Screening of 31 Lactic Acid Bacteria Strains Identified Levilactobacillus brevis NCTC 13768 as a High-Yield GABA Producer
Abstract
1. Introduction
2. Materials and Methods
2.1. Micro-Organisms
2.2. Screening for GABA Overproducing Strain
2.3. Dynamics of Growth and GABA Production by L. brevis NCTC 13768
2.4. High-Performance Liquid Chromatography Analysis of GABA and GA
2.5. Real-Time qPCR Assay
2.6. Statistical Analysis
3. Results
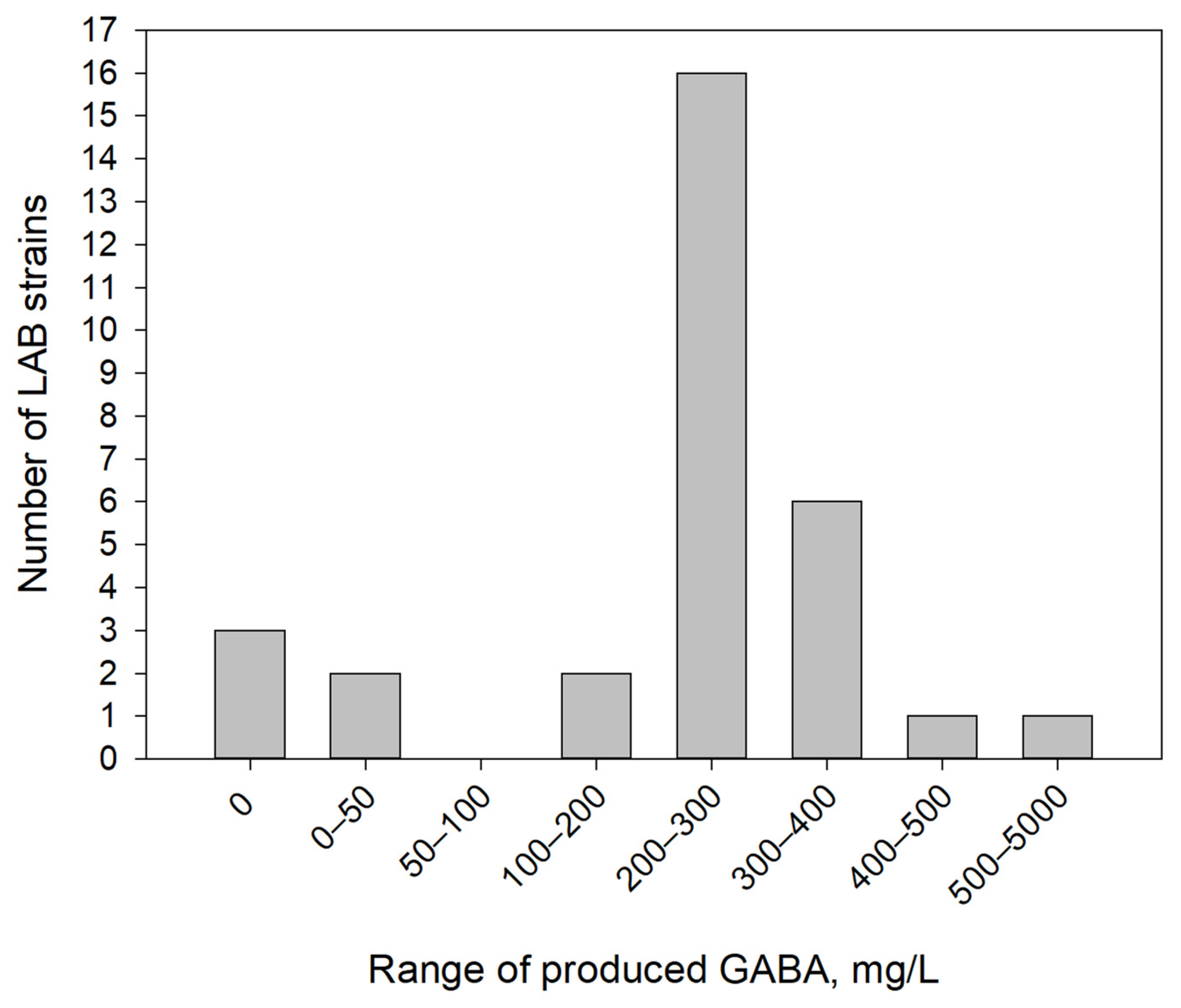
3.1. GABA-Producing Capacity of Investigated Strains
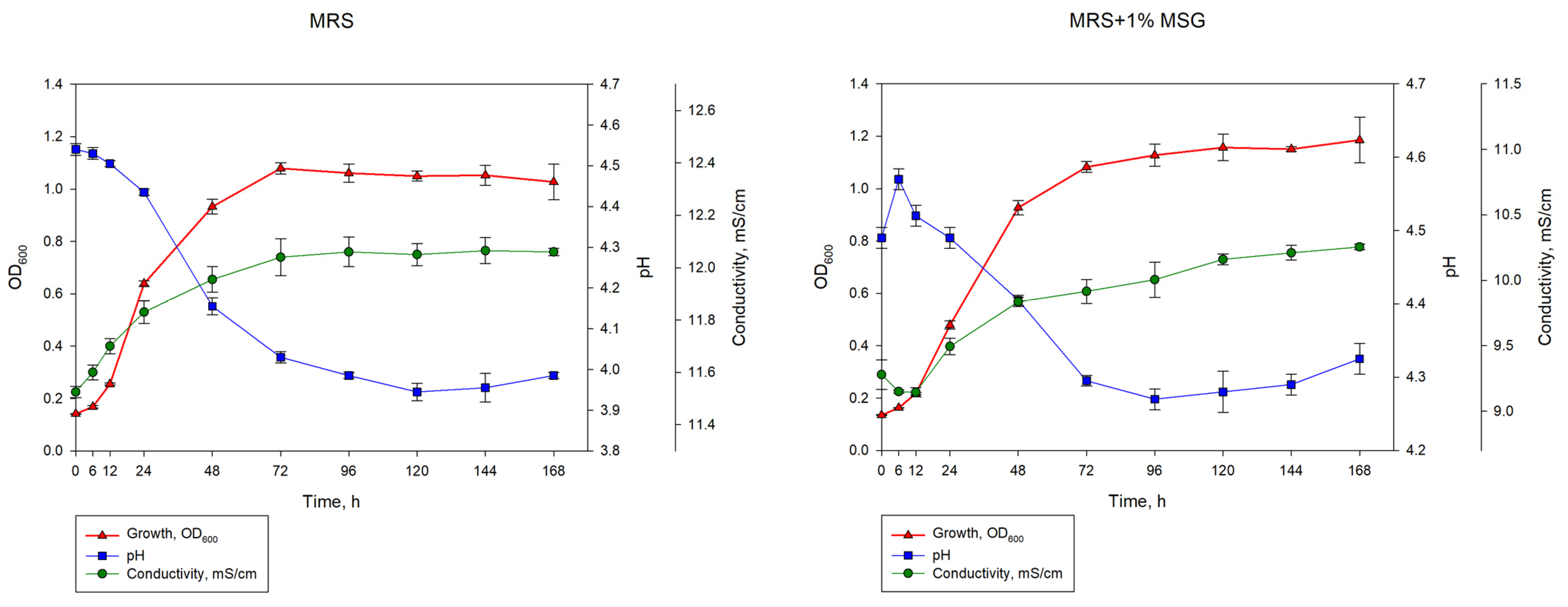
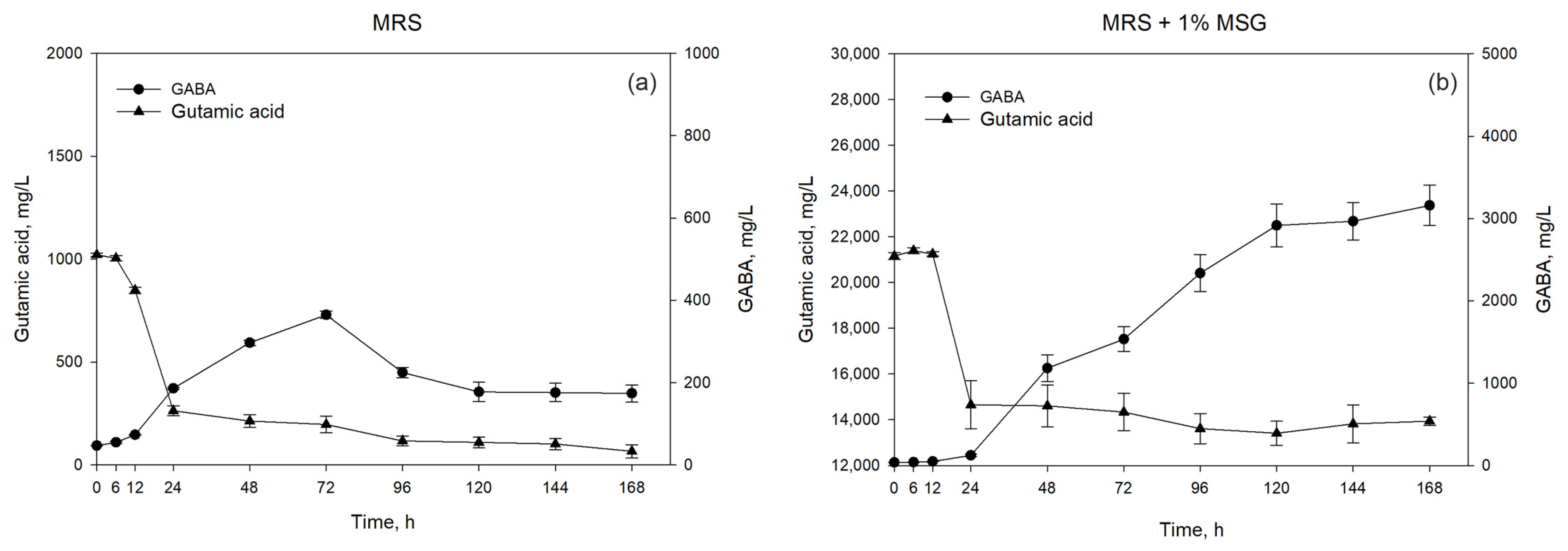
3.2. Dynamics of Growth and GABA Production by L. brevis NCTC 13768
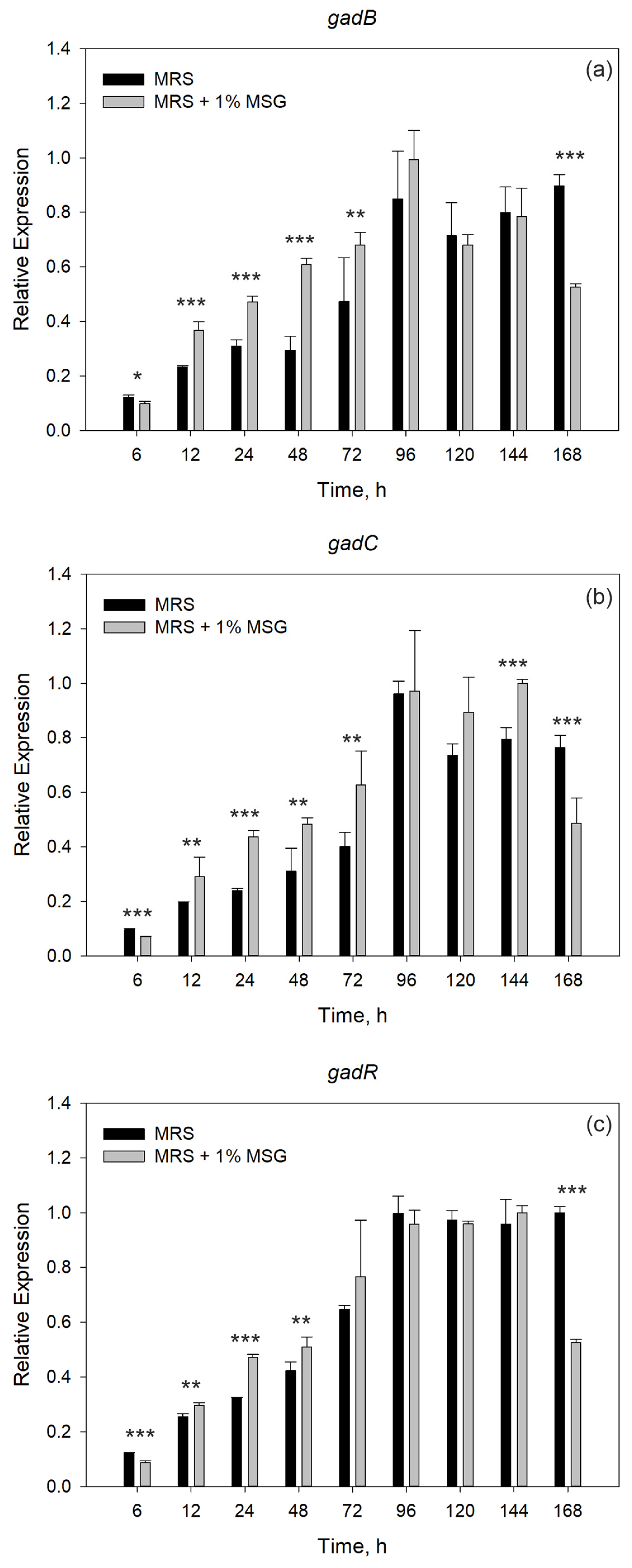
3.3. Gene Expression Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GABA | Gamma-aminobutyric acid |
| GA | Glutamic acid |
| MRS | De Man–Rogosa–Sharpe |
| HPLC | High-performance liquid chromatography |
| ATCC | American Type Culture Collection |
| GAD | Glutamic acid decarboxylase |
| LAB | Lactic acid bacteria |
| GRAS | Generally Recognized as Safe |
| MSG | Monosodium glutamate |
| NBIMCC | National Bank for Industrial Micro-organisms and Cell Cultures |
| BAS | Bulgarian Academy of Sciences |
| OD | Optical density |
References
