Non-Invasive Assessment of Hepatic Fibrosis by Elastic Measurement of Liver Using Magnetic Resonance Tagging Images
Abstract
:1. Introduction
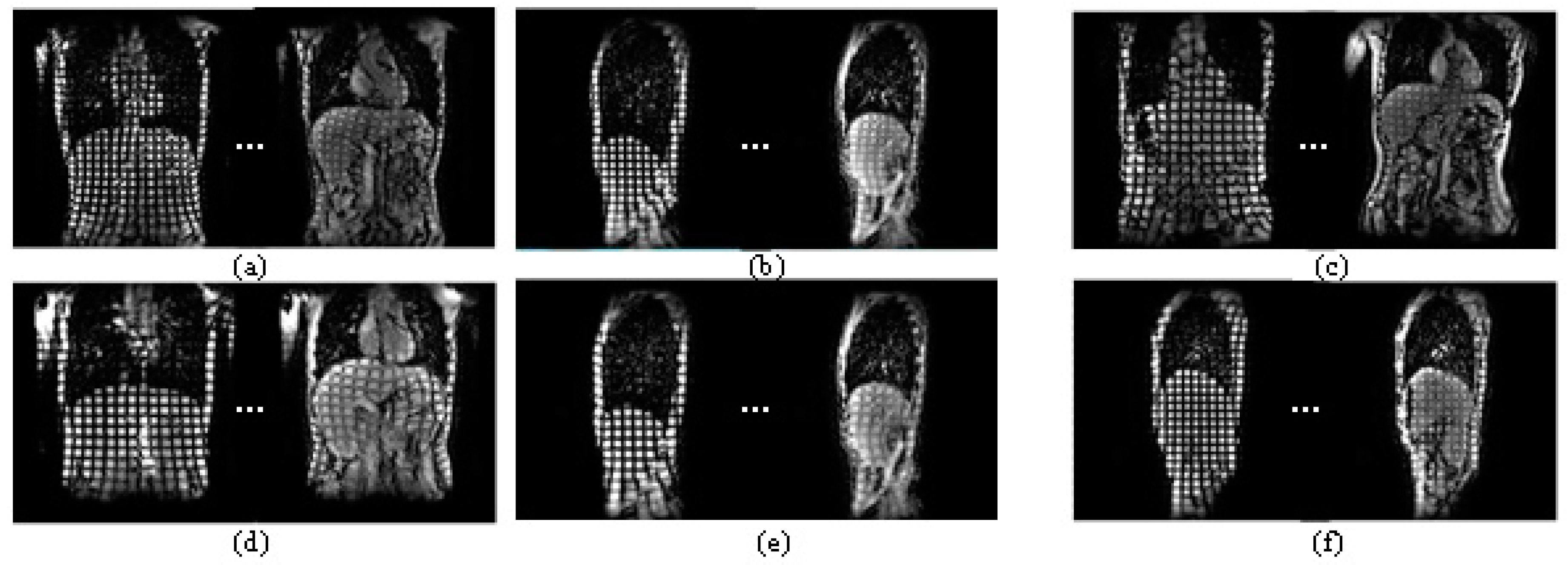
2. Experimental Materials
3. Methods
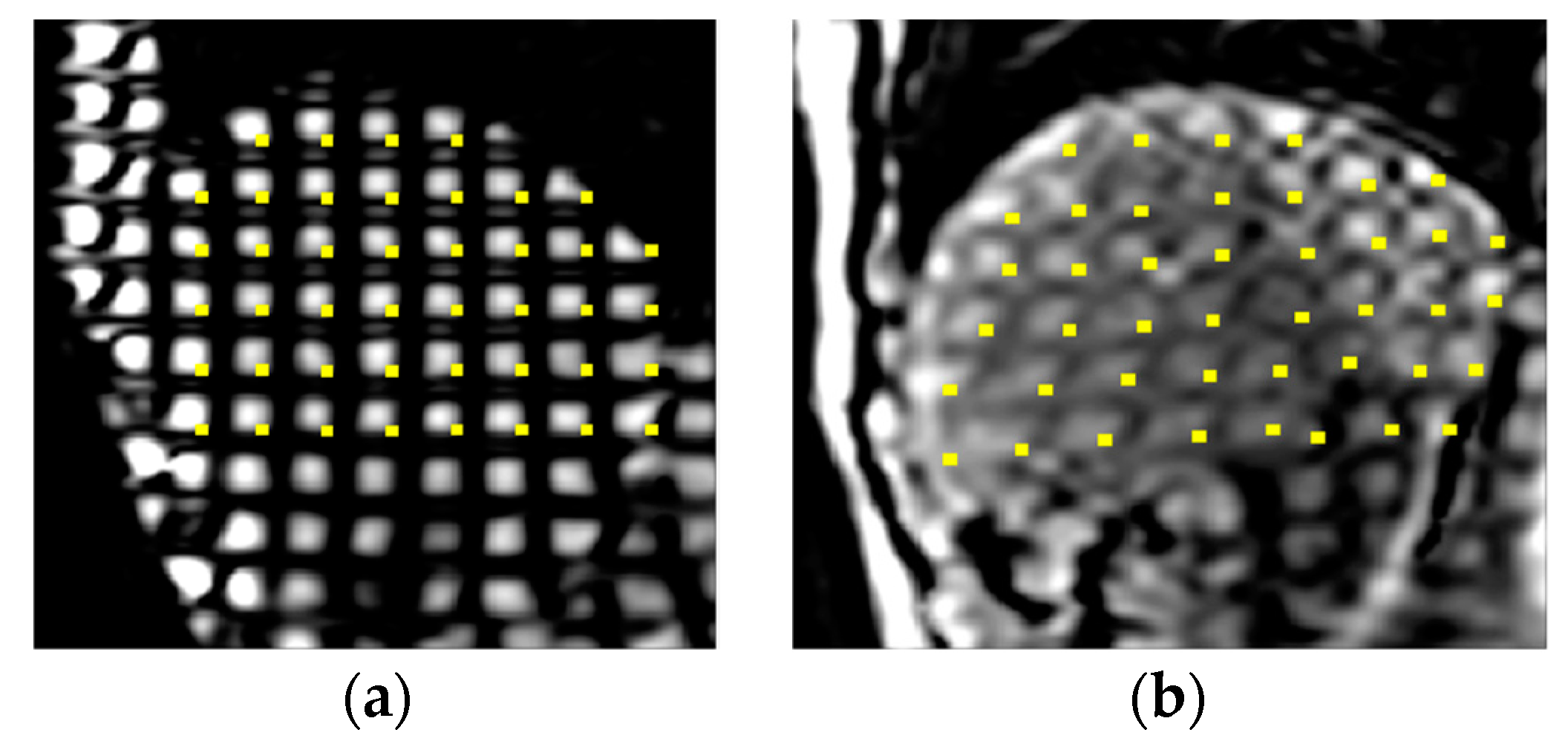
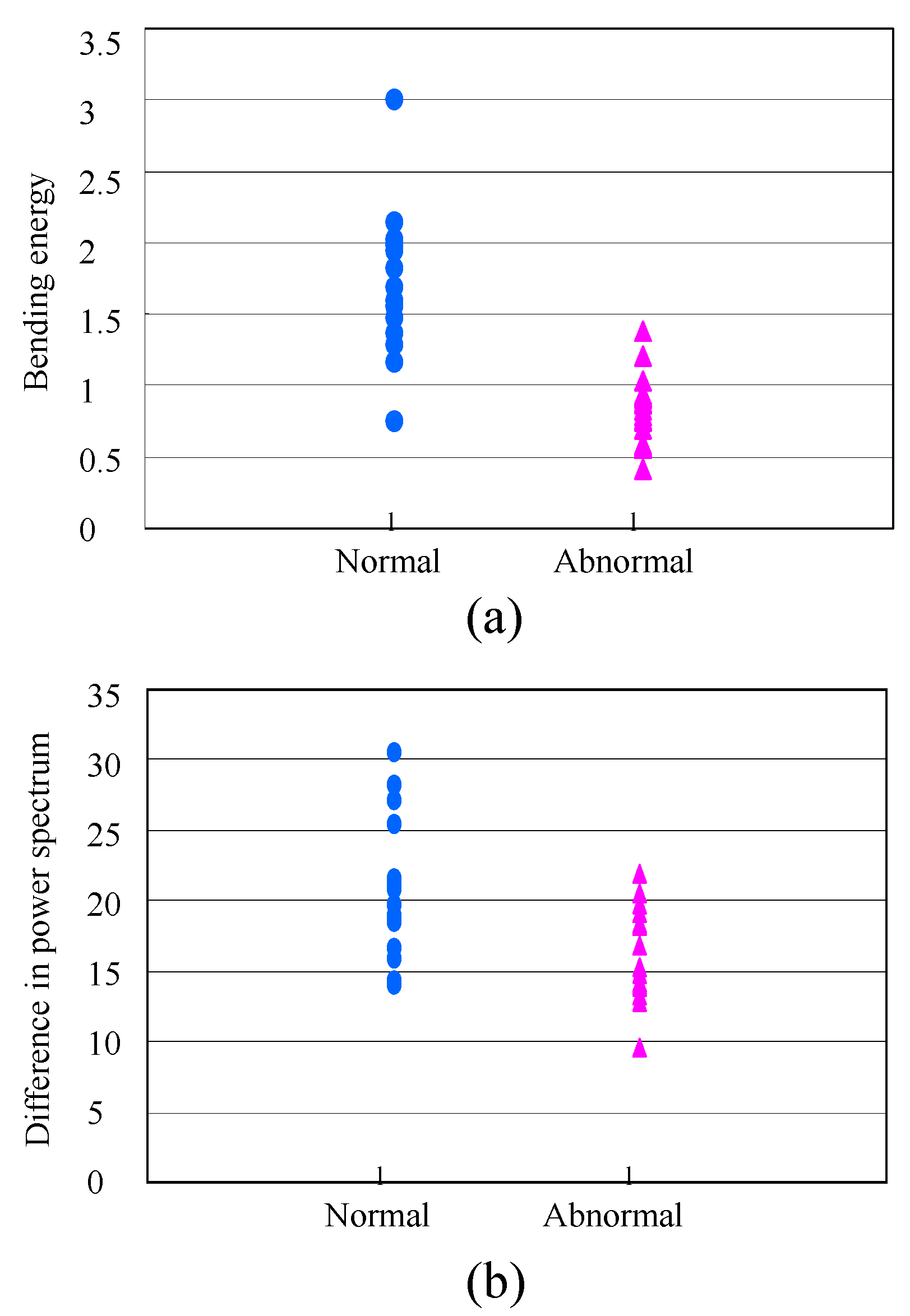
3.1. Calculation of Bending Energy Based on Thin-Plate Spline (TPS) Method in the Spatial Domain
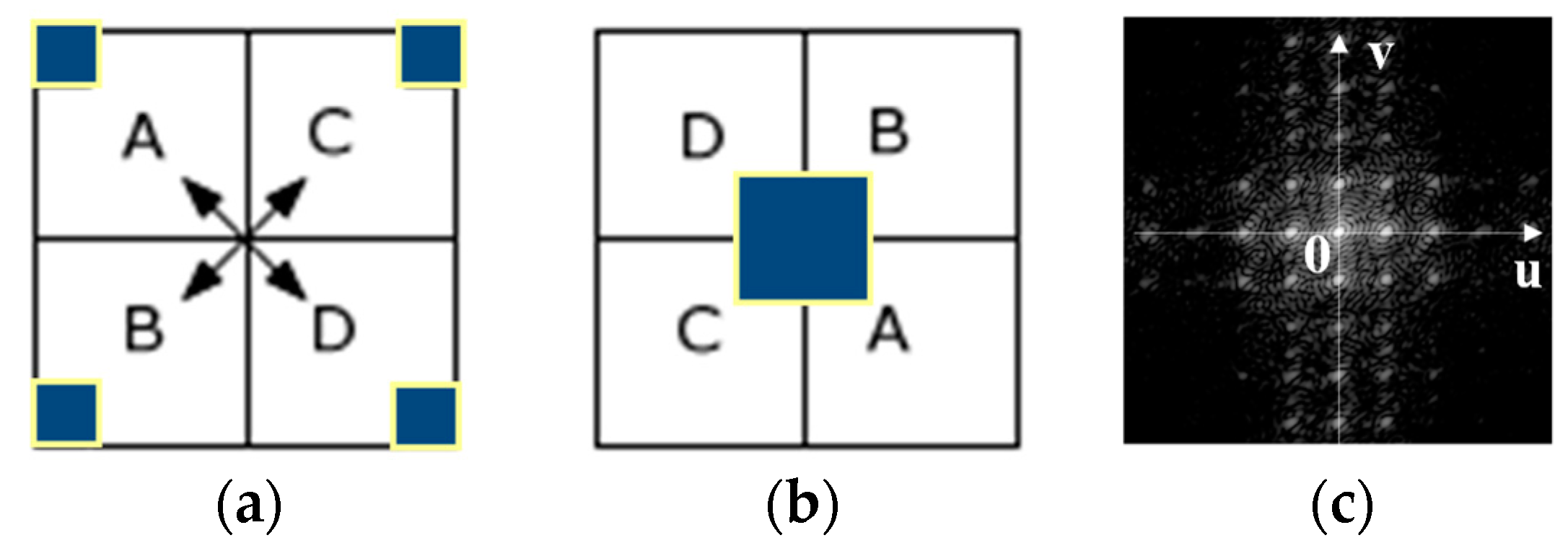
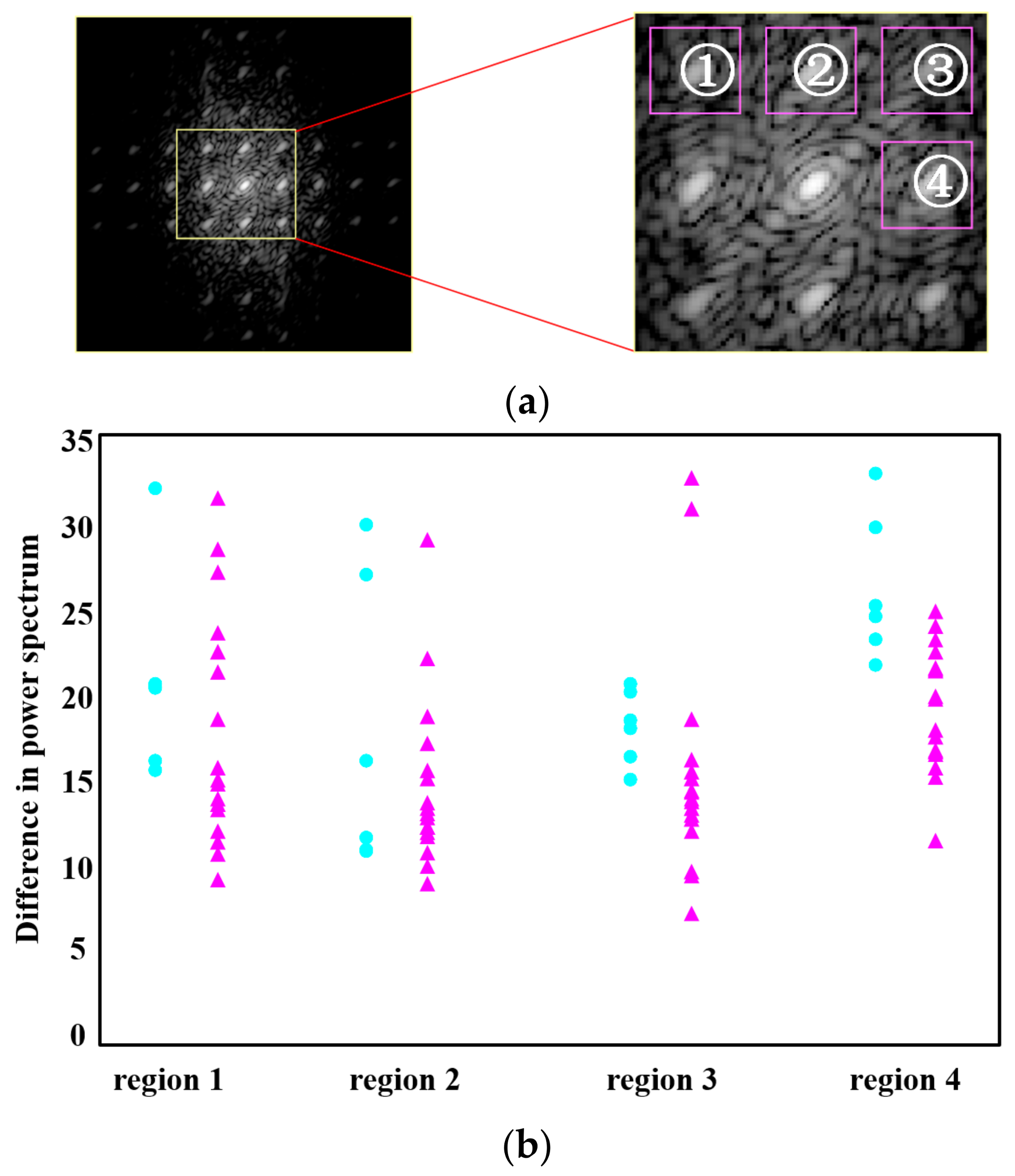
3.2. Calculation of the Difference in the Power Spectral Values of the Tags by FFT in the Frequency Domain
3.3. Classification Method
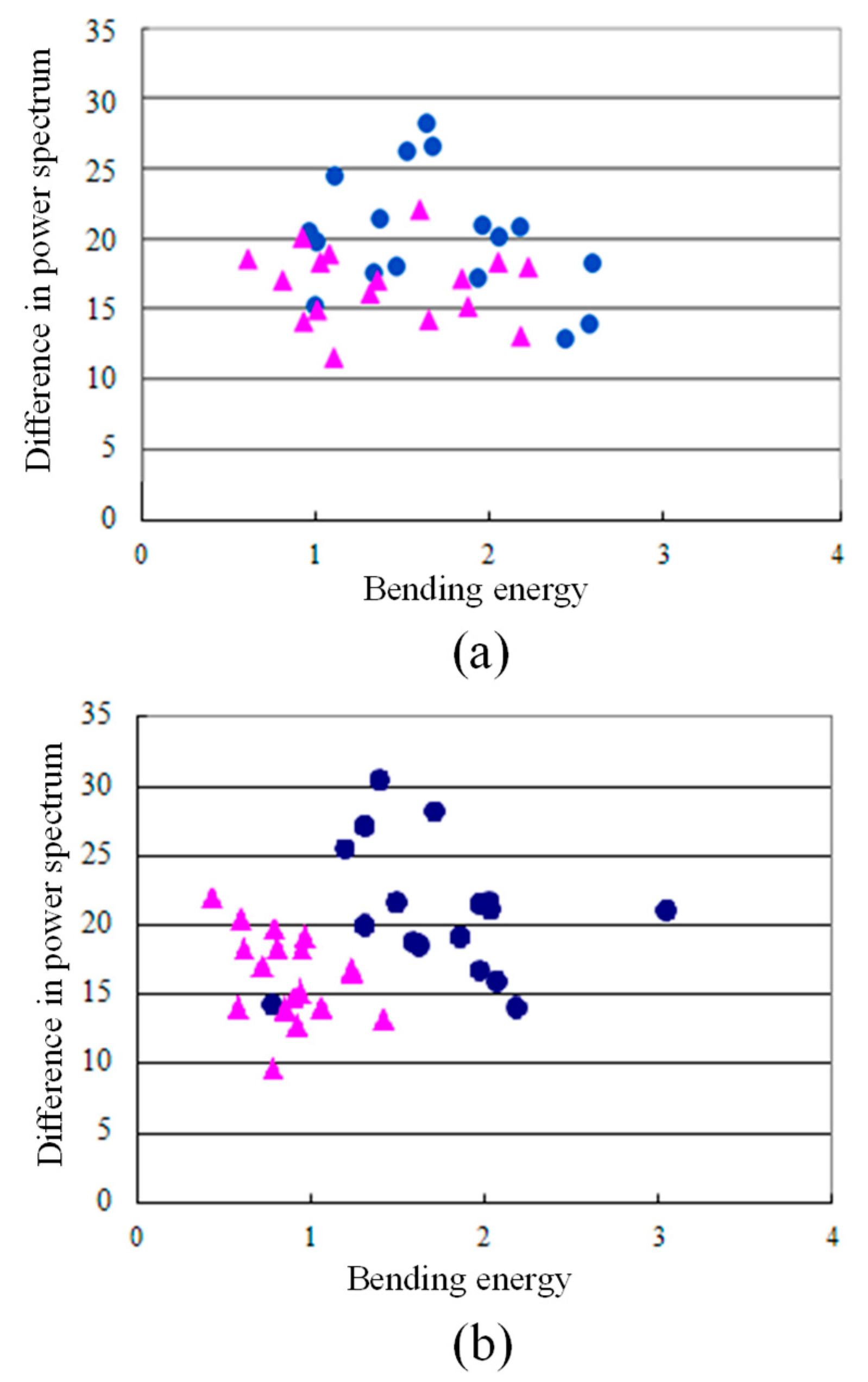
4. Results and Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- El-Serag, H.B.; Mason, A.C. Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 1999, 340, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Yatsuhashi, H.; Yano, M. Natural history of chronic hepatitis C. J. Gastroenterol. Hepatol. 2000, 15, 111–116. [Google Scholar] [CrossRef]
