Diagnosis of Sarcoidosis Through Supervised Ensemble Method and GenAI-Based Data Augmentation: An Intelligent Diagnostic Tool
Abstract
1. Introduction
2. Literature Review
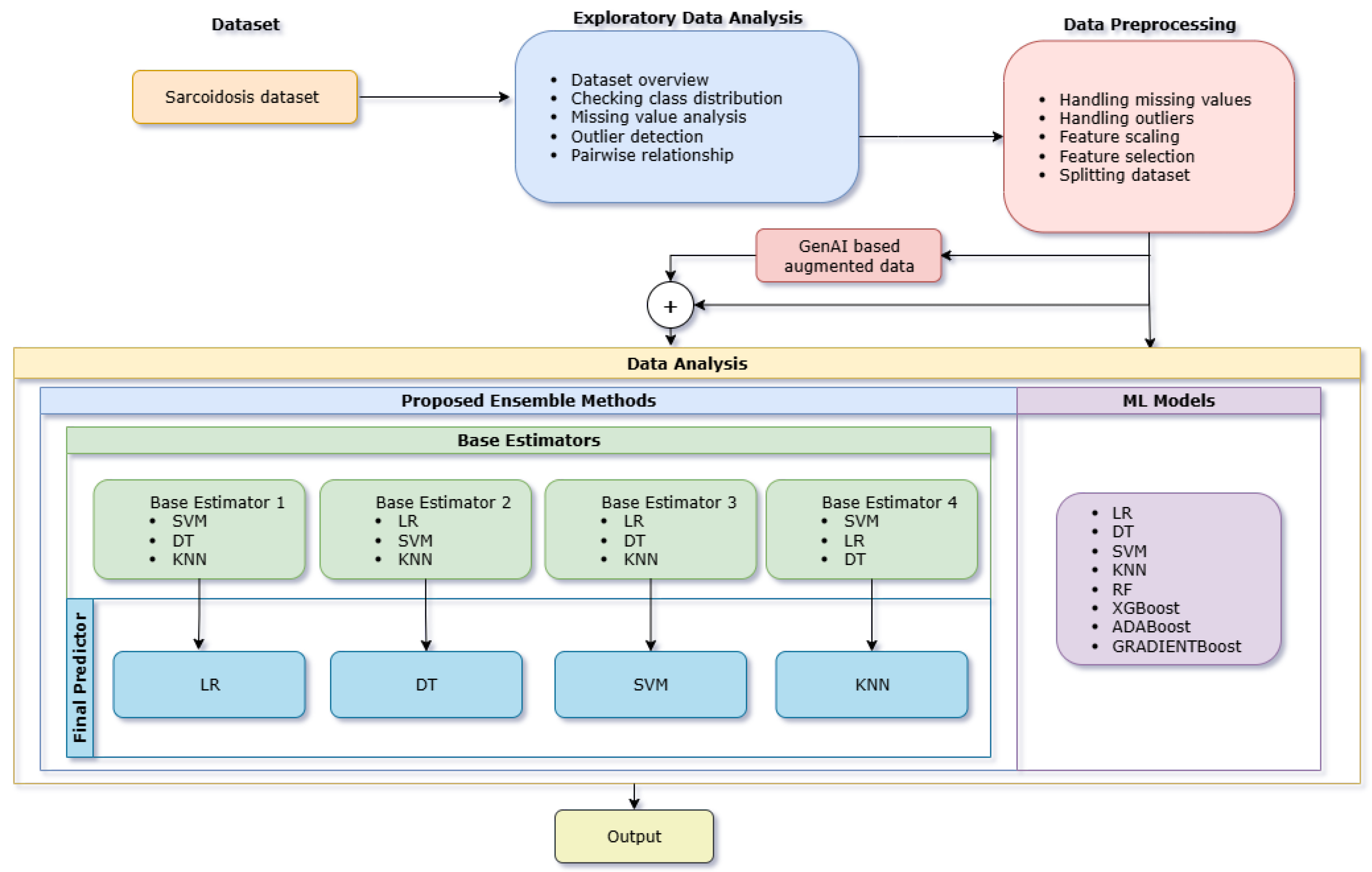
3. Materials and Methods
3.1. Data Wrangling
3.2. Exploratory Data Analysis (EDA)
3.3. Data Preprocessing
3.3.1. Handling Missing Values
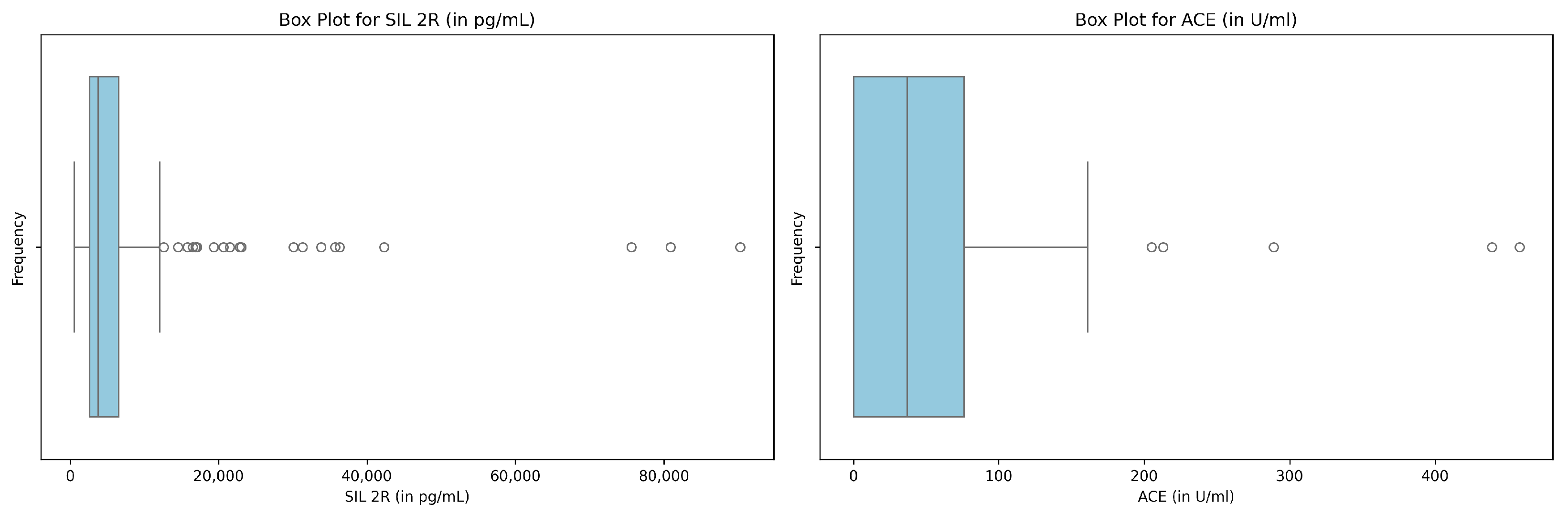
3.3.2. Handling Outliers
3.3.3. Feature Scaling
3.3.4. Feature Selection
3.3.5. Synthetic Data Generation Using CTGAN
3.3.6. Splitting Dataset
3.4. Data Analysis
- Model Combination 1:
- –
- Base Estimators: Decision Tree, Support Vector Machine, k-Nearest Neighbors
- –
- Final predictor: Logistic Regression
- Model Combination 2:
- –
- Base Estimators: Logistic Regression, Support Vector Machine, k-Nearest Neighbors
- –
- Final predictor: Decision Tree Classifier
- Model Combination 3:
- –
- Base Estimators: Logistic Regression, Decision Tree, k-Nearest Neighbors
- –
- Final predictor: Support Vector Classifier
- Model Combination 4:
- –
- Base Estimators: Logistic Regression, Decision Tree, Support Vector Machine
- –
- Final predictor: k-Nearest Neighbors
4. Results and Discussion
- Evaluation on the Original Dataset: Performance validation under authentic real-world conditions.
- Supplementary Experiments on Synthetic Data (81 data points): Analysis of model robustness and adaptability across diverse synthetic datasets in addition to the original dataset.
- Extended Experiments on Large-scale Synthetic Data (1000 data points): Examination of scalability, consistency, and generalization in extensive simulated environments.
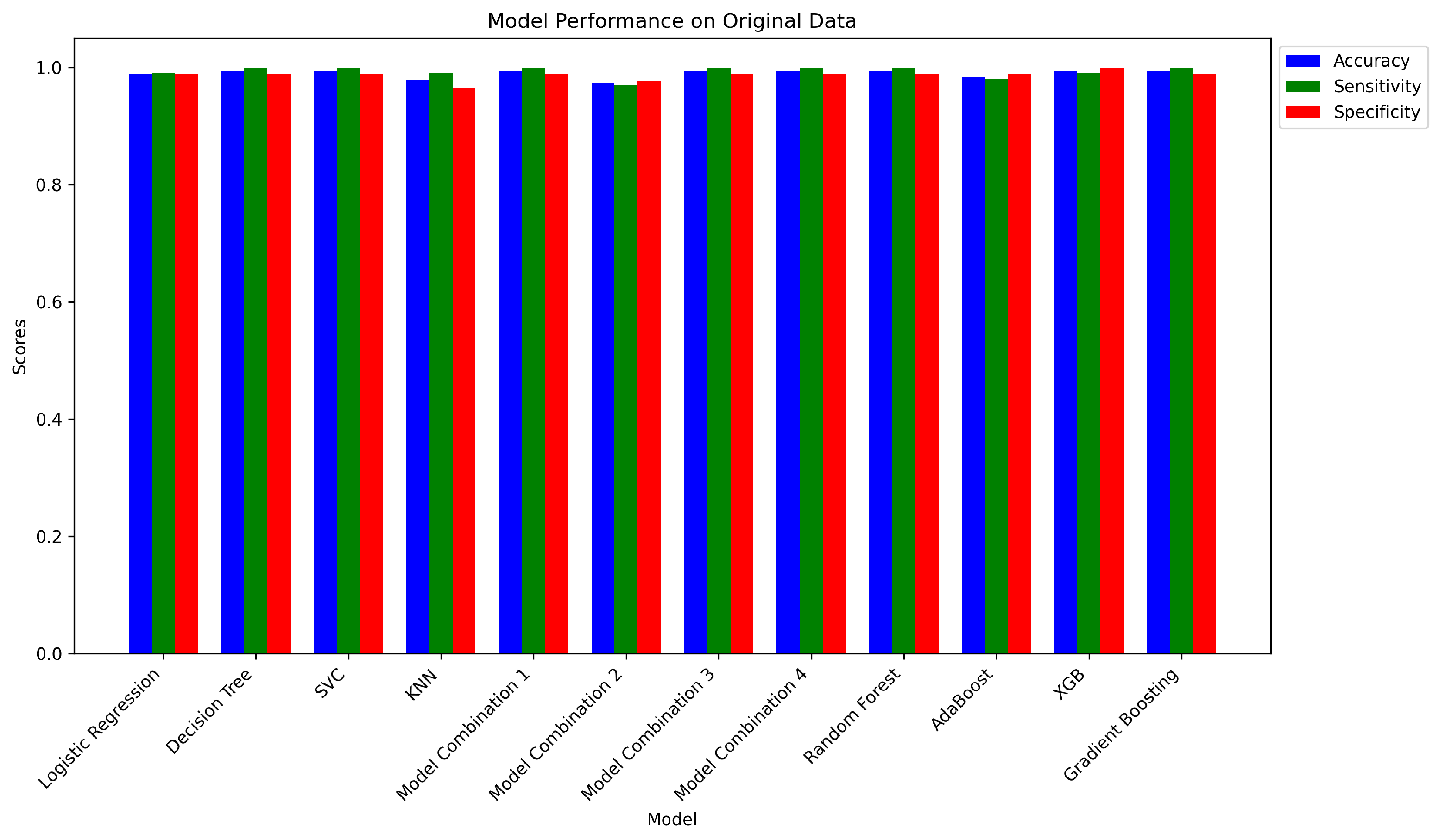
4.1. Evaluation on the Original Dataset
- SIL 2R (in pg/mL): Mean = , Standard Deviation = 1.00
- ACE (in U/mL): Mean = , Standard Deviation = 1.00