- Wu, Q.L.; Shah, N.P. High γ-aminobutyric acid production from lactic acid bacteria: Emphasis on Lactobacillus brevis as a functional dairy starter. Crit. Rev. Food Sci. Nutr. 2017, 57, 3661–3672. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of Gamma-Aminobutyric Acid from Lactic Acid Bacteria: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef] [PubMed]
- Ghit, A.; Assal, D.; Al-Shami, A.S.; Hussein, D.E.E. GABAA receptors: Structure, function, pharmacology, and related disorders. J. Genet. Eng. Biotechnol. 2021, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.; Newland, J.; Faull, R.L.M.; Kwakowsky, A. Cation-Chloride Cotransporters KCC2 and NKCC1 as Therapeutic Targets in Neurological and Neuropsychiatric Disorders. Molecules 2023, 28, 1344. [Google Scholar] [CrossRef]
- Kesika, P.; Suganthy, N.; Sivamaruthi, B.S.; Chaiyasut, C. Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer’s disease. Life Sci. 2021, 264, 118627. [Google Scholar] [CrossRef]
- Almutairi, S.; Sivadas, A.; Kwakowsky, A. The Effect of Oral GABA on the Nervous System: Potential for Therapeutic Intervention. Nutraceuticals 2024, 4, 241–259. [Google Scholar] [CrossRef]
- Diez-Gutiérrez, L.; San Vicente, L.; Barrón, L.J.R.; del Carmen Villarán, M.; Chávarri, M. Gamma-aminobutyric acid and probiotics: Multiple health benefits and their future in the global functional food and nutraceuticals market. J. Funct. Foods 2020, 64, 103669. [Google Scholar] [CrossRef]