- Wynn, T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Saadeh, S.; Cammell, G.; Carey, W.D.; Younossi, Z.; Barnes, D.; Easley, K. The role of liver biopsy in chronic hepatitis C. Hepatology 2001, 33, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Poynard, T.; Ratziu, V.; Bedossa, P. Appropriateness of liver biopsy. Can. J. Gastroenterol. 2000, 14, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.A.; Sheth, S.G.; Chopra, S. Liver biopsy. N. Engl. J. Med. 2001, 344, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Regev, A.; Berho, M.; Jeffers, L.J.; Milikowski, C.; Molina, E.G.; Pyrsopoulos, N.T.; Feng, Z.Z.; Reddy, K.R.; Schiff, E.R. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am. J. Gastroenterol. 2002, 97, 2614–2618. [Google Scholar] [CrossRef] [PubMed]
- Muthupillai, R.; Lomas, D.J.; Rossman, P.J.; Greenleaf, J.F.; Manduca, A.; Ehman, R.L. Magnetic resonance elastography by direct visualization of propagating acoustic strain waves. Science 1995, 269, 1854–1857. [Google Scholar] [CrossRef] [PubMed]
- Rouviere, O.; Yin, M.; Dresner, M.A.; Rossman, P.J.; Burgart, L.J.; Fidler, J.L.; Ehman, R.L. MR elastography of the liver: Preliminary results. Radiology 2006, 240, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Huwart, L.; Sempoux, C.; Salameh, N.; Jamart, J.; Annet, L.; Sinkus, R.; Peeters, F.; ter