4.1.1. Independent Models on Original Dataset
4.1.2. Ensemble Model on Original Dataset
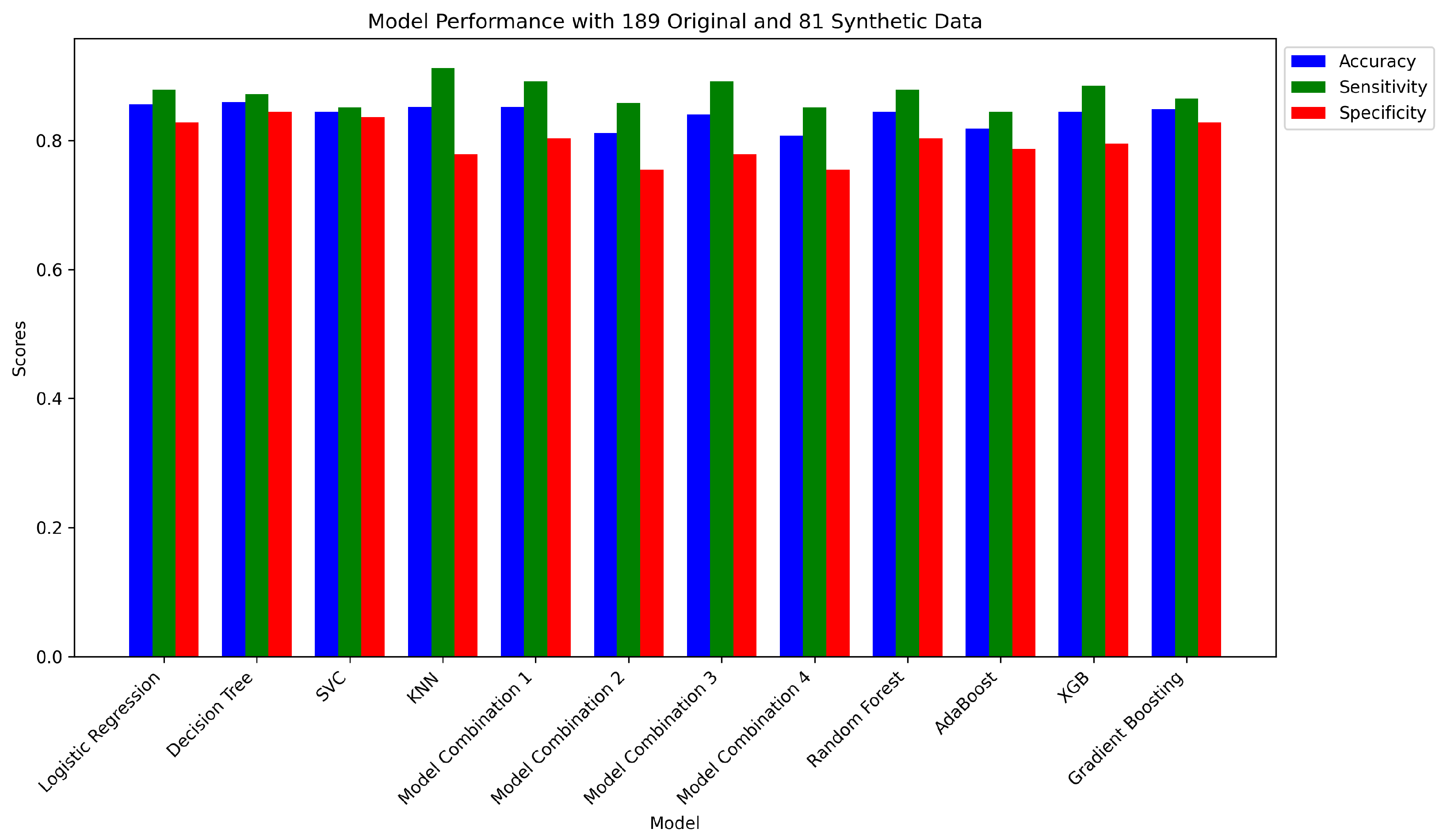
4.2. Supplementary Experiments on Synthetic Data (81 Data Points)
4.2.1. Independent Models on Synthetic Data (81 Data Points)
4.2.2. Ensemble Method on Synthetic Data (81 Data Points)
4.3. Extended Experiments on Large-Scale Synthetic Data (1000 Data Points)
4.3.1. Independent Models on Large-Scale Synthetic Data (1000 Data Points)
4.3.2. Ensemble Method on Large-Scale Synthetic Data (1000 Data Points)
4.3.3. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carreras-Puigvert, J.; Spjuth, O. Artificial intelligence for high content imaging in drug discovery. Curr. Opin. Struct. Biol. 2024, 87, 102842. [Google Scholar] [CrossRef]
- Pariser, A.R.; Gahl, W.A. Important role of translational science in rare disease innovation, discovery, and drug development. J. Gen. Intern. Med. 2014, 29, 804–807. [Google Scholar] [CrossRef]
- Bargagli, E.; Prasse, A. Sarcoidosis: A review for the internist. Intern. Emerg. Med. 2018, 13, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ramón, R.; Gaitán-Valdizán, J.J.; González-Mazón, I.; Sánchez-Bilbao, L.; Martín-Varillas, J.L.; Martínez-López, D.; Demetrio-Pablo, R.; González-Vela, M.C.; Ferraz-Amaro, I.; Castañeda, S.; et al. Systemic treatment in sarcoidosis: Experience over two decades. Eur. J. Intern. Med. 2023, 108, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Sève, P.; Pacheco, Y.; Durupt, F.; Jamilloux, Y.; Gerfaud-Valentin, M.; Isaac, S.; Boussel, L.; Calender, A.; Androdias, G.; Valeyre, D.; et al. Sarcoidosis: A clinical overview from symptoms to diagnosis. Cells 2021, 10, 766. [Google Scholar] [CrossRef]
- Decherchi, S.; Pedrini, E.; Mordenti, M.; Cavalli, A.; Sangiorgi, L. Opportunities and challenges for machine learning in rare diseases. Front. Med. 2021, 8, 747612. [Google Scholar] [CrossRef]
