Comparative Evaluation of Cow and Goat Milk Samples Utilizing Non-Destructive Techniques and Chemometric Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Lyophilization of the Milk Samples
2.2. Color Evaluation, Microscopic Image Analysis, and Statistical Assessment
2.3. ATR-FTIR Spectroscopy of the Lyophilized Milk Samples
2.4. Univariate Statistical Analysis
2.5. Chemometrics and Multivariate Statistical Analysis
3. Results and Discussion
3.1. Nutritional Information and Color Parameters of Milk Samples
3.2. Assessment of Milk Samples Using Texture Analysis of the Microscopic Images
3.3. ATR-FTIR Spectra Evaluation of Milk Samples
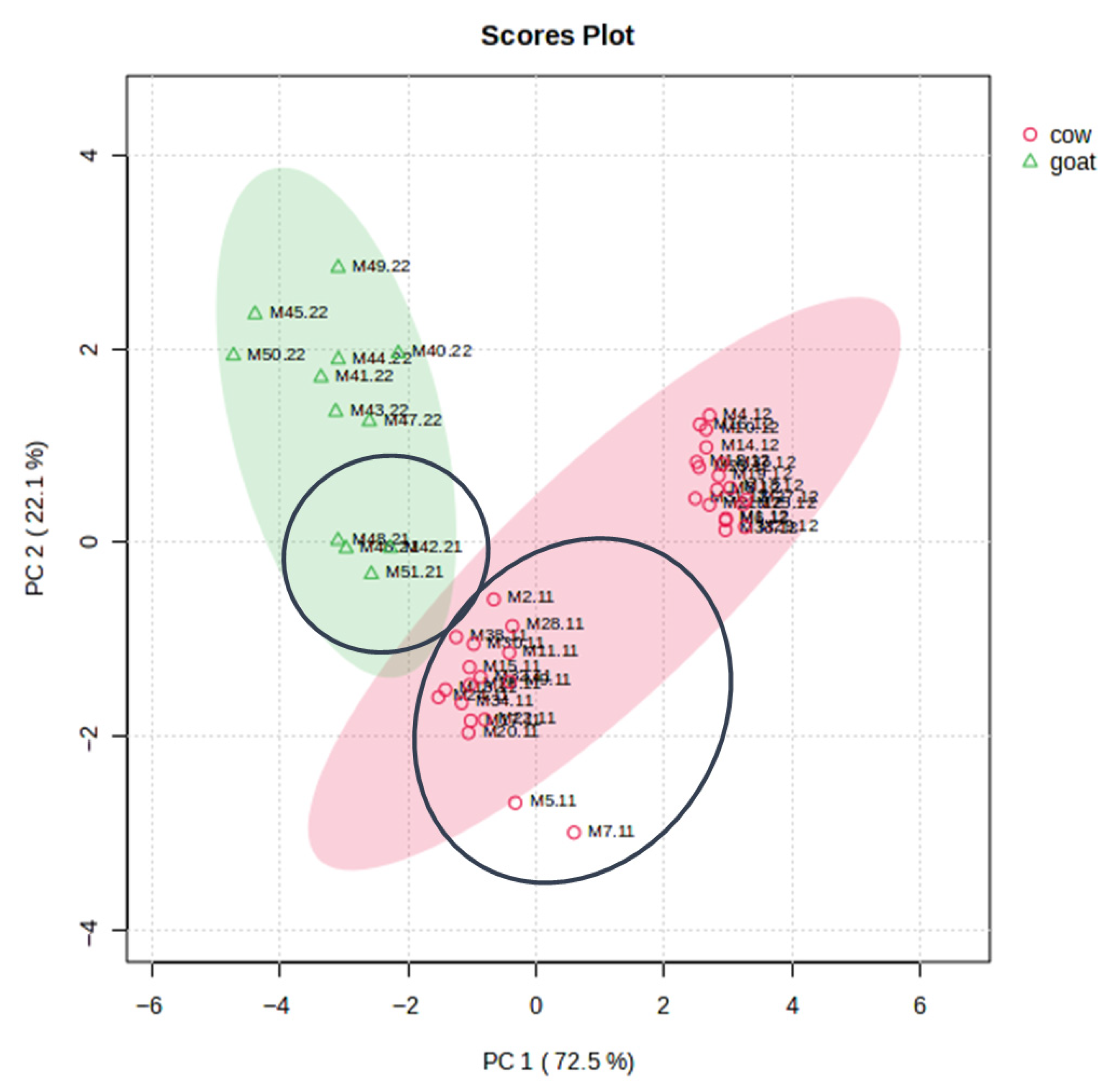
3.4. Multivariate Statistical Analysis and Marker Validation
3.5. Marker Validation
3.5.1. Comparative Analysis of Cow and Goat Milk
3.5.2. Comparative Analysis of Cow Whole and Goat Whole Milk
3.5.3. Comparative Analysis of Cow Light and Goat Light Milk
- (i)
- 1396–1400 cm−1 (AUC = 1.000)—CH3 bending modes, potentially linked to residual phospholipids
- (ii)
- 1440–1450 cm−1 (AUC = 0.99)—CH2 from saturated fatty acids
- (iii)
- 2854 cm−1 (AUC = 0.99)—CH2 symmetric stretching from lipid chains
- (iv)
- 1064, 1382, 1638–1645, 1741–1745, and 777 cm−1—AUCs ranging from 0.79 to 0.96
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanjulián, L.; Fernández-Rico, S.; González-Rodríguez, N.; Cepeda, A.; Miranda, J.M.; Fente, C.; Lamas, A.; Regal, P. The Role of Dairy in Human Nutrition: Myths and Realities. Nutrients 2025, 17, 646. [Google Scholar] [CrossRef]
- Shawky, E.; Nahar, L.; Nassief, S.M.; Sarker, S.D.; Ibrahim, R.S. Dairy Products Authentication with Biomarkers: A Comprehensive Critical Review. Trends Food Sci. Technol. 2024, 147, 104445. [Google Scholar] [CrossRef]
- Mozart, G.G.; Köptcke, F.B.N.; Pinto, L.A.; Moebus, V.F.; Tamy, W.P.; Aronovich, M.; Keller, L.A.M. Enhancement of Dairy Cow Milk Quality with Probiotic and Inorganic Selenium Supplementation. Dairy 2024, 5, 336–345. [Google Scholar] [CrossRef]
- Organization for Economic Co-operation and Development; Food and Agriculture Organization of the United Nations. OECD-FAO Agricultural Outlook 2019–2028; OECD-FAO Agricultural Outlook; OECD: Paris, France, 2019; ISBN 978-92-64-31245-6. [Google Scholar]
- Anusha Siddiqui, S.; Mahmood Salman, S.H.; Ali Redha, A.; Zannou, O.; Chabi, I.B.; Oussou, K.F.; Bhowmik, S.; Nirmal, N.P.; Maqsood, S. Physicochemical and Nutritional Properties of Different Non-Bovine Milk and Dairy Products: A Review. Int. Dairy J. 2024, 148, 105790. [Google Scholar] [CrossRef]
- Eurostat. (2023). Milk and milk product production statistics. In Agricultural production—Livestock and meat. European Commission. Available online: https://ec.europa.eu/eurostat (accessed on 1 June 2025).
