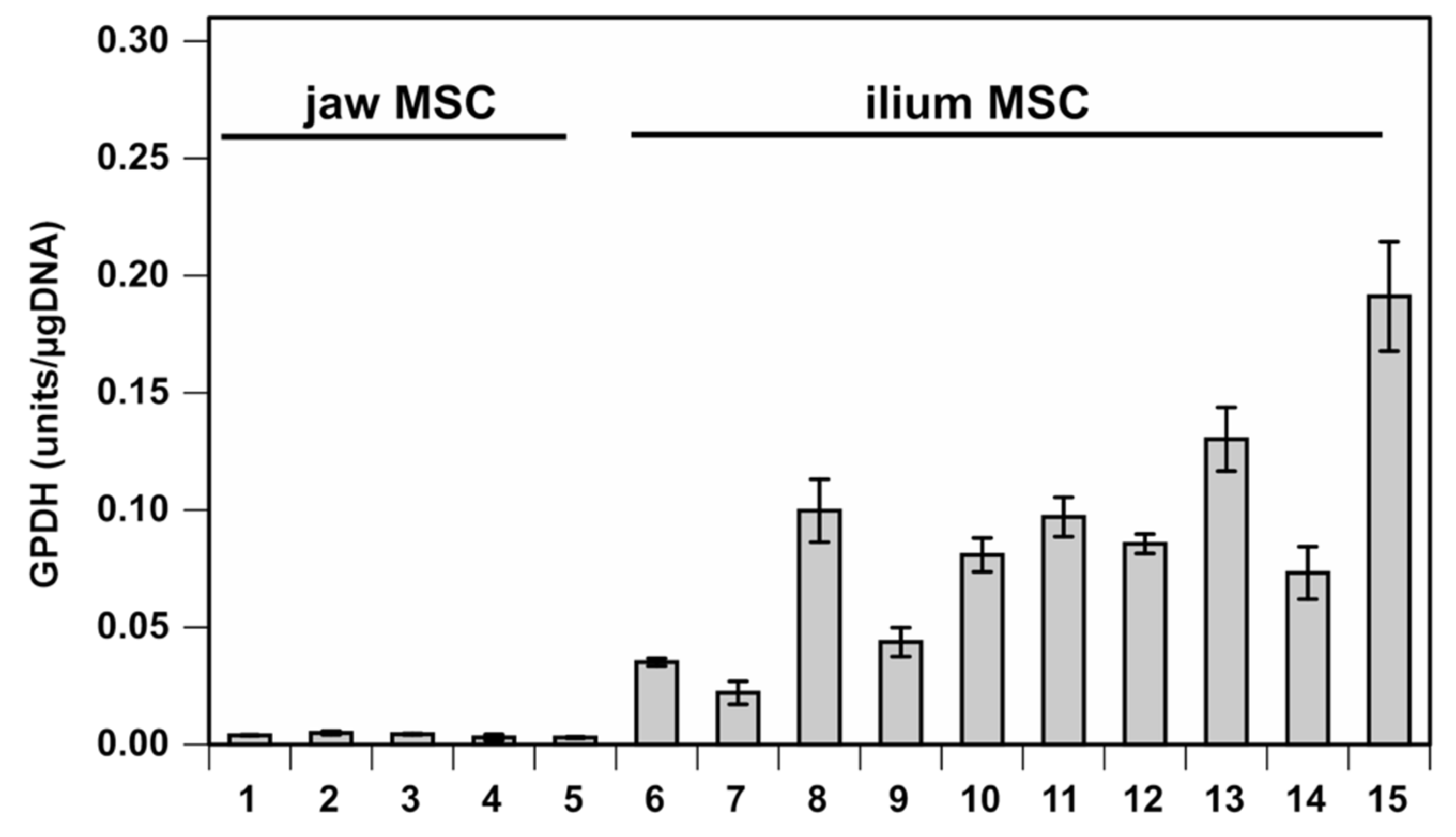
3.1. Correlational Analysis between Gene Expression before Differentiation and GPDH Activity after Adipogenic Differentiation
3.2. Combined Evaluation of mRNA Expression of Candidate Genes
3.3. Effects of Anti-ITGA5 Antibody on Adipogenic Differentiation of Ilium MSCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, X.; Gao, L.; Han, Y.; Xing, M.; Zhao, C.; Peng, J.; Chang, J. Silicon-Enhanced Adipogenesis and Angiogenesis for Vascularized Adipose Tissue Engineering. Adv. Sci. 2018, 5, 1800776. [Google Scholar] [CrossRef] [PubMed]
- Janderova, L.; McNeil, M.; Murrell, A.N.; Mynatt, R.L.; Smith, S.R. Human mesenchymal stem cells as an in vitro model for human adipogenesis. Obes. Res. 2003, 11, 65–74. [Google Scholar] [CrossRef]
- Samsonraj, R.M.; Raghunath, M.; Nurcombe, V.; Hui, J.H.; van Wijnen, A.J.; Cool, S.M. Concise Review: Multifaceted Characterization of Human Mesenchymal Stem Cells for Use in Regenerative Medicine. Stem Cells Transl. Med. 2017, 6, 2173–2185. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Park, S.N.; Suh, H. Adipose tissue engineering using mesenchymal stem cells attached to injectable PLGA spheres. Biomaterials 2005, 26, 5855–5863. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Suardita, K.; Ishii, M.; Sugiyama, M.; Igarashi, A.; Oda, R.; Nishimura, M.; Saito, M.; Nakagawa, K.; Yamanaka, K.; et al. Alveolar bone marrow as a cell source for regenerative medicine: Differences between alveolar and iliac bone marrow stromal cells. J. Bone Miner. Res. 2005, 20, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Bearden, R.N.; Huggins, S.S.; Cummings, K.J.; Smith, R.; Gregory, C.A.; Saunders, W.B. In-vitro characterization of canine multipotent stromal cells isolated from synovium, bone marrow, and adipose tissue: A donor-matched comparative study. Stem Cell Res. Ther. 2017, 8, 218. [Google Scholar] [CrossRef] [PubMed]
- Kanawa, M.; Igarashi, A.; Fujimoto, K.; Higashi, Y.; Kurihara, H.; Sugiyama, M.; Saskianti, T.; Kato, Y.; Kawamoto, T. Genetic Markers Can Predict Chondrogenic Differentiation Potential in Bone Marrow-Derived Mesenchymal Stromal Cells. Stem Cells Int. 2018, 2018, 9530932. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Liu, F.L.; Sytwu, H.K.; Tsai, C.Y.; Chang, D.M. CD146+ mesenchymal stem cells display greater therapeutic potential than CD146- cells for treating collagen-induced arthritis in mice. Stem Cell Res. Ther. 2016, 7, 23. [Google Scholar] [CrossRef]
- Shi, Y.; He, G.; Lee, W.C.; McKenzie, J.A.; Silva, M.J.; Long, F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017, 8, 2043. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, A.; Segoshi, K.; Sakai, Y.; Pan, H.; Kanawa, M.; Higashi, Y.; Sugiyama, M.; Nakamura, K.; Kurihara, H.; Yamaguchi, S.; et al. Selection of common markers for bone marrow stromal cells from various bones using real-time RT-PCR: Effects of passage number and donor age. Tissue Eng. 2007, 13, 2405–2417. [Google Scholar] [CrossRef]