Beek, L.C.; Horsmans, Y.; van Beers, B.E. Liver fibrosis: Noninvasive assessment with MR elastography versus aspartate aminotransferase-to-platelet ratio index. Radiology 2007, 245, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Lucidarme, O.; Baleston, F.; Cadi, M.; Bellin, M.F.; Charlotte, F.; Ratziu, V.; Grenier, P.A. Non-invasive detection of liver fibrosis: Is superparamagnetic iron oxide particle-enhanced MR imaging a contributive technique? Eur. Radiol. 2002, 13, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, D.A.; Behling, C.A.; Alpert, E.; Hassanein, T.I.; Sirlin, C.B. Liver fibrosis: Noninvasive diagnosis with double contrast material-enhanced MR imaging. Radiology 2006, 239, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Taouli, B.; Tolia, A.J.; Losada, M.; Babb, J.S.; Chan, E.S.; Bannan, M.A.; Tobias, H. Diffusion-weighted MRI for quantification of liver fibrosis: Preliminary experience. Am. J. Roentgenol. 2007, 189, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Friedrich-Rust, M.; Ong, M.F.; Herrmann, E.; Dries, V.; Samaras, P.; Zeuzem, S.; Sarrazin, C. Real-time elastography for noninvasive assessment of liver fibrosis in chronic viral hepatitis. Am. J. Roentgenol. 