- Pappa, E.C.; Kondyli, E.; Sotirakoglou, K.; Bosnea, L.; Mataragas, M.; Allouche, L.; Tsiplakou, E.; Pappas, A.C. Farmers Profile and Characterization of Sheep and Goat Dairy Chain in Northwestern Greece. Sustainability 2021, 13, 833. [Google Scholar] [CrossRef]
- Wang, L.; Wu, T.; Zhang, Y.; Yang, K.; He, Y.; Deng, K.; Liang, C.; Gu, Y. Comparative Studies on the Nutritional and Physicochemical Properties of Yoghurts from Cows’, Goats’, and Camels’ Milk Powder. Int. Dairy J. 2023, 138, 105542. [Google Scholar] [CrossRef]
- Prosser, C.G. Compositional and Functional Characteristics of Goat Milk and Relevance as a Base for Infant Formula. J. Food Sci. 2021, 86, 257–265. [Google Scholar] [CrossRef]
- Turkmen, N. The Nutritional Value and Health Benefits of Goat Milk Components. In Nutrients in Dairy and their Implications on Health and Disease; Elsevier: Amsterdam, The Netherlands, 2017; pp. 441–449. ISBN 978-0-12-809762-5. [Google Scholar]
- Nayik, G.A.; Jagdale, Y.D.; Gaikwad, S.A.; Devkatte, A.N.; Dar, A.H.; Ansari, M.J. Nutritional Profile, Processing and Potential Products: A Comparative Review of Goat Milk. Dairy 2022, 3, 622–647. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Mao, X. Composition and Interfacial Properties Play Key Roles in Different Lipid Digestion between Goat and Cow Milk Fat Globules in Vitro. Food Chem. 2022, 374, 131538. [Google Scholar] [CrossRef]
- Visoka, Y.; Majadi, M.; Kovacs, Z.; Gecaj, R.M. Utilizing Near-Infrared Spectroscopy for Discriminant Analysis of Goat Milk Composition across Diverse Breeds and Lactation Seasons. Biol. Life Sci. Forum 2023, 26, 64. [Google Scholar] [CrossRef]
- Kailasapathy, K. Chemical Composition, Physical, and Functional Properties of Milk and Milk Ingredients. In Dairy Processing and Quality Assurance; Chandan, R.C., Kilara, A., Shah, N.P., Eds.; Wiley: Hoboken, NJ, USA, 2015; pp. 77–105. ISBN 978-1-118-81031-6. [Google Scholar]
- Pereira, C.G.; Luiz, L.C.; Bell, M.J.V.; Anjos, V. Near And Mid Infrared Spectroscopy To Assess Milk Products Quality: A Review Of Recent Applications. J. Dairy Res. Technol. 2020, 3, 1–10. [Google Scholar] [CrossRef]
- Balan, B.; Dhaulaniya, A.S.; Jamwal, R.; Amit; Sodhi, K.K.; Kelly, S.; Cannavan, A.; Singh, D.K. Application of Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FTIR) Spectroscopy Coupled with Chemometrics for Detection and Quantification of Formalin in Cow Milk. Vib. Spectrosc. 2020, 107, 103033. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Rahaman, M.M.; Yao, Y.; Ma, P.; Zhang, J.; Zhao, X.; Jiang, T.; Grzegorzek, M. A Comprehensive Review of Image Analysis Methods for Microorganism Counting: From Classical Image Processing to Deep Learning Approaches. Artif. Intell. Rev. 2022, 55, 2875–2944. [Google Scholar] [CrossRef] [PubMed]