- Pencheva, D.; Teneva, D.; Denev, P. Validation of HPLC Method for Analysis of Gamma-Aminobutyric and Glutamic Acids in Plant Foods and Medicinal Plants. Molecules 2023, 28, 84. [Google Scholar] [CrossRef]
- Liu, W.J.; Pang, H.L.; Zhang, H.P.; Cai, Y.M. Biodiversity of lactic acid bacteria. In Lactic Acid Bacteria. Fundamentals and Practice, 1st ed.; Zhang, H.P., Cai, Y.M., Eds.; Springer Publishing: New York, NY, USA, 2014; pp. 103–203. [Google Scholar] [CrossRef]
- Abdelhamid, A.G.; El-Dougdoug, N.K. Controlling foodborne pathogens with natural antimicrobials by biological control and antivirulence strategies. Heliyon 2020, 6, e05020. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, X.; Fu, J.; Wang, S.; Chen, Y.; Chang, K.; Li, H. Substrate sustained release-based high efficacy biosynthesis of GABA by Lactobacillus brevis NCL912. Microb. Cell Factories 2018, 17, 80. [Google Scholar] [CrossRef]
- Gangaraju, D.S.; Murty, V.R.; Prapulla, S.G. Probiotic-mediated biotransformation of monosodium glutamate to γ-aminobutyric acid: Differential production in complex and minimal media and kinetic modelling. Ann. Microbiol. 2014, 64, 229–237. [Google Scholar] [CrossRef]