- Figueira, A.; Vaz, B. Survey on synthetic data generation, evaluation methods and GANs. Mathematics 2022, 10, 2733. [Google Scholar] [CrossRef]
- Livieris, I.E.; Alimpertis, N.; Domalis, G.; Tsakalidis, D. An evaluation framework for synthetic data generation models. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations, Corfu, Greece, 27–30 June 2024; Springer: Cham, Switherland, 2024; pp. 320–335. [Google Scholar]
- Cao, L.; Wu, H.; Liu, Y. Value of CT spectral imaging in the differential diagnosis of sarcoidosis and Hodgkin’s lymphoma based on mediastinal enlarged lymph node: A STARD compliant article. Medicine 2022, 101, e31502. [Google Scholar] [CrossRef]
- LaValley, M.P. Logistic regression. Circulation 2008, 117, 2395–2399. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Y.; Ying, L. Decision tree methods: Applications for classification and prediction. Shanghai Arch. Psychiatry 2015, 27, 130. [Google Scholar]
- Abdullah, D.M.; Abdulazeez, A.M. Machine learning applications based on SVM classification a review. Qubahan Acad. J. 2021, 1, 81–90. [Google Scholar] [CrossRef]
- Guo, G.; Wang, H.; Bell, D.; Bi, Y.; Greer, K. KNN model-based approach in classification. In Proceedings of the On The Move to Meaningful Internet Systems 2003: CoopIS, DOA, and ODBASE: OTM Confederated International Conferences, CoopIS, DOA, and ODBASE 2003, Catania, Sicily, Italy, 3–7 November 2003; Proceedings. Springer: Berlin/Heidelberg, Germany, 2003; pp. 986–996. [Google Scholar]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Zounemat-Kermani, M.; Batelaan, O.; Fadaee, M.; Hinkelmann, R. Ensemble machine learning paradigms in hydrology: A review. J. Hydrol. 2021, 598, 126266. [Google Scholar] [CrossRef]