- Boruczkowska, H.; Boruczkowski, T.; Bronkowska, M.; Prajzner, M.; Rytel, E. Comparison of Colour Measurement Methods in the Food Industry. Processes 2025, 13, 1268. [Google Scholar] [CrossRef]
- Kardas, M.; Rakuła, M.; Kołodziejczyk, A.; Staśkiewicz-Bartecka, W. Consumer Preferences, Sensory Evaluation, and Color Analysis of Beetroot and Tomato Juices: Implications for Product Development and Marketing in Health-Promoting Beverages. Foods 2024, 13, 4059. [Google Scholar] [CrossRef]
- Agiomavriti, A.-A.; Nikolopoulou, M.P.; Bartzanas, T.; Chorianopoulos, N.; Demestichas, K.; Gelasakis, A.I. Spectroscopy-Based Methods and Supervised Machine Learning Applications for Milk Chemical Analysis in Dairy Ruminants. Chemosensors 2024, 12, 263. [Google Scholar] [CrossRef]
- Kharbach, M.; Alaoui Mansouri, M.; Taabouz, M.; Yu, H. Current Application of Advancing Spectroscopy Techniques in Food Analysis: Data Handling with Chemometric Approaches. Foods 2023, 12, 2753. [Google Scholar] [CrossRef]
- Cirak, O.; Icyer, N.C.; Durak, M.Z. Rapid Detection of Adulteration of Milks from Different Species Using Fourier Transform Infrared Spectroscopy (FTIR). J. Dairy Res. 2018, 85, 222–225. [Google Scholar] [CrossRef]
- Vinciguerra, L.; Marcelo, M.; Motta, T.; Meneghini, L.; Bergold, A.; Ferrão, M. Chemometric Tools and FTIR-ATR Spectroscopy Applied in Milk Adulterated with Cheese Whey. Química Nova 2019, 42, 249–254. [Google Scholar] [CrossRef]
- Tsiaka, T.; Kritsi, E.; Bratakos, S.M.; Sotiroudis, G.; Petridi, P.; Savva, I.; Christodoulou, P.; Strati, I.F.; Zoumpoulakis, P.; Cavouras, D.; et al. Quality Assessment of Ground Coffee Samples from Greek Market Using Various Instrumental Analytical Methods, In Silico Studies and Chemometrics. Antioxidants 2023, 12, 1184. [Google Scholar] [CrossRef]
- Christodoulou, P.; Ladika, G.; Tsiantas, K.; Kritsi, E.; Tsiaka, T.; Cavouras, D.; Zoumpoulakis, P.; Sinanoglou, V.J. Quality Assessment of Greenhouse-Cultivated Cucumbers (Cucumis sativus) during Storage Using Instrumental and Image Analyses. Appl. Sci. 2024, 14, 8676. [Google Scholar] [CrossRef]