- Kanawa, M.; Igarashi, A.; Ronald, V.S.; Higashi, Y.; Kurihara, H.; Sugiyama, M.; Saskianti, T.; Pan, H.; Kato, Y. Age-dependent decrease in the chondrogenic potential of human bone marrow mesenchymal stromal cells expanded with fibroblast growth factor-2. Cytotherapy 2013, 15, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Warnke, I.; Goralczyk, R.; Fuhrer, E.; Schwager, J. Dietary constituents reduce lipid accumulation in murine C3H10 T1/2 adipocytes: A novel fluorescent method to quantify fat droplets. Nutr. Metab. (Lond.) 2011, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Girish, V.; Vijayalakshmi, A. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer 2004, 41, 47. [Google Scholar] [PubMed]
- Wang, H.; Li, J.; Zhang, X.; Ning, T.; Ma, D.; Ge, Y.; Xu, S.; Hao, Y.; Wu, B. Priming integrin alpha 5 promotes the osteogenic differentiation of human periodontal ligament stem cells due to cytoskeleton and cell cycle changes. J. Proteom. 2018, 179, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Stopp, S.; Bornhauser, M.; Ugarte, F.; Wobus, M.; Kuhn, M.; Brenner, S.; Thieme, S. Expression of the melanoma cell adhesion molecule in human mesenchymal stromal cells regulates proliferation, differentiation, and maintenance of hematopoietic stem and progenitor cells. Haematologica 2013, 98, 505–513. [Google Scholar] [CrossRef][Green Version]
- Liu, F.; Zhu, C.; Huang, X.; Cai, J.; Wang, H.; Wang, X.; He, S.; Liu, C.; Yang, X.; Zhang, Y.; et al. A low level of GPR37 is associated with human hepatocellular carcinoma progression and poor patient survival. Pathol. Res. Pract. 2014, 210, 885–892. [Google Scholar] [CrossRef]
- Granchi, C. ATP citrate lyase (ACLY) inhibitors: An anti-cancer strategy at the crossroads of glucose and lipid metabolism. Eur. J. Med. Chem. 2018, 157, 1276–1291. [Google Scholar] [CrossRef]
- Kardon, J.R.; Vale, R.D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009, 10, 854–865. [Google Scholar] [CrossRef]
- Xiong, G.; Deng, L.; Zhu, J.; Rychahou, P.G.; Xu, R. Prolyl-4-hydroxylase alpha subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 2014, 14, 1. [Google Scholar] [CrossRef]
- Li, X.; Yang, Q.; Yu, H.; Wu, L.; Zhao, Y.; Zhang, C.; Yue, X.; Liu, Z.; Wu, H.; Haffty, B.G.; et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 2014, 5, 788–801. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, W.; Jia, X.; Luo, R.; Tan, Y.; Zhu, X.; Yang, X.; Wang, X.; Wang, K. Transcriptional repression of CDKN2D by PML/RARalpha contributes to the altered proliferation and differentiation block of acute promyelocytic leukemia cells. Cell Death Dis. 2014, 5, e1431. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.P.; Liu, C.L.; Chen, M.J.; Chien, M.N.; Leung, C.H.; Lin, C.H.; Hsu, Y.C.; Lee, J.J. CD74 expression and its therapeutic potential in thyroid carcinoma. Endocr. Relat. Cancer 2015, 22, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Lee, M.W.; Yoo, K.H.; Lee, T.H.; Kim, H.J.; Jang, I.K.; Chun, Y.H.; Kim, H.J.; Park, S.J.; Lee, S.H.; et al. Gene expression profiles of human adipose tissue-derived mesenchymal stem cells are modified by cell culture density. PLoS ONE 2014, 9, e83363. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Wang, J.; Yan, W.; Cui, Y.; Chen, Z.; Gao, X.; Wen, X.; Chen, J. GLUT1 regulates cell glycolysis and proliferation in prostate cancer. Prostate 2018, 78, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Katsurada, A.; Fukuda, H.; Iritani, N. Effects of dietary nutrients on substrate and effector levels of lipogenic enzymes, and lipogenesis from tritiated water in rat liver. Biochim. Biophys. Acta 1986, 878, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, Y.; Sekiya, I.; Yagishita, K.; Muneta, T. Comparison of human stem cells derived from various mesenchymal tissues: Superiority of synovium as a cell source. Arthritis Rheum. 2005, 52, 2521–2529. [Google Scholar] [CrossRef] [PubMed]
- Mohamed-Ahmed, S.; Fristad, I.; Lie, S.A.; Suliman, S.; Mustafa, K.; Vindenes, H.; Idris, S.B. Adipose-derived and bone marrow mesenchymal stem cells: A donor-matched comparison. Stem Cell Res. Ther. 2018, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Woo, D.H.; Hwang, H.S.; Shim, J.H. Comparison of adult stem cells derived from multiple stem cell niches. Biotechnol. Lett. 2016, 38, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Secunda, R.; Vennila, R.; Mohanashankar, A.M.; Rajasundari, M.; Jeswanth, S.; Surendran, R. Isolation, expansion and characterisation of mesenchymal stem cells from human bone marrow, adipose tissue, umbilical cord blood and matrix: A comparative study. Cytotechnology 2015, 67, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Patrick, C.W., Jr. Adipose tissue engineering: The future of breast and soft tissue reconstruction following tumor resection. Semin. Surg. Oncol. 2000, 19, 302–311. [Google Scholar] [CrossRef]
- Entenmann, G.; Hauner, H. Relationship between replication and differentiation in cultured human adipocyte precursor cells. Am. J. Physiol. 1996, 270, C1011–C1016. [Google Scholar] [CrossRef] [PubMed]

| Symbol | Gene Name | Correlation (r) |
|---|---|---|
| ITGA5 | integrin subunit alpha 5 | 0.826 *** |
| MCAM | melanoma cell adhesion molecule | 0.812 *** |
| GPR37 | G protein-coupled receptor 37 | 0.782 ** |
| PSMC5 | proteasome 26S subunit, ATPase 5 | 0.769 ** |
| ACLY | ATP citrate lyase | 0.737 ** |
| DNCI1 | dynein cytoplasmic 1 intermediate chain 1 | 0.732 ** |
| P4HA2 | prolyl 4-hydroxylase subunit alpha 2 | 0.720 ** |
| LIF | leukemia inhibitory factor | 0.708 ** |