- Zhuang, K.; Jiang, Y.; Feng, X.; Li, L.; Dang, F.; Zhang, W.; Man, C. Transcriptomic response to GABA-producing Lactobacillus plantarum CGMCC 1.2437T induced by L-MSG. PLoS ONE 2018, 13, e0199021. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: Microbial endocrinology in the design and use of probiotics. BioEssays 2011, 33, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Mancini, A.; Carafa, I.; Franciosi, E.; Franciosi, E.; Nardin, T.; Bottari, B.; Larcher, R.; Tuohy, K. In vitro probiotic characterization of high GABA producing strain Lactobacillus brevis DSM 32386 isolated from traditional “wild” Alpine cheese. Ann. Microbiol. 2019, 69, 1435–1443. [Google Scholar] [CrossRef]
- Liu, H.; Liu, D.; Zhang, C.; Niu, H.; Xin, X.; Yi, H.; Liu, D.; Zhang, J. Whole-genome analysis, evaluation and regulation of in vitro and in vivo GABA production from Levilactobacillus brevis YSJ3. Int. J. Food Microbiol. 2024, 421, 110787. [Google Scholar] [CrossRef]
- Dovom, M.R.E.; Najafi, M.B.H.; Vosough, P.R.; Norouzi, N.; Nezhad, S.J.E.; Mayo, B. Screening of lactic acid bacteria strains isolated from Iranian traditional dairy products for GABA production and optimization by response surface methodology. Sci. Rep. 2023, 13, 440. [Google Scholar] [CrossRef]