- Kritsi, E.; Ladika, G.; Stavropoulou, N.A.; Oikonomakou, M.; Ioannou, A.-G.; Christodoulou, P.; Konteles, S.J.; Cavouras, D.; Sinanoglou, V.J. Evaluation of the Quality Changes in Three Commercial Pastourma Samples during Refrigerated Storage Using Physicochemical, Microbiological, and Image Analyses Combined with Chemometrics. Foods 2024, 13, 1017. [Google Scholar] [CrossRef]
- Moatsou, G.; Park, Y.W. Goat Milk Products: Types of Products, Manufacturing Technology, Chemical Composition, and Marketing. In Handbook of Milk of Non-Bovine Mammals; Park, Y.W., Haenlein, G.F.W., Wendorff, W.L., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 84–150. ISBN 978-1-119-11027-9. [Google Scholar]
- Foroutan, A.; Guo, A.C.; Vazquez-Fresno, R.; Lipfert, M.; Zhang, L.; Zheng, J.; Badran, H.; Budinski, Z.; Mandal, R.; Ametaj, B.N.; et al. Chemical Composition of Commercial Cow’s Milk. J. Agric. Food Chem. 2019, 67, 4897–4914. [Google Scholar] [CrossRef]
- Felice, V.D.; Owens, R.A.; Kennedy, D.; Hogan, S.A.; Lane, J.A. Comparative Structural and Compositional Analyses of Cow, Buffalo, Goat and Sheep Cream. Foods 2021, 10, 2643. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Han, H.; Wang, T.; Li, H.; Qian, Y.; Zhu, M.; Jia, Q.; Qiu, J. Comparative Analysis of the Fatty Acid Profiles in Goat Milk during Different Lactation Periods and Their Interactions with Volatile Compounds and Metabolites. Food Chem. 2024, 460, 140427. [Google Scholar] [CrossRef]
- Roy, D.; Ye, A.; Moughan, P.J.; Singh, H. Composition, Structure, and Digestive Dynamics of Milk From Different Species—A Review. Front. Nutr. 2020, 7, 577759. [Google Scholar] [CrossRef]
- Muñoz-Salinas, F.; Andrade-Montemayor, H.M.; De La Torre-Carbot, K.; Duarte-Vázquez, M.Á.; Silva-Jarquin, J.C. Comparative Analysis of the Protein Composition of Goat Milk from French Alpine, Nubian, and Creole Breeds and Holstein Friesian Cow Milk: Implications for Early Infant Nutrition. Animals 2022, 12, 2236. [Google Scholar] [CrossRef]
- Chauhan, S.; Powar, P.; Mehra, R. A Review on Nutritional Advantages and Nutraceutical Properties of Cow and Goat Milk. Int. J. Appl. Res. 2021, 7, 101–105. [Google Scholar] [CrossRef]
- Milovanovic, B.; Djekic, I.; Miocinovic, J.; Djordjevic, V.; Lorenzo, J.M.; Barba, F.J.; Mörlein, D.; Tomasevic, I. What Is the Color of Milk and Dairy Products and How Is It Measured? Foods 2020, 9, 1629. [Google Scholar] [CrossRef]
- Chudy, S.; Bilska, A.; Kowalski, R.; Teichert, J. Colour of Milk and Milk Products in CIE L*a*b* Space. Med. Weter. 2020, 76, 77–81. [Google Scholar] [CrossRef]