- Poudel, S. A study of disease diagnosis using machine learning. Med. Sci. Forum 2022, 10, 8. [Google Scholar]
- Hurvitz, N.; Azmanov, H.; Kesler, A.; Ilan, Y. Establishing a second-generation artificial intelligence-based system for improving diagnosis, treatment, and monitoring of patients with rare diseases. Eur. J. Hum. Genet. 2021, 29, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Huang, H.; Zhang, S.; Zhang, D.; Shi, Q.; Liu, J.; Guo, J. Artificial intelligence in pancreatic cancer. Theranostics 2022, 12, 6931. [Google Scholar] [CrossRef] [PubMed]
- Eurelings, L.E.; Miedema, J.R.; Dalm, V.A.; van Daele, P.L.; van Hagen, P.M.; van Laar, J.A.; Dik, W.A. Sensitivity and specificity of serum soluble interleukin-2 receptor for diagnosing sarcoidosis in a population of patients suspected of sarcoidosis. PLoS ONE 2019, 14, e0223897. [Google Scholar] [CrossRef]
- Sánchez Fernández, I.; Yang, E.; Calvachi, P.; Amengual-Gual, M.; Wu, J.Y.; Krueger, D.; Northrup, H.; Bebin, M.E.; Sahin, M.; Yu, K.H.; et al. Deep learning in rare disease. Detection of tubers in tuberous sclerosis complex. PLoS ONE 2020, 15, e0232376. [Google Scholar] [CrossRef]
- Osipov, N.; Kudryavtsev, I.; Spelnikov, D.; Rubinstein, A.; Belyaeva, E.; Kulpina, A.; Kudlay, D.; Starshinova, A. Differential diagnosis of tuberculosis and sarcoidosis by immunological features using machine learning. Diagnostics 2024, 14, 2188. [Google Scholar] [CrossRef]
- Dai, Q.; Sherif, A.A.; Jin, C.; Chen, Y.; Cai, P.; Li, P. Machine learning predicting mortality in sarcoidosis patients admitted for acute heart failure. Cardiovasc. Digit. Health J. 2022, 3, 297–304. [Google Scholar] [CrossRef]
- van der Sar, I.G.; van Jaarsveld, N.; Spiekerman, I.A.; Toxopeus, F.J.; Langens, Q.L.; Wijsenbeek, M.S.; Dauwels, J.; Moor, C.C. Evaluation of different classification methods using electronic nose data to diagnose sarcoidosis. J. Breath Res. 2023, 17, 047104. [Google Scholar] [CrossRef]