- Siragusa, S.; De Angelis, M.; Di Cagno, R.; Rizzello, C.G.; Coda, R.; Gobbetti, M. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 2007, 73, 7283–7290. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, Q.; Tan, X.; Zhang, S.; Zeng, L.; Tang, J.; Xiang, W. Characterization of γ-aminobutyric acid (GABA)-producing Saccharomyces cerevisiae and coculture with Lactobacillus plantarum for mulberry beverage brewing. J. Biosci. Bioeng. 2020, 129, 447–453. [Google Scholar] [CrossRef]
- Hwang, E.; Park, J.-Y. Isolation and characterization of gamma-aminobutyric acid (GABA)-producing lactic acid bacteria from kimchi. Curr. Top. Lact. Acid Bact. Probiotics 2020, 6, 64–69. [Google Scholar] [CrossRef]
- Ly, D.; Mayrhofer, S.; Agung Yogeswara, I.B.; Nguyen, T.-H.; Domig, K.J. Identification, Classification and Screening for γ-Amino-butyric Acid Production in Lactic Acid Bacteria from Cambodian Fermented Foods. Biomolecules 2019, 9, 768. [Google Scholar] [CrossRef]
- Petrova, P.; Ivanov, I.; Tsigoriyna, L.; Valcheva, N.; Vasileva, E.; Parvanova-Mancheva, T.; Arsov, A.; Petrov, K. Traditional Bulgarian Dairy Products: Ethnic Foods with Health Benefits. Microorganisms 2021, 9, 480. [Google Scholar] [CrossRef]
- National Bank for Industrial Microorganisms and Cell Cultures (NBIMCC). Available online: https://www.nbimcc.org/www_2020/en/ (accessed on 8 July 2025).