- Cheong, F.C.; Xiao, K.; Grier, D.G. Technical Note: Characterizing Individual Milk Fat Globules with Holographic Video Microscopy. J. Dairy Sci. 2009, 92, 95–99. [Google Scholar] [CrossRef]
- Sinanoglou, V.J.; Tsiaka, T.; Aouant, K.; Mouka, E.; Ladika, G.; Kritsi, E.; Konteles, S.J.; Ioannou, A.-G.; Zoumpoulakis, P.; Strati, I.F.; et al. Quality Assessment of Banana Ripening Stages by Combining Analytical Methods and Image Analysis. Appl. Sci. 2023, 13, 3533. [Google Scholar] [CrossRef]
- Lu, N.; Wang, J.; Chen, Z.; Zhang, X.; Chen, C.; Wang, S. The Effect of Adding Phospholipids before Homogenization on the Properties of Milk Fat Globules. LWT 2021, 146, 111659. [Google Scholar] [CrossRef]
- Julmohammad, N.; Suhaini, I.K.M.; Govindasamy, T.; Tan, E.; Soloi, S.; Julmohamad, N.; Akanda, M.J.H. Quantification of Adulterant Residues in UHT Milk Products Using ATR-FTIR Spectroscopy Coupled with Multivariate Analysis. ARD 2024, 115, 1–13. [Google Scholar] [CrossRef]
- Nandiyanto, A.B.D.; Oktiani, R.; Ragadhita, R. How to Read and Interpret FTIR Spectroscope of Organic Material. Indones. J. Sci. Technol. 2019, 4, 97. [Google Scholar] [CrossRef]
- Nicolaou, N.; Xu, Y.; Goodacre, R. Fourier Transform Infrared Spectroscopy and Multivariate Analysis for the Detection and Quantification of Different Milk Species. J. Dairy Sci. 2010, 93, 5651–5660. [Google Scholar] [CrossRef]
- Stocco, G.; Dadousis, C.; Pazzola, M.; Vacca, G.M.; Dettori, M.L.; Mariani, E.; Cipolat-Gotet, C. Prediction Accuracies of Cheese-Making Traits Using Fourier-Transform Infrared Spectra in Goat Milk. Food Chem. 2023, 403, 134403. [Google Scholar] [CrossRef]
- Sinanoglou, V.J.; Cavouras, D.; Xenogiannopoulos, D.; Proestos, C.; Zoumpoulakis, P. Quality Assessment of Pork and Turkey Hams Using FT-IR Spectroscopy, Colorimetric, and Image Analysis. Foods 2018, 7, 152. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Christodoulou, P.; Kritsi, E.; Ladika, G.; Tsafou, P.; Tsiantas, K.; Tsiaka, T.; Zoumpoulakis, P.; Cavouras, D.; Sinanoglou, V.J. Estimation of Tomato Quality During Storage by Means of Image Analysis, Instrumental Analytical Methods, and Statistical Approaches. Appl. Sci. 2025, 15, 7936. [Google Scholar] [CrossRef]
- Conceição, D.; Gonçalves, B.-H.; Da Hora, F.; Faleiro, A.; Santos, L.; Ferrão, S. Use of FTIR-ATR Spectroscopy Combined with Multivariate Analysis as a Screening Tool to Identify Adulterants in Raw Milk. J. Braz. Chem. Soc. 2018, 30, 780–785. [Google Scholar] [CrossRef]