- Eckstein, J.; Moghadasi, N.; Körperich, H.; Akkuzu, R.; Sciacca, V.; Sohns, C.; Sommer, P.; Berg, J.; Paluszkiewicz, J.; Burchert, W.; et al. Machine-learning-based diagnostics of cardiac sarcoidosis using multi-chamber wall motion analyses. Diagnostics 2023, 13, 2426. [Google Scholar] [CrossRef]
- Bobbio, E.; Eldhagen, P.; Polte, C.L.; Hjalmarsson, C.; Karason, K.; Rawshani, A.; Darlington, P.; Kullberg, S.; Sörensson, P.; Bergh, N.; et al. Clinical Outcomes and Predictors of Long-Term Survival in Patients With and Without Previously Known Extracardiac Sarcoidosis Using Machine Learning: A Swedish Multicenter Study. J. Am. Heart Assoc. 2023, 12, e029481. [Google Scholar] [CrossRef] [PubMed]
- Attar, H.; Solyman, A.; Deif, M.A.; Hafez, M.; Kasem, H.M.; Mohamed, A.E.F. Machine Learning Model Based on Gary-Level Co-occurrence Matrix for Chest Sarcoidosis Diagnosis. In Proceedings of the 2023 2nd International Engineering Conference on Electrical, Energy, and Artificial Intelligence (EICEEAI), Zarqa, Jordan, 27–28 December 2023; pp. 1–8. [Google Scholar]
- Mushari, N.A.; Soultanidis, G.; Duff, L.; Trivieri, M.G.; Fayad, Z.A.; Robson, P.; Tsoumpas, C. An assessment of PET and CMR radiomic features for detection of cardiac sarcoidosis. Front. Nucl. Med. 2024, 4, 1324698. [Google Scholar] [CrossRef]
- Eurelings, L.E.M.; Miedema, J.R.; Dalm, V.A.S.H.; van Daele, P.L.A.; van Hagen, P.M.; van Laar, J.A.M.; Dik, W.A. Dataset of Sarcoidosis and Non-Sarcoidosis Patients. 2019. Available online: https://figshare.com/articles/dataset/Sensitivity_and_specificity_of_serum_soluble_interleukin-2_receptor_for_diagnosing_sarcoidosis_in_a_population_of_patients_suspected_of_sarcoidosis/9996461?file=18029612 (accessed on 20 October 2024).