- NCBI Taxonomy Browser. Levilactobacillus brevis. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1580&lvl=3&keep=1&srchmode=1&unlock&mod=1&log_op=modifier_toggle (accessed on 8 July 2025).
- Danova, S.; Yankov, D.; Dobreva, L.; Dobreva, A.; Armenova, N.; Apostolov, A.; Mileva, M. Postbiotics Production of Candidate-Probiotic Lactiplantibacillus plantarum AC131 with Renewable Bio Resources. Life 2023, 13, 2006. [Google Scholar] [CrossRef]
- Dobreva, L.; Atanasova, N.; Donchev, P.; Krumova, E.; Abrashev, R.; Karakirova, Y.; Mladenova, R.; Tolchkov, V.; Ralchev, N.; Dishliyska, V.; et al. Candidate-Probiotic Lactobacilli and Their Postbiotics as Health-Benefit Promoters. Microorganisms 2024, 12, 1910. [Google Scholar] [CrossRef]
- Teneva-Angelova, T.; Beshkova, D. Non-traditional sources for isolation of lactic acid bacteria. Ann. Microbiol. 2016, 66, 449–459. [Google Scholar] [CrossRef]
- Yogeswara, I.B.A.; Kittibunchakul, S.; Rahayu, E.S.; Domig, K.J.; Haltrich, D.; Nguyen, T.H. Microbial Production and Enzymatic Biosynthesis of γ-Aminobutyric Acid (GABA) Using Lactobacillus plantarum FNCC 260 Isolated from Indonesian Fermented Foods. Processes 2021, 9, 22. [Google Scholar] [CrossRef]
- Yunes, R.; Poluektova, E.; Dyachkova, M.; Klimina, K.; Kovtun, A.; Averina, O.; Orlova, V.; Danilenko, V. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 2016, 42, 197–204. [Google Scholar] [CrossRef]
- Cataldo, P.G.; Urquiza Martínez, M.P.; Villena, J.; Kitazawa, H.; Saavedra, L.; Hebert, E.M. Comprehensive characterization of γ-aminobutyric acid (GABA) production by Levilactobacillus brevis CRL 2013: Insights from physiology, genomics, and proteomics. Front. Microbiol. 2024, 15, 1408624. [Google Scholar] [CrossRef]
- Banerjee, S.; Poore, M.; Gerdes, S.; Nedveck, D.; Lauridsen, L.; Kristensen, H.T.; Jensen, H.M.; Byrd, F.M.; Ouwehand, A.C.; Patterson, E.; et al. Transcriptomics reveal different metabolic strategies for acid resistance and gamma-aminobutyric acid (GABA) production in select Levilactobacillus brevis strains. Microb. Cell Fact. 2021, 20, 173. [Google Scholar] [CrossRef]
- Cataldo, P.G.; Villegas, J.M.; Savoy De Giori, G.; Saavedra, L.; Hebert, E.M. Enhancement of γ-aminobutyric acid (GABA) production by Lactobacillus brevis CRL 2013 based on carbohydrate fermentation. Int. J. Food Microbiol. 2020, 333, 108792–108799. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, M.; Lu, W.; Zhang, C.; Chen, D.; Shah, N.P.; Xiao, C. Optimizing Levilactobacillus brevis NPS-QW 145 Fermentation for Gamma-Aminobutyric Acid (GABA) Production in Soybean Sprout Yogurt-like Product. Foods 2023, 12, 977. [Google Scholar] [CrossRef]
- Li, H.; Qiu, T.; Huang, G.; Cao, Y. Production of gamma-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb. Cell Fact. 2010, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Wu, O.; Shah, N.P. Restoration of GABA production machinery in Lactobacillus brevis by accessible carbohydrates, anaerobiosis and early acidification. Food Microbiol. 2018, 69, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Sarkisyan, V.A.; Kochetkova, A.A.; Bessonov, V.V.; Isakov, V.A.; Nikityuk, D.B. Estimation of gammaaminobutyric acid intake from the human diet. Vopr. Pitan. 2024, 93, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Natural and Non-Prescription Health Products Directorate (NNHPD) Natural Health Products Ingredients Database. 4-Aminobutanoic Acid. Group 7: Ingredients with Relaxation Action. 2025. Available online: https://webprod.hc-sc.gc.ca/nhpid-bdipsn/atReq?atid=fonc.cognitive.func2&lang=eng (accessed on 16 September 2025).