- Hong, T.; Yin, J.-Y.; Nie, S.-P.; Xie, M.-Y. Applications of Infrared Spectroscopy in Polysaccharide Structural Analysis: Progress, Challenge and Perspective. Food Chem. X 2021, 12, 100168. [Google Scholar] [CrossRef] [PubMed]
- Advanced Dairy Chemistry: Volume 1A: Proteins: Basic Aspects, 4th ed.; McSweeney, P.L.H., Fox, P.F., Eds.; Springer: Boston, MA, USA, 2013; ISBN 978-1-4614-4713-9. [Google Scholar]
- Ye, M.P.; Zhou, R.; Shi, Y.R.; Chen, H.C.; Du, Y. Effects of Heating on the Secondary Structure of Proteins in Milk Powders Using Mid-Infrared Spectroscopy. J. Dairy Sci. 2017, 100, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hong, T.; Li, Z.; Shen, G.; Gu, Y.; Han, J. A Comparison of Milk Fat Globule Membranes and Whey Proteomes: New Insight into Variation Nutrient Differences between Buffalo, Cow, Goat, and Yak. Food Chem. 2023, 429, 136845. [Google Scholar] [CrossRef] [PubMed]
- Tarapoulouzi, M.; Kokkinofta, R.; Theocharis, C.R. Chemometric Analysis Combined with FTIR Spectroscopy of Milk and Halloumi Cheese Samples According to Species’ Origin. Food Sci. Nutr. 2020, 8, 3262–3273. [Google Scholar] [CrossRef]
- Jaiswal, P.; Jha, S.N.; Borah, A.; Gautam, A.; Grewal, M.K.; Jindal, G. Detection and quantification of soymilk in cow–buffalo milk using Attenuated Total Reflectance Fourier Transform Infrared spectroscopy (ATR–FTIR). Food Chem. 2015, 168, 41–47. [Google Scholar] [CrossRef]

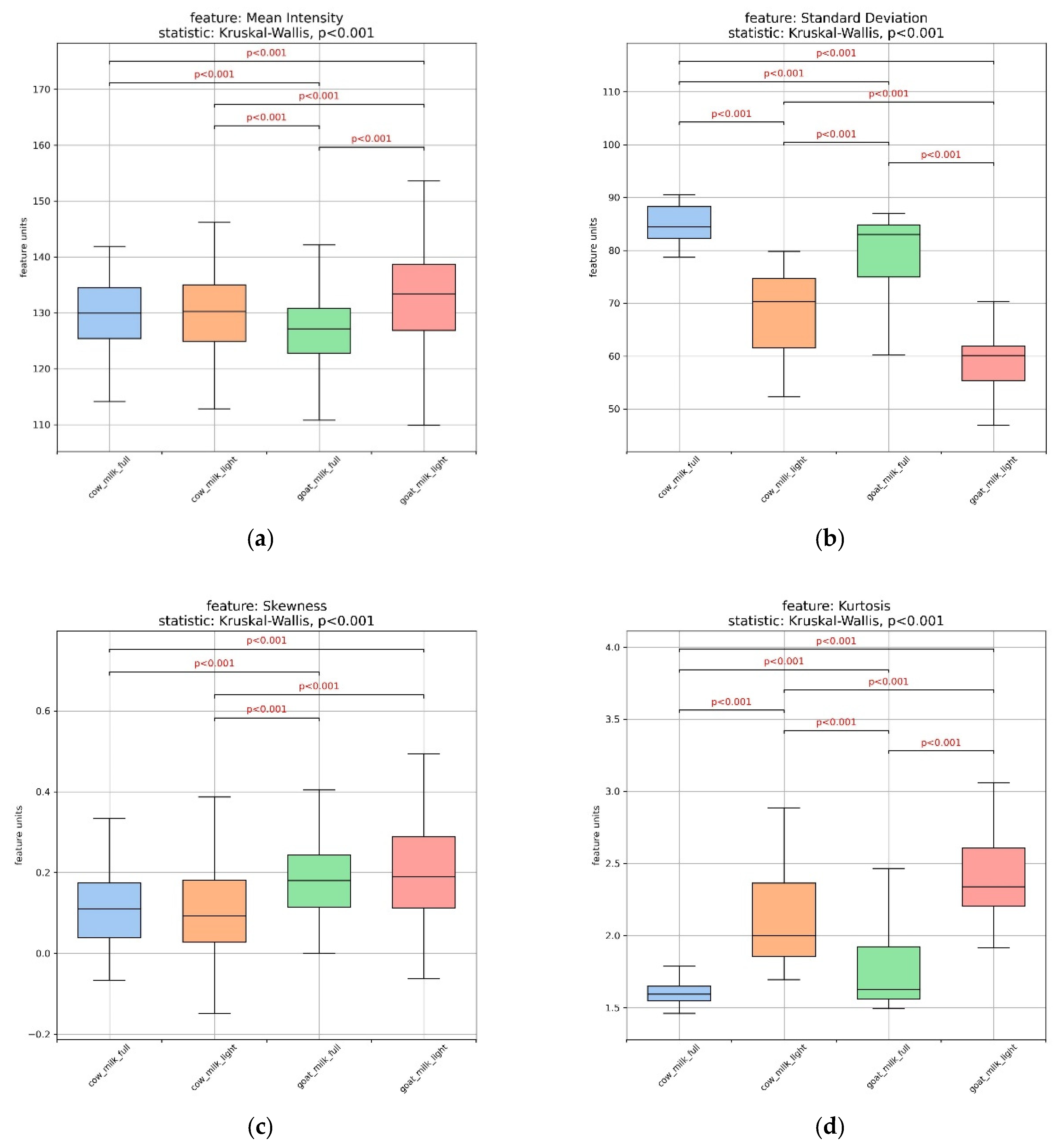

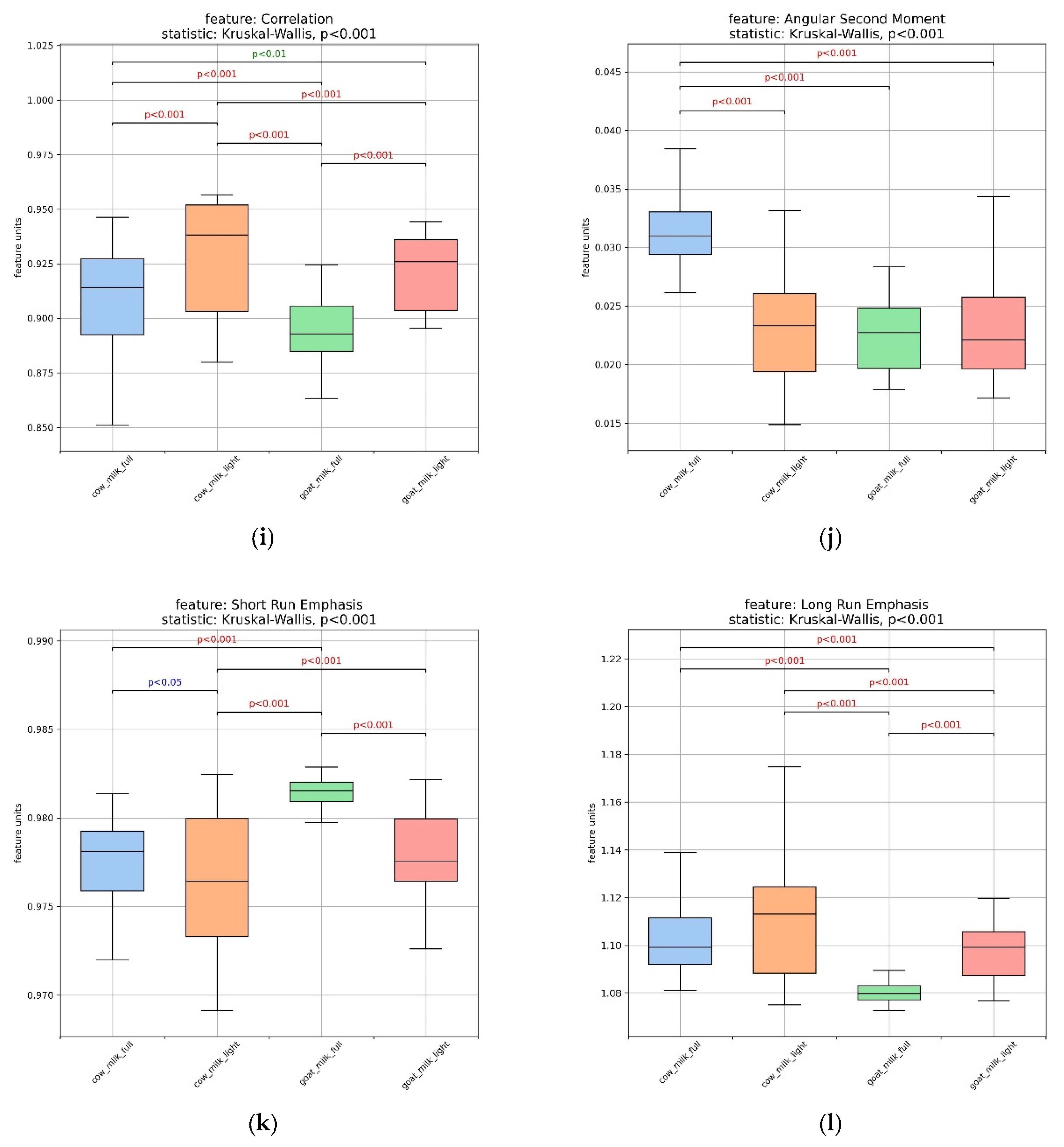
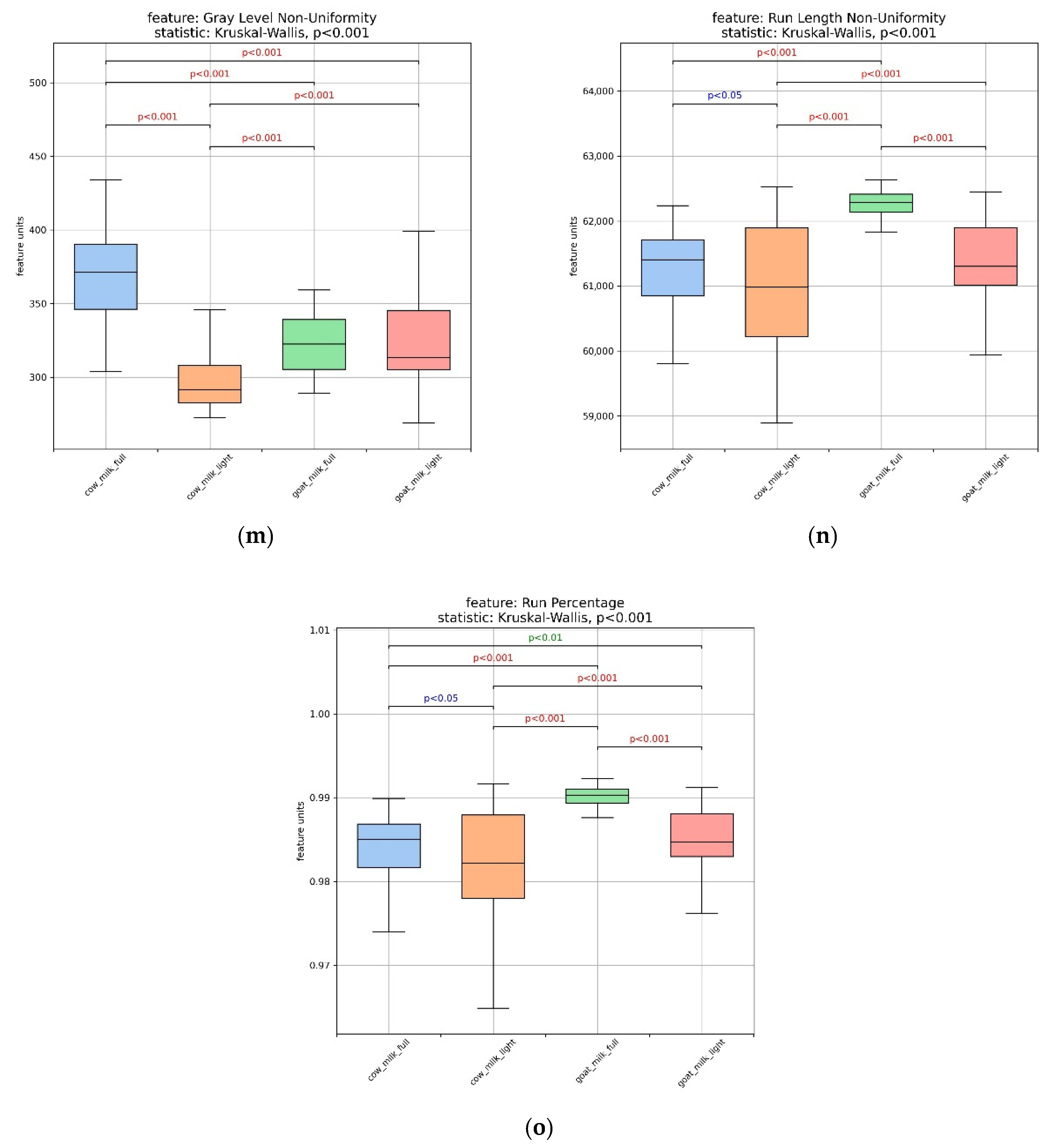
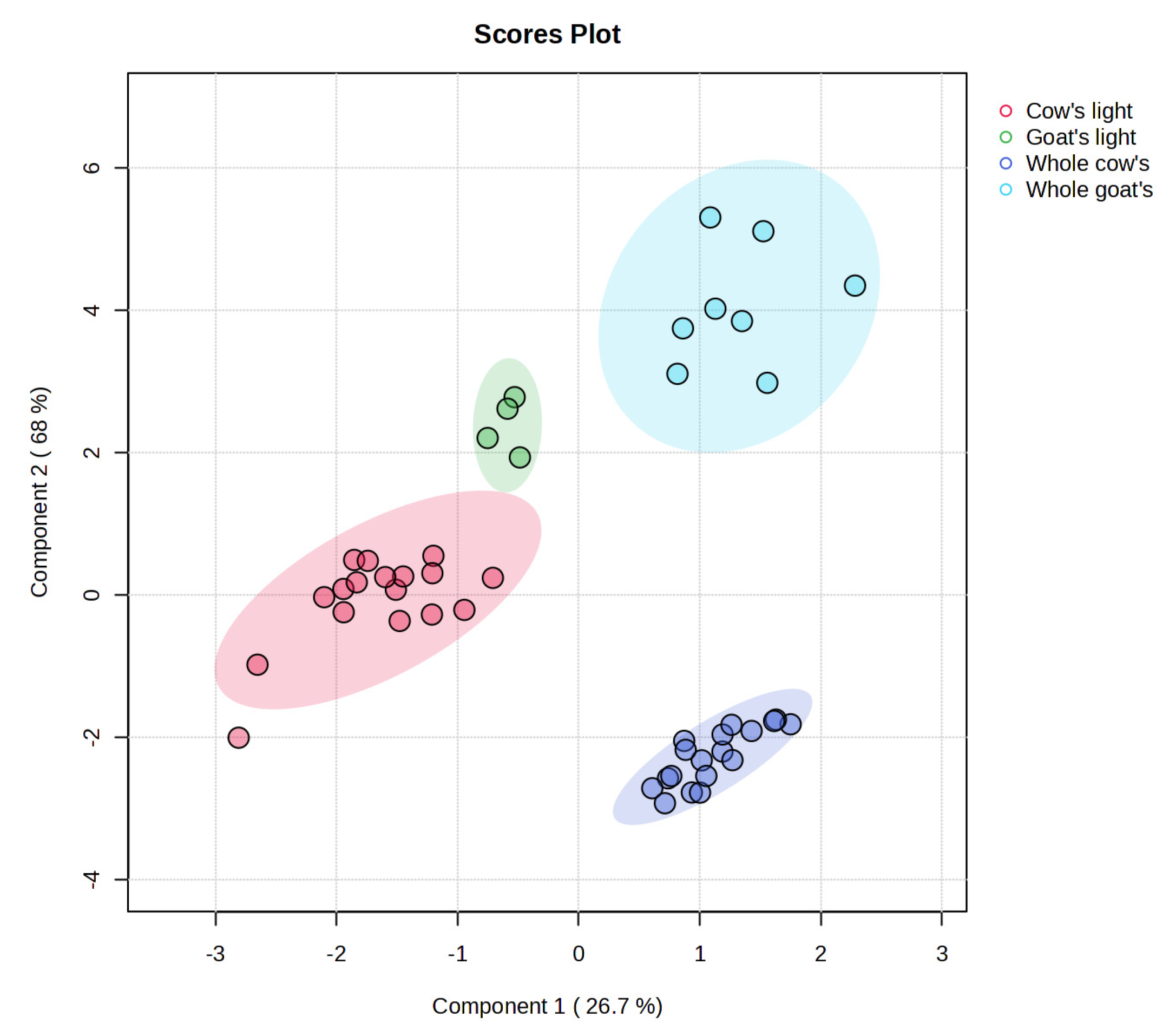
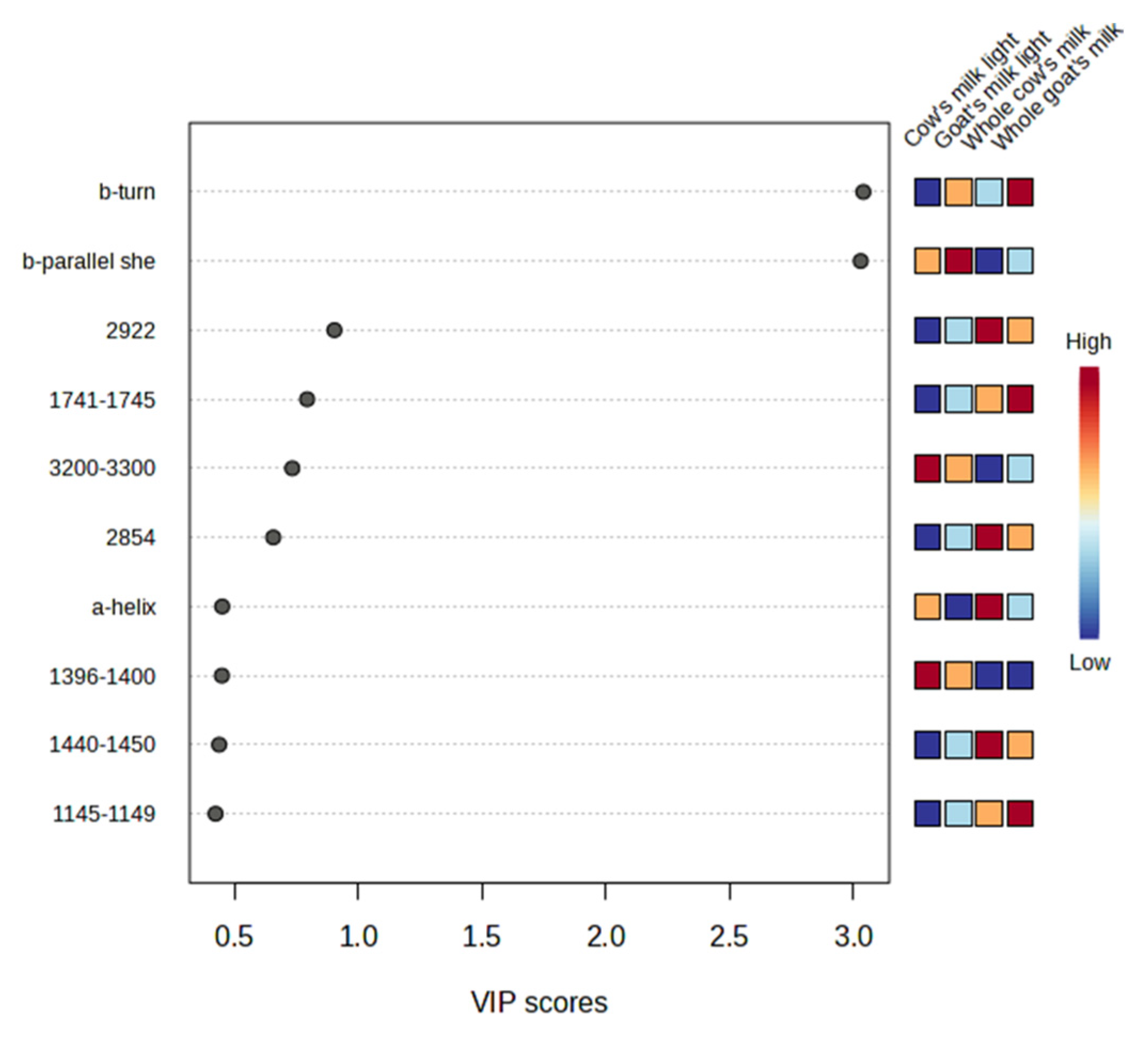
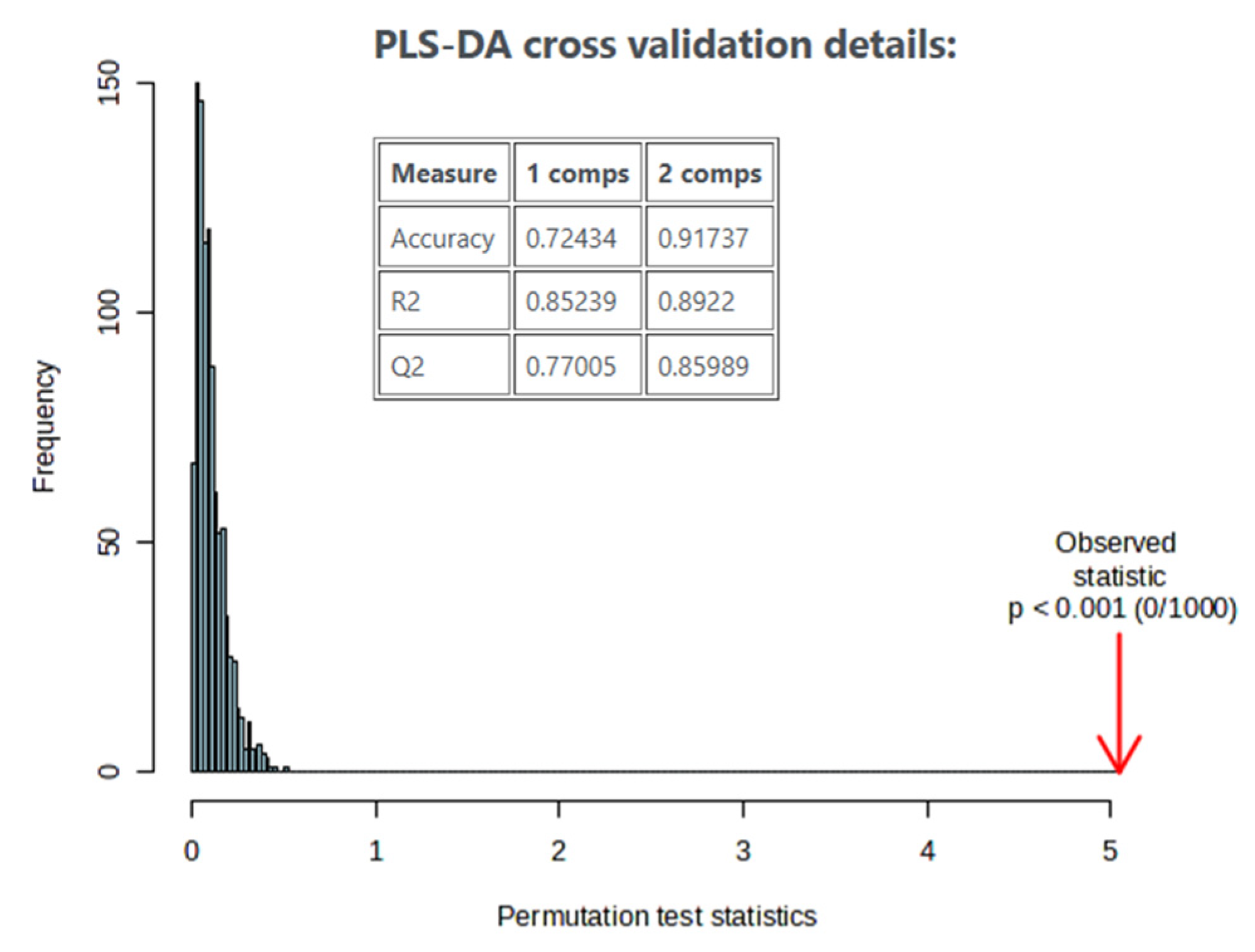
| Composition (g/100 g) | Cow’s Milk Light (17 Samples) | Whole Cow’s Milk (18 Samples) | Goat’s Milk Light (4 Samples) | Whole Goat’s Milk (8 Samples) |
|---|---|---|---|---|
| Total fat | 1.4 ± 0.3a * | 3.6 ± 0.1b | 1.7 ± 0.1a | 3.7 ± 0.2b |
| Proteins | 3.4 ± 0.1ac | 3.3 ± 0.1a | 3.7 ± 0.1b | 3.6 ± 0.1bc |
| Carbohydrates | 4.8 ± 0.1a | 4.7 ± 0.1ab | 4.7 ± 0.1ab | 4.5 ± 0.1b |
| Sugars | 4.8 ± 0.1a | 4.7 ± 0.1a | 4.4 ± 0.1b | 4.4 ± 0.1b |
| Salt | 0.11 ± 0.02a | 0.10 ± 0.02a | 0.08 ± 0.00b | 0.07 ± 0.01b |
| Color Parameters | Cow’s Milk Light (17 Samples) | Whole Cow’s Milk (18 Samples) | Goat’s Milk Light (4 Samples) | Whole Goat’s Milk (8 Samples) |
|---|---|---|---|---|
| L* (Lightness) | 75.67 ± 5.55a * | 78.91 ± 6.94a | 73.23 ± 7.89a | 74.24 ± 5.02a |
| a* (red–green) | −3.26 ± 0.61a | −2.50 ± 0.45b | −3.07 ± 0.48ab | −2.33 ± 0.36b |
| b* (yellow–blue) | 3.52 ± 0.38a | 5.33 ± 0.72b | 4.79 ± 0.26b | 4.92 ± 0.54b |