| ZNF185 | zinc finger protein 185 with LIM domain | 0.708 ** |
| CDKN2D | cyclin dependent kinase inhibitor 2D | 0.703 ** |
| DPYSL3 | dihydropyrimidinase like 3 | 0.664 ** |
| INPP5E | inositol polyphosphate-5-phosphatase E | 0.650 ** |
| UBE2C | ubiquitin conjugating enzyme E2 C | 0.612 * |
| E2F1 | E2F transcription factor 1 | 0.608 * |
| CCNB1 | cyclin B1 | 0.598 * |
| CD74 | CD74 antigen | 0.561 * |
| COL7A1 | collagen type VII alpha 1 chain | 0.561 * |
| AURKB | aurora kinase B | 0.556 * |
| AMD1 | adenosylmethionine decarboxylase 1 | 0.545 * |
| CDC20 | cell division cycle 20 | 0.526 * |
| SLC2A1 | solute carrier family 2 member 1 | 0.526 * |
| MCM7 | minichromosome maintenance complex component 7 | 0.517 * |
| Gene | Relative mRNA Levels | |
|---|---|---|
| Jaw | Ilium | |
| CDKN2D | 1.00 ± 0.16 | 22.09 ± 4.09 ** |
| CD74 | 1.00 ± 0.41 | 18.02 ± 5.05 ** |
| MCAM | 1.00 ± 0.64 | 13.44 ± 2.94 ** |
| DNCI1 | 1.00 ± 0.23 | 4.52 ± 0.61 *** |
| GPR37 | 1.00 ± 0.65 | 4.32 ± 0.83 ** |
| P4HA2 | 1.00 ± 0.43 | 4.31 ± 0.91 ** |
| ACLY | 1.00 ± 0.15 | 4.14 ± 0.49 *** |
| SLC2A1 | 1.00 ± 0.17 | 3.78 ± 0.44 *** |
| ITGA5 | 1.00 ± 0.13 | 3.72 ± 0.42 *** |
| LIF | 1.00 ± 0.39 | 3.66 ± 0.53 ** |
| AURKB | 1.00 ± 0.26 | 3.09 ± 0.70 * |
| E2F1 | 1.00 ± 0.12 | 2.38 ± 0.40 ** |
| COL7A1 | 1.00 ± 0.23 | 2.21 ± 0.42 * |
| UBE2C | 1.00 ± 0.20 | 2.15 ± 0.45 * |
| CDC20 | 1.00 ± 0.23 | 2.02 ± 0.40 * |
| PSMC5 | 1.00 ± 0.08 | 1.98 ± 0.31 * |
| MCM7 | 1.00 ± 0.10 | 1.81 ± 0.27 * |
| INPP5E | 1.00 ± 0.24 | 1.79 ± 0.26 * |
| ZNF185 | 1.00 ± 0.06 | 1.79 ± 0.69 |
| CCNB1 | 1.00 ± 0.16 | 1.69 ± 0.36 |
| DPYSL3 | 1.00 ± 0.17 | 1.35 ± 0.16 |
| AMD1 | 1.00 ± 0.11 | 1.22 ± 0.16 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanawa, M.; Igarashi, A.; Fujimoto, K.; Ronald, V.S.; Higashi, Y.; Kurihara, H.; Kato, Y.; Kawamoto, T. Potential Marker Genes for Predicting Adipogenic Differentiation of Mesenchymal Stromal Cells. Appl. Sci. 2019, 9, 2942. https://doi.org/10.3390/app9142942
Kanawa M, Igarashi A, Fujimoto K, Ronald VS, Higashi Y, Kurihara H, Kato Y, Kawamoto T. Potential Marker Genes for Predicting Adipogenic Differentiation of Mesenchymal Stromal Cells. Applied Sciences. 2019; 9(14):2942. https://doi.org/10.3390/app9142942
Chicago/Turabian StyleKanawa, Masami, Akira Igarashi, Katsumi Fujimoto, Veronica Sainik Ronald, Yukihito Higashi, Hidemi Kurihara, Yukio Kato, and Takeshi Kawamoto. 2019. "Potential Marker Genes for Predicting Adipogenic Differentiation of Mesenchymal Stromal Cells" Applied Sciences 9, no. 14: 2942. https://doi.org/10.3390/app9142942
APA StyleKanawa, M., Igarashi, A., Fujimoto, K., Ronald, V. S., Higashi, Y., Kurihara, H., Kato, Y., & Kawamoto, T. (2019). Potential Marker Genes for Predicting Adipogenic Differentiation of Mesenchymal Stromal Cells. Applied Sciences, 9(14), 2942. https://doi.org/10.3390/app9142942