2007, 188, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, L.; Shi, X.Y.; Pan, W.Q.; Bai, Y.F.; Ai, H. Real-time elastography with a novel quantitative technology for assessment of liver fibrosis in chronic hepatitis B. Eur. J. Radiol. 2012, 81, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Li, W.G.; Fujita, H.; Kanematsu, M.; Hara, T.; Zhou, X.R.; Kondo, H.; Hoshi, H. Automatic segmentation of hepatic tissue and 3D volume analysis of cirrhosis in multi-detector row CT scans and MR imaging. IEICE Trans. Inf. Syst. 2004, 87, 2138–2147. [Google Scholar]
- Goshima, S.; Kanematsu, M.; Kobayashi, T.; Furukawa, T.; Zhang, X.; Fujita, H.; Watanabe, H.; Kondo, H.; Moriyama, N.; Bae, K.T. Staging hepatic fibrosis: Computer-aided analysis of hepatic contours on gadolinium ethoxybenzyl diethylenetriaminepentaacetic acid-enhanced hepatocyte-phase magnetic resonance imaging. Hepatology 2012, 55, 328–329. [Google Scholar] [CrossRef] [PubMed]
- Li, W.D.; Zeng, Y.F.; Zhang, X.J.; Huang, Y.; Long, L.L.; Fujita, H. Staging the hepatic fibrosis on CT images: Optimizing the slice thickness and texture features. In Proceedings of the IEEE 2011 International Symposium on Bioelectronics and Bioinformatics, Suzhou, China, 3–5 November 2011; pp. 267–270. [Google Scholar]
- Watanabe, H.; Kanematsu, M.; Kitagawa, T.; Suzuki, Y.; Kondo, H.; Goshima, S.; Kajita, K.; Bae, K.T.; Hirose, Y.; Miotani, S.; et al. MR elastography of the liver at 3 T with cine-tagging and bending energy analysis: Preliminary results. Eur. Radiol. 2010, 20, 2381–2389. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, W.; Wakabayashi, T.; Kimura, F.; Tsuruoka, S.; Sekioka, K. Automatic extraction of SPAMM grids in left ventricular myocardium from MR tagging images. In Proceedings of the 2009 International Workshop on Regional Innovation Studies: Biomedical Engineering, Mie, Japan, 8 October 2009; pp. 25–29. [Google Scholar]
- Zerhouni, E.A.; Parish, D.M.; Rogers, W.J.; Yang, A.; Shapiro, E.P. Human heart: Tagging with MR imaging—A method for noninvasive assessment of myocardialmotion. Radiology 1988, 169, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Inaba, T.; Kinosada, Y.; Kawasaki, S.; Obata, H.; Tokuda, M. Analysis