| h (hue angle) | 133.68 ± 11.72a | 115.46 ± 4.53b | 122.52 ± 2.53a | 115.29 ± 1.48b |
| Spectra Bands (cm−1) | Cow’s Milk Light (17 Samples) | Whole Cow’s Milk (18 Samples) | Goat’s Milk Light (4 Samples) | Whole Goat’s Milk (8 Samples) |
|---|---|---|---|---|
| 3200–3300 | 0.786 ± 0.018a | 0.651 ± 0.054b | 0.749 ± 0.034a | 0.676 ± 0.045b |
| 2922 | 0.412 ± 0.044a | 0.583 ± 0.035b | 0.474 ± 0.009c | 0.569 ± 0.019b |
| 2854 | 0.115 ± 0.017a | 0.213 ± 0.023b | 0.151 ± 0.008c | 0.195 ± 0.011b |
| 1741–1745 | 0.189 ± 0.024a | 0.308 ± 0.030b | 0.232 ± 0.011c | 0.315 ± 0.036b |
| 1638–1645 | 0.437 ± 0.016a | 0.405 ± 0.013b | 0.489 ± 0.011c | 0.441 ± 0.005a |
| 1535–1545 | 0.197 ± 0.014ab | 0.184 ± 0.010a | 0.209 ± 0.008b | 0.195 ± 0.006ab |
| 1440–1470 | 0.054 ± 0.009a | 0.097 ± 0.007b | 0.078 ± 0.003c | 0.092 ± 0.005b |
| 1396–1400 | 0.041 ± 0.005a | - | 0.021 ± 0.002b | - |
| 1370–1380 | 0.011 ± 0.001a | 0.029 ± 0.004b | 0.021 ± 0.001c | 0.049 ± 0.006d |
| 1280–1300 | 0.013 ± 0.001a | 0.012 ± 0.002a | 0.011 ± 0.002a | 0.011 ± 0.001a |
| 1242–1245 | 0.056 ± 0.005a | 0.051 ± 0.004a | 0.057 ± 0.003a | 0.056 ± 0.005a |
| 1145–1149 | 0.067 ± 0.008a | 0.091 ± 0.007b | 0.086 ± 0.004b | 0.092 ± 0.004b |
| 1064 | 0.025 ± 0.003a | 0.037 ± 0.005b | 0.026 ± 0.002a | 0.035 ± 0.002b |
| 1026–1028 | 0.557 ± 0.040a | 0.544 ± 0.028a | 0.453 ± 0.017b | 0.475 ± 0.018b |
| 891 | 0.041 ± 0.004a | 0.040 ± 0.005a | 0.045 ± 0.002a | 0.046 ± 0.004a |
| 777 | 0.045 ± 0.005a | 0.038 ± 0.003a | 0.041 ± 0.003a | 0.039 ± 0.002a |
| 700 | 0.030 ± 0.001a | 0.029 ± 0.002a | 0.030 ± 0.001a | 0.028 ± 0.001a |
| 538–542 | 0.045 ± 0.005a | 0.046 ± 0.004a | 0.053 ± 0.002a | 0.049 ± 0.002a |
| Secondary Structure of Proteins (%) | Cow’s Milk Light (17 Samples) | Whole Cow’s Milk (18 Samples) | Goat’s Milk Light (4 Samples) | Whole Goat’s Milk (8 Samples) |
|---|---|---|---|---|
| β-parallel sheet 1610–1642 cm−1 | 42.45 ± 0.63ac * | 37.43 ± 0.29b | 42.80 ± 0.35a | 41.54 ± 0.83c |
| random coil 1642–1650 cm−1 | 26.86 ± 0.37a | 29.17 ± 0.45b | 25.78 ± 0.46c | 24.57 ± 0.55d |
| α-helix 1650–1660 cm−1 | 22.02 ± 0.59a | 24.52 ± 0.40b | 20.25 ± 0.46c | 20.32 ± 1.15c |
| β-turn 1660–1680 cm−1 | 8.66 ± 0.88a | 8.88 ± 0.48a | 11.16 ± 0.30b | 13.56 ± 0.86c |
| Cow Milk vs. Goat Milk | ||
|---|---|---|
| Features | AUC | p-Values |
| β-turn | 1 | 6.44 × 10−17 |
| random coil | 1 | 3.07 × 10−10 |
| α-helix | 0.99 | 8.06 × 10−9 |
| 891 cm−1 | 0.87 | 4.28 × 10−6 |
| 1382 cm−1 | 0.83 | 4.96 × 10−6 |
| 538–542 cm−1 | 0.8 | 1.33 × 10−3 |