- Templ, M. Enhancing precision in large-scale data analysis: An innovative robust imputation algorithm for managing outliers and missing values. Mathematics 2023, 11, 2729. [Google Scholar] [CrossRef]
- Aguinis, H.; Gottfredson, R.K.; Joo, H. Best-practice recommendations for defining, identifying, and handling outliers. Organ. Res. Methods 2013, 16, 270–301. [Google Scholar] [CrossRef]
- McHugh, M.L. The chi-square test of independence. Biochem. Medica 2013, 23, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Vergara, J.R.; Estévez, P.A. A review of feature selection methods based on mutual information. Neural Comput. Appl. 2014, 24, 175–186. [Google Scholar] [CrossRef]
- Rückstieß, T.; Osendorfer, C.; Van Der Smagt, P. Sequential feature selection for classification. In Proceedings of the AI 2011: Advances in Artificial Intelligence: 24th Australasian Joint Conference, Perth, Australia, 5–8 December 2011; Proceedings 24. Springer: Berlin/Heidelberg, Germany, 2011; pp. 132–141. [Google Scholar]
- Jaiswal, J.K.; Samikannu, R. Application of random forest algorithm on feature subset selection and classification and regression. In Proceedings of the 2017 World Congress on Computing and Communication Technologies (WCCCT), Tiruchirappalli, India, 2–4 February 2017; pp. 65–68. [Google Scholar]
| Sl. No. | Attribute Name | Role | Type |
|---|---|---|---|
| 1 | Sex | Feature | Categorical |
| 2 | Diagnosis | Target | Categorical |
| 3 | Histology | Feature | Categorical |
| 4 | X-Thorax | Feature | Categorical |
| 5 | CT | Feature | Categorical |
| 6 | SPECT | Feature | Categorical |
| 7 | SRS | Feature | Categorical |
| 8 | PET | Feature | Categorical |
| 9 | Lung Involvement | Feature | Categorical |
| 10 | Eye Involvement | Feature | Categorical |
| 11 | Neurological Involvement | Feature | Categorical |
| 12 | Skin Involvement | Feature | Categorical |
| 13 | SIL 2R | Feature | Continuous |
| 14 | ACE | Feature | Continuous |
| Sl. No. | Attribute Name | Missing Values | Missing Type |
|---|---|---|---|
| 1 | Sex | 0 | NA |
| 2 | Diagnosis | 0 | NA |
| 3 | Histology | 83 | MAR |
| 4 | X-Thorax | 33 | MAR |
| 5 | CT | 75 | MAR |
| 6 | SPECT | 157 | MAR |
| 7 | SRS | 188 | MCAR |
| 8 | PET | 150 | MAR |
| 9 | Lung Involvement | 86 | MCAR |
| 10 | Eye Involvement | 86 | MCAR |
| 11 | Neurological Involvement | 86 | MCAR |
| 12 | Skin Involvement | 86 | MCAR |
| 13 | SIL 2R | 0 | NA |
| 14 | ACE | 0 | NA |
| PC | EV | Top Contributing Features |
|---|---|---|
| PC1 | 0.3608 | Histology, CT, SIL 2R, X-thorax, lung involvement, ACE, eye involvement |
| PC2 | 0.1313 | Neurological involvement, sex, skin involvement, eye involvement, SIL 2R, ACE, Histology |
| PC3 | 0.1209 | Eye involvement, ACE, X-thorax, lung involvement, skin involvement, CT, SIL 2R |
| PC4 | 0.0949 | Neurological involvement, skin involvement, sex, X-thorax, eye involvement, lung involvement, SIL 2R |
| PC5 | 0.0778 | Sex, skin involvement, lung involvement, neurological involvement, X-thorax, ACE, eye involvement |