of left ventricular wall motion using a magnetic resonance tagging technique: Measurement of circumferential elongation of ventricular Wall in Patients with DCM. Trans. Jpn. Soc. Med. Biol. Eng. 2003, 41, 136–139. (In Japanese) [Google Scholar]
- Watanabe, H.; Kanematsu, M.; Kondo, H.; Goshima, S.; Kitagawa, T.; Fujita, H. MR elastography of the liver with cine-tagging and physical bending energy analysis using 3T MRI: Phantom study and preliminary clinical application. In Proceedings of the 94th Scientific Assembly and Annual Meeting of the Radiological Society of North America, Chicago, IL, USA, 30 November–5 December 2008; p. 828. [Google Scholar]
- Axel, L.; Dougherty, L. MR imaging of motion with spatial modulation of magnetization. Radiology 1989, 171, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Bookstein, F.L. Principal warps: Thin-plate splines and the decomposition of deformations. IEEE Trans. Pattern Anal. Mach. Intell. 1989, 11, 567–585. [Google Scholar]
- Miotani, S.; Zhou, X.G.; Kitagawa, T.; Hara, T.; Fujita, H.; Yokoyama, R.; Watanae, H.; Kanematsu, M.; Hoshi, H. Initial Examination of Automatic Calculation Method of Non-Rigid Deformity of the Liver in MR Tagging Images; IEICE Technical Report; IEICE: Minato-ku, Tokyo, 2009; Volume 109, pp. 213–218. (In Japanese) [Google Scholar]
- Hosseini, H.S.; Safabakhsh, R. TASOM: A New Time Adaptive Self-Organizing Map. IEEE Trans. Syst. Man Cybern. 2003, 33, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Goshima, S.; Zhou, X.G.; Hara, T.; Kanematsu, M.; Fujita, H.; Furukawa, T. Quantitative staging the hepatic Fibrosis: Computer-aided diagnosis by shape, texture, volume, and elasticity analysis. In Proceedings of the Radiological Society of North America 2011 Scientific Assembly and Annual Meeting, Chicago, IL, USA, 26 November–2 December 2011. [Google Scholar]
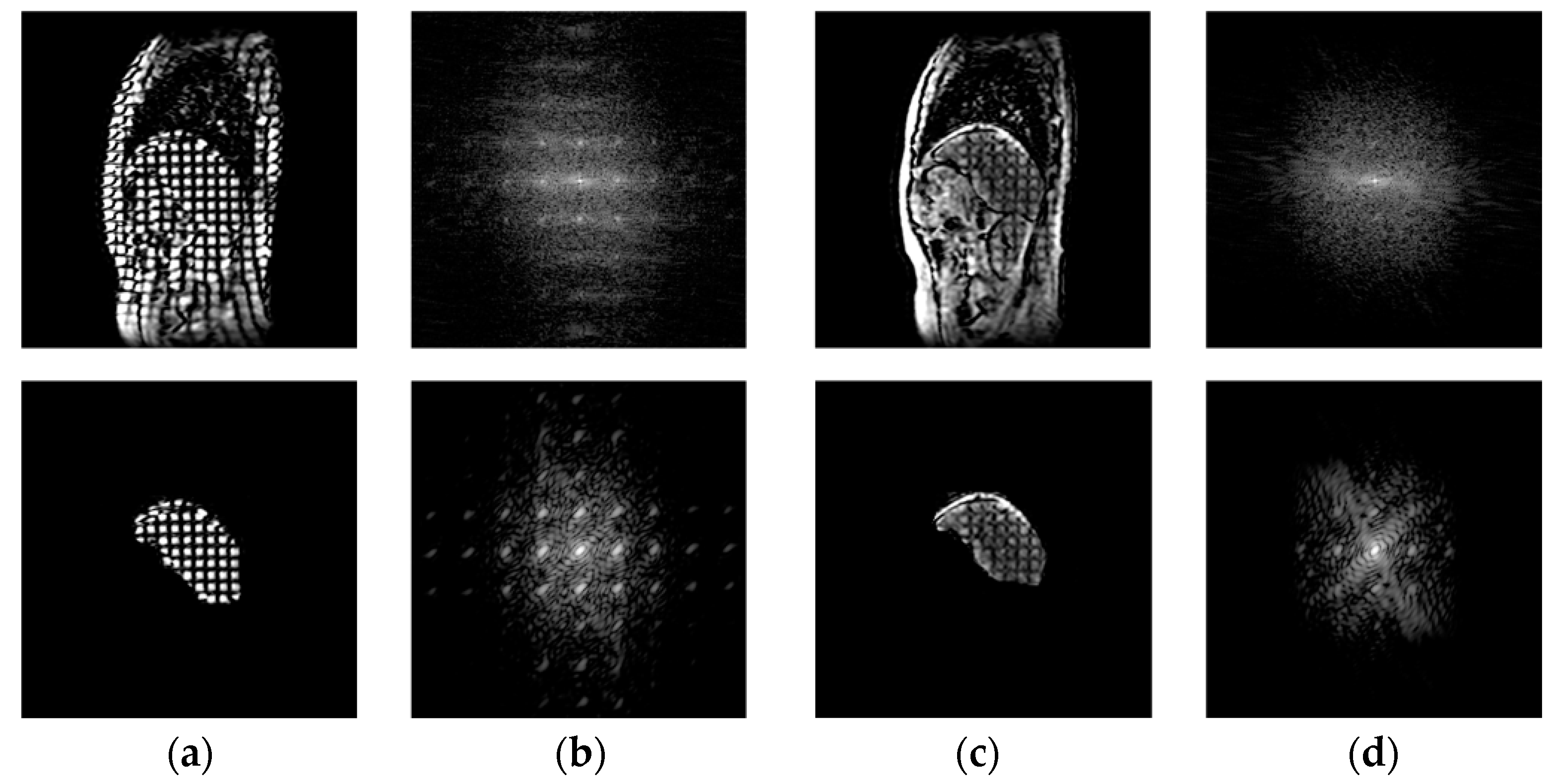
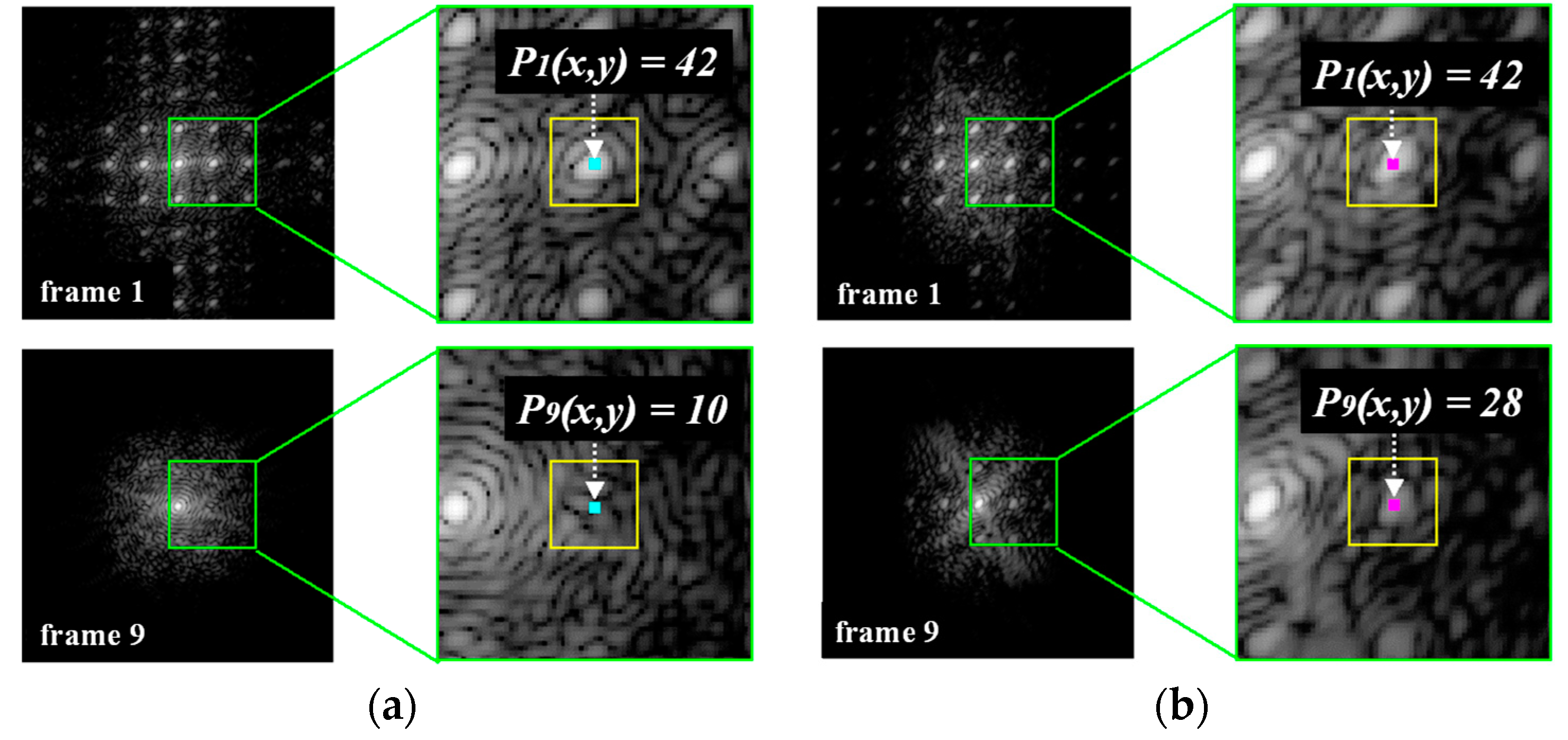
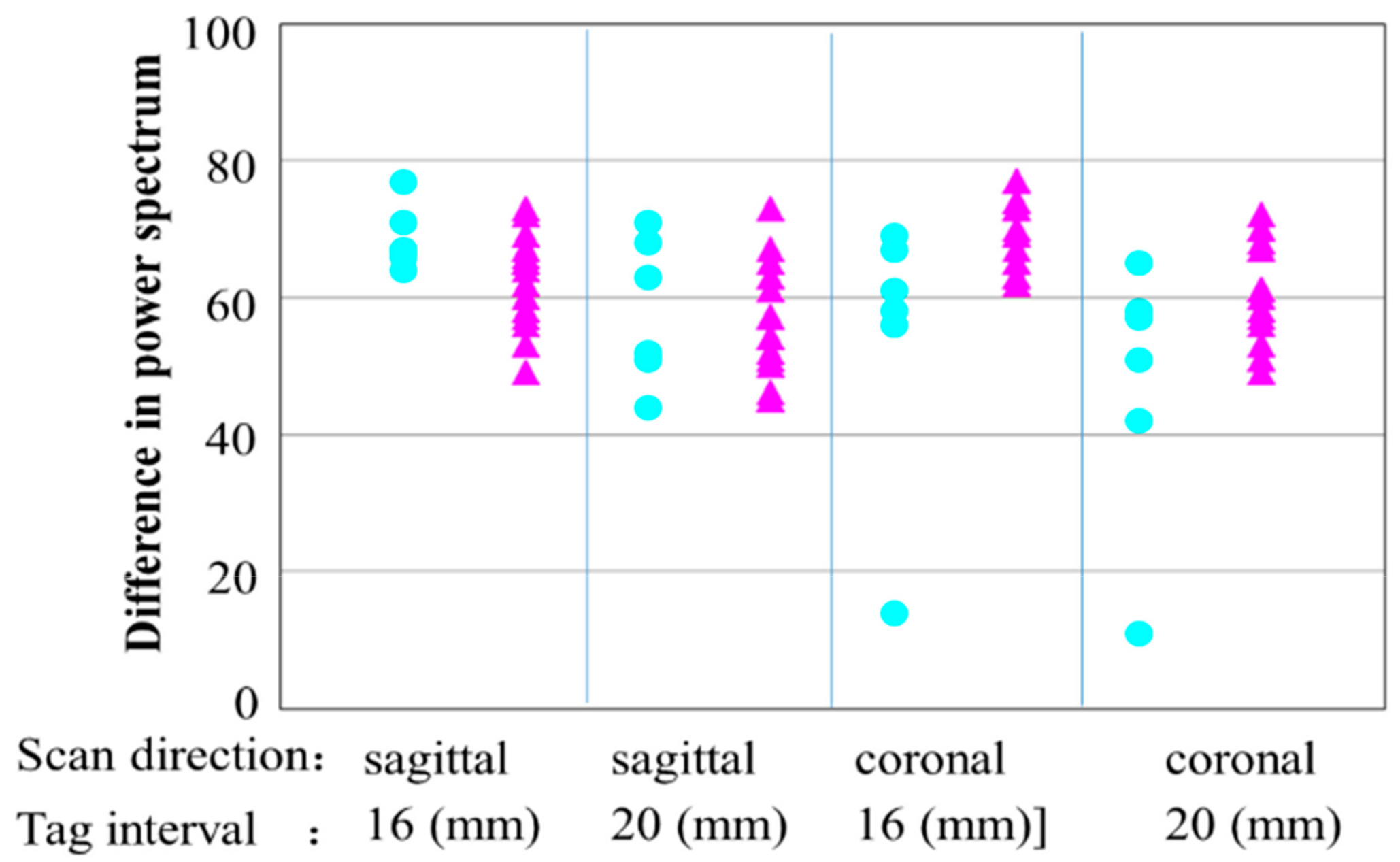
| Average DPS Values | Normal Liver | Abnormal Liver | Difference between Two Groups | |
|---|---|---|---|---|
| Type of MR Tagging Images | ||||
| 16 (mm), sagittal | 68.7 | 62.8 | 5.9 | |
| 20 (mm), sagittal | 58.2 | 55.4 | 2.8 | |
| 16 (mm), coronal | 54.2 | 69.2 | 5.0 | |
| 20 (mm), coronal | 47.3 | 60.7 | 3.4 | |