| 1242–1245 cm−1 | 0.78 | 9.89 × 10−3 |
| 1311–1313 cm−1 | 0.78 | 1.08 × 10−3 |
| 1145–1149 cm−1 | 0.74 | 5.64 × 10−3 |
| Cow Whole Milk vs. Goat Whole Milk | ||
|---|---|---|
| Features | AUC | p-Values |
| 1382 cm−1 | 1 | 2.41 × 10−7 |
| β-parallel sheet | 1 | 5.16 × 10−16 |
| random coil | 1 | 1.36 × 10−17 |
| α-helix | 1 | 5.33 × 10−13 |
| β-turn | 1 | 1.91 × 10−15 |
| 1242–1245 cm−1 | 0.81 | 6.30 × 10−3 |
| 1311–1313 cm−1 | 0.8 | 1.18 × 10−2 |
| 1440–1450 cm−1 | 0.73 | 4.63 × 10−2 |
| Cow Light Milk vs. Goat Light Milk | ||
|---|---|---|
| Features | AUC | p-Values |
| 891 cm−1 | 1 | 1.03 × 10−2 |
| 1026–1028 cm−1 | 1 | 3.81 × 10−12 |
| 1145–1149 cm−1 | 1 | 3.08 × 10−6 |
| 1396–1400 cm−1 | 1 | 1.48 × 10−7 |
| α-helix | 1 | 2.27 × 10−5 |
| β-turn | 1 | 2.61 × 10−5 |
| 1440–1450 cm−1 | 0.99 | 3.49 × 10−5 |
| 2854 cm−1 | 0.99 | 2.02 × 10−2 |
| random coil | 0.97 | 7.18 × 10−5 |
| 1064 cm−1 | 0.96 | 4.66 × 10−4 |
| 1382 cm−1 | 0.94 | 1.96 × 10−3 |
| 538–542 cm−1 | 0.93 | 7.15 × 10−3 |
| 1741–1745 cm−1 | 0.93 | 2.77 × 10−3 |
| 1638–1645 cm−1 | 0.91 | 1.00 × 10−2 |
| 777 cm−1 | 0.79 | 1.42 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatzimichail, K.; Ladika, G.; Christodoulou, P.; Bartzis, V.; Konteles, S.J.; Lazou, A.E.; Kritsi, E.; Cavouras, D.; Sinanoglou, V.J. Comparative Evaluation of Cow and Goat Milk Samples Utilizing Non-Destructive Techniques and Chemometric Approaches. Appl. Sci. 2025, 15, 10883. https://doi.org/10.3390/app152010883
Chatzimichail K, Ladika G, Christodoulou P, Bartzis V, Konteles SJ, Lazou AE, Kritsi E, Cavouras D, Sinanoglou VJ. Comparative Evaluation of Cow and Goat Milk Samples Utilizing Non-Destructive Techniques and Chemometric Approaches. Applied Sciences. 2025; 15(20):10883. https://doi.org/10.3390/app152010883
Chicago/Turabian StyleChatzimichail, Kyriaki, Georgia Ladika, Paris Christodoulou, Vasileios Bartzis, Spyros J. Konteles, Andriana E. Lazou, Eftichia Kritsi, Dionisis Cavouras, and Vassilia J. Sinanoglou. 2025. "Comparative Evaluation of Cow and Goat Milk Samples Utilizing Non-Destructive Techniques and Chemometric Approaches" Applied Sciences 15, no. 20: 10883. https://doi.org/10.3390/app152010883
APA StyleChatzimichail, K., Ladika, G., Christodoulou, P., Bartzis, V., Konteles, S. J., Lazou, A. E., Kritsi, E., Cavouras, D., & Sinanoglou, V. J. (2025). Comparative Evaluation of Cow and Goat Milk Samples Utilizing Non-Destructive Techniques and Chemometric Approaches. Applied Sciences, 15(20), 10883. https://doi.org/10.3390/app152010883