| PC6 | 0.0663 | ACE, eye involvement, X-thorax, CT, Histology, skin involvement, neurological involvement |
| PC7 | 0.0540 | Lung involvement, eye involvement, Histology, CT, X-thorax, ACE, neurological involvement |
| Sl. No. | Feature | Chi2 Score | RFE Ranking |
|---|---|---|---|
| 1 | Histology | 85.386139 | 1 |
| 2 | CT | 64.842416 | 2 |
| 3 | lung involvement | 39.032516 | 3 |
| 4 | eye involvement | 19.967687 | 4 |
| 5 | skin involvement | 13.940594 | 5 |
| 6 | X-thorax | 86.750969 | 6 |
| 7 | neurological involvement | 5.227723 | 7 |
| 8 | sex | 0.637596 | 8 |
| 9 | ACE | 677.198346 | 9 |
| 10 | SIL 2R | 158,436.830482 | 10 |
| Sl. No. | Feature | Importance |
|---|---|---|
| 1 | Histology | 0.432447 |
| 2 | CT | 0.251656 |
| 3 | SIL 2R | 0.128611 |
| 4 | X-thorax | 0.094628 |
| 5 | lung involvement | 0.045384 |
| 6 | eye involvement | 0.022870 |
| 7 | ACE | 0.018926 |
| 8 | skin involvement | 0.003525 |
| 9 | sex | 0.001856 |
| 10 | neurological involvement | 0.000097 |
| Sl. No. | Feature | MI Score |
|---|---|---|
| 1 | Histology | 0.622457 |
| 2 | CT | 0.468798 |
| 3 | SIL 2R | 0.332907 |
| 4 | X-thorax | 0.262964 |
| 5 | lung involvement | 0.160474 |
| 6 | eye involvement | 0.115752 |
| 7 | skin involvement | 0.074443 |
| 8 | neurological involvement | 0.041284 |
| 9 | ACE | 0.000544 |
| 10 | sex | 0.000000 |
| Sl. No. | Feature |
|---|---|
| 1 | sex |
| 2 | Histology |
| 3 | CT |
| 4 | lung involvement |
| 5 | neurological involvement |
| Model | Accuracy | Sensitivity | Specificity | Confusion Matrix | |
|---|---|---|---|---|---|
| 1 | Logistic Regression | 0.9894 | 0.9901 | 0.9886 | [[87, 1], [1, 100]] |
| 2 | Decision Tree Classifier | 0.9947 | 1.0000 | 0.9886 | [[87, 1], [0, 101]] |
| 3 | SVC | 0.9947 | 1.0000 | 0.9886 | [[87, 1], [0, 101]] |
| 4 | K-Neighbors Classifier | 0.9788 | 0.9901 | 0.9659 | [[85, 3], [1, 100]] |
| Model | Accuracy | Sensitivity | Specificity | Confusion Matrix | |
|---|---|---|---|---|---|
| 1 | Model Combination 1 | 0.9947 | 1.0000 | 0.9886 | [[87, 1], [0, 101]] |
| 2 | Model Combination 2 | 0.9735 | 0.9703 | 0.9773 | [[86, 2], [3, 98]] |
| 3 | Model Combination 3 | 0.9947 | 1.0000 | 0.9886 | [[87, 1], [0, 101]] |
| 4 | Model Combination 4 | 0.9947 | 1.0000 | 0.9886 | [[87, 1], [0, 101]] |
| 5 | Random Forest Classifier | 0.9947 | 1.0000 | 0.9886 | [[87, 1], [0, 101]] |
| 6 | AdaBoost Classifier | 0.9841 | 0.9802 | 0.9886 | [[87, 1], [2, 99]] |
| 7 | XGB Classifier | 0.9947 | 0.9901 | 1.0000 | [[88, 0], [1, 100]] |
| 8 | Gradient Boosting Classifier | 0.9947 | 1.0000 | 0.9886 | [[87, 1], [0, 101]] |
| No. | Model | Accuracy | Sensitivity | Specificity | Confusion Matrix |
|---|---|---|---|---|---|
| 1 | Logistic Regression | 0.8556 | 0.8784 | 0.8279 | [[101, 21], [18, 130]] |
| 2 | Decision Tree Classifier | 0.8593 | 0.8716 | 0.8443 | [[103, 19], [19, 129]] |
| 3 | SVC | 0.8444 | 0.8514 | 0.8361 | [[102, 20], [22, 126]] |
| 4 | K-Neighbors Classifier | 0.8519 | 0.9122 | 0.7787 | [[95, 27], [13, 135]] |
| Model | Accuracy | Sensitivity | Specificity | Confusion Matrix | |
|---|---|---|---|---|---|