| Gold Standard | Normal Liver | Abnormal Liver | |
|---|---|---|---|
| SOM | |||
| Normal liver | 16 | 0 | |
| Abnormal liver | 1 | 17 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhou, X.; Hara, T.; Fujita, H. Non-Invasive Assessment of Hepatic Fibrosis by Elastic Measurement of Liver Using Magnetic Resonance Tagging Images. Appl. Sci. 2018, 8, 437. https://doi.org/10.3390/app8030437
Zhang X, Zhou X, Hara T, Fujita H. Non-Invasive Assessment of Hepatic Fibrosis by Elastic Measurement of Liver Using Magnetic Resonance Tagging Images. Applied Sciences. 2018; 8(3):437. https://doi.org/10.3390/app8030437
Chicago/Turabian StyleZhang, Xuejun, Xiangrong Zhou, Takeshi Hara, and Hiroshi Fujita. 2018. "Non-Invasive Assessment of Hepatic Fibrosis by Elastic Measurement of Liver Using Magnetic Resonance Tagging Images" Applied Sciences 8, no. 3: 437. https://doi.org/10.3390/app8030437
APA StyleZhang, X., Zhou, X., Hara, T., & Fujita, H. (2018). Non-Invasive Assessment of Hepatic Fibrosis by Elastic Measurement of Liver Using Magnetic Resonance Tagging Images. Applied Sciences, 8(3), 437. https://doi.org/10.3390/app8030437