- Hasegawa, M.; Yamane, D.; Funato, K.; Yoshida, A.; Sambongi, Y. Gamma-aminobutyric acid fermentation with date residue by a lactic acid bacterium, Lactobacillus brevis. J. Biosci. Bioeng. 2018, 125, 316–319. [Google Scholar] [CrossRef]
- Youssef, H.A.I.; Vitaglione, P.; Ferracane, R.; Abuqwider, J.; Mauriello, G. Evaluation of GABA Production by Alginate-Microencapsulated Fresh and Freeze-Dried Bacteria Enriched with Monosodium Glutamate during Storage in Chocolate Milk. Microorganisms 2023, 11, 2648. [Google Scholar] [CrossRef]
- Tanamool, V.; Hongsachart, P.; Soemphol, W. Screening and characterisation of gamma-aminobutyric acid (GABA) producing lactic acid bacteria isolated from Thai fermented fish (Plaa-som) in Nong Khai and its application in Thai fermented vegetables (Som-pak). Food Sci. Technol, Camp. 2020, 40, 483–490. [Google Scholar] [CrossRef]
- Valenzuela, J.A.; Flórez, A.B.; Vázquez, L.; Vasek, O.M.; Mayo, B. Production of γ-aminobutyric acid (GABA) by lactic acid bacteria strains isolated from traditional, starter-free dairy products made of raw milk. Benef. Microbes 2019, 10, 579–587. [Google Scholar] [CrossRef]
- Wu, C.-H.; Hsueh, Y.-H.; Kuo, J.-M.; Liu, S.-J. Characterization of a Potential Probiotic Lactobacillus brevis RK03 and Efficient Production of γ-Aminobutyric Acid in Batch Fermentation. Int. J. Mol. Sci. 2018, 19, 143. [Google Scholar] [CrossRef]
- Sezgin, E.; Tekin, B. Molecular evolution and population genetics of glutamate decarboxylase acid resistance pathway in lactic acid bacteria. Front. Genet. 2023, 14, 1027156. [Google Scholar] [CrossRef]
- Icer, M.A.; Sarikaya, B.; Kocyigit, E.; Atabilen, B.; Çelik, M.N.; Capasso, R.; Ağagündüz, D.; Budán, F. Contributions of Gamma-Aminobutyric Acid (GABA) Produced by Lactic Acid Bacteria on Food Quality and Human Health: Current Applications and Future Prospects. Foods 2024, 13, 2437. [Google Scholar] [CrossRef]
- Pizzi, A.; Parolin, C.; Gottardi, D.; Ricci, A.; Parpinello, G.P.; Lanciotti, R.; Patrignani, F.; Vitali, B. A Novel GABA-Producing Levilactobacillus brevis Strain Isolated from Organic Tomato as a Promising Probiotic. Biomolecules 2025, 15, 979. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Liu, X.; Cao, Y. gadA gene locus in Lactobacillus brevis NCL912 and its expression during fed-batch fermentation. FEMS Microbiol. Lett. 2013, 349, 108–116. [Google Scholar] [CrossRef]