| 1 | Model Combination 1 | 0.8519 | 0.8919 | 0.8033 | [[98, 24], [16, 132]] |
| 2 | Model Combination 2 | 0.8111 | 0.8581 | 0.7541 | [[92, 30], [21, 127]] |
| 3 | Model Combination 3 | 0.8407 | 0.8919 | 0.7787 | [[95, 27], [16, 132]] |
| 4 | Model Combination 4 | 0.8074 | 0.8514 | 0.7541 | [[92, 30], [22, 126]] |
| 5 | Random Forest Classifier | 0.8444 | 0.8784 | 0.8033 | [[98, 24], [18, 130]] |
| 6 | AdaBoost Classifier | 0.8185 | 0.8446 | 0.7869 | [[96, 26], [23, 125]] |
| 7 | XGB Classifier | 0.8444 | 0.8851 | 0.7951 | [[97, 25], [17, 131]] |
| 8 | Gradient Boosting Classifier | 0.8481 | 0.8649 | 0.8279 | [[101, 21], [20, 128]] |
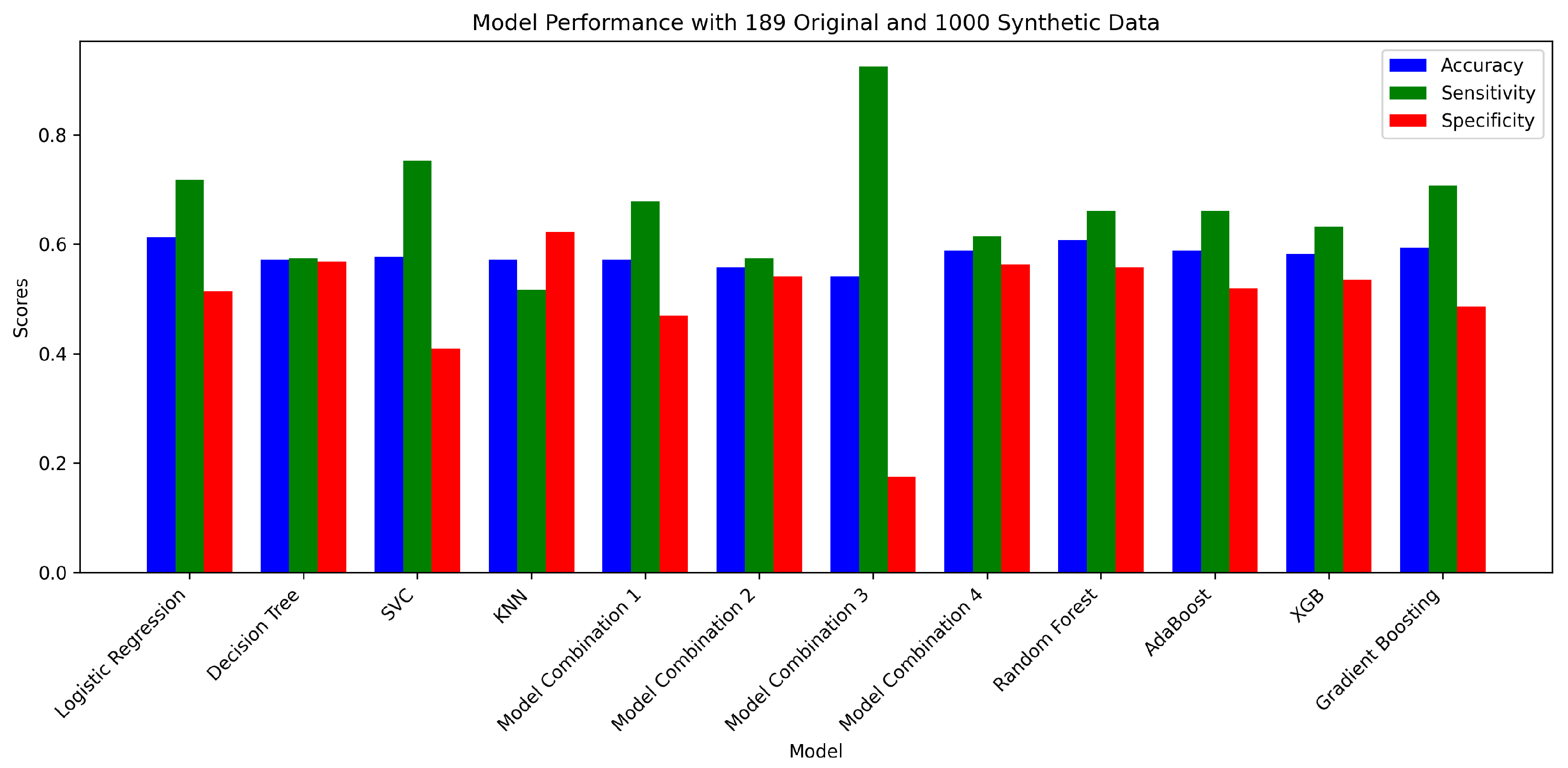
| No. | Model | Accuracy | Sensitivity | Specificity | Confusion Matrix |
|---|---|---|---|---|---|
| 1 | Logistic Regression | 0.6134 | 0.7184 | 0.5137 | [[94, 89], [49, 125]] |
| 2 | Decision Tree Classifier | 0.5714 | 0.5747 | 0.5683 | [[104, 79], [74, 100]] |
| 3 | SVC | 0.5770 | 0.7529 | 0.4098 | [[75, 108], [43, 131]] |
| 4 | K-Neighbors Classifier | 0.5714 | 0.5172 | 0.6230 | [[114, 69], [84, 90]] |
| Model | Accuracy | Sensitivity | Specificity | Confusion Matrix | |
|---|---|---|---|---|---|
| 1 | Model Combination 1 | 0.5714 | 0.6782 | 0.4699 | [[86, 97], [56, 118]] |
| 2 | Model Combination 2 | 0.5574 | 0.5747 | 0.5410 | [[99, 84], [74, 100]] |
| 3 | Model Combination 3 | 0.5406 | 0.9253 | 0.1749 | [[32, 151], [13, 161]] |
| 4 | Model Combination 4 | 0.5882 | 0.6149 | 0.5628 | [[103, 80], [67, 107]] |
| 5 | Random Forest Classifier | 0.6078 | 0.6609 | 0.5574 | [[102, 81], [59, 115]] |
| 6 | AdaBoost Classifier | 0.5882 | 0.6609 | 0.5191 | [[95, 88], [59, 115]] |
| 7 | XGB Classifier | 0.5826 | 0.6322 | 0.5355 | [[98, 85], [64, 110]] |
| 8 | Gradient Boosting Classifier | 0.5938 | 0.7069 | 0.4863 | [[89, 94], [51, 123]] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, S.; Abubakar, A.A.; Shetty, R.; Bijur, G.; Shetty, N.K.; Kumar, A.P. Diagnosis of Sarcoidosis Through Supervised Ensemble Method and GenAI-Based Data Augmentation: An Intelligent Diagnostic Tool. Appl. Sci. 2025, 15, 12213. https://doi.org/10.3390/app152212213
Rai S, Abubakar AA, Shetty R, Bijur G, Shetty NK, Kumar AP. Diagnosis of Sarcoidosis Through Supervised Ensemble Method and GenAI-Based Data Augmentation: An Intelligent Diagnostic Tool. Applied Sciences. 2025; 15(22):12213. https://doi.org/10.3390/app152212213
Chicago/Turabian StyleRai, Shwetha, Adam Azman Abubakar, Roopashri Shetty, Gururaj Bijur, Nakul K. Shetty, and Archana Praveen Kumar. 2025. "Diagnosis of Sarcoidosis Through Supervised Ensemble Method and GenAI-Based Data Augmentation: An Intelligent Diagnostic Tool" Applied Sciences 15, no. 22: 12213. https://doi.org/10.3390/app152212213
APA StyleRai, S., Abubakar, A. A., Shetty, R., Bijur, G., Shetty, N. K., & Kumar, A. P. (2025). Diagnosis of Sarcoidosis Through Supervised Ensemble Method and GenAI-Based Data Augmentation: An Intelligent Diagnostic Tool. Applied Sciences, 15(22), 12213. https://doi.org/10.3390/app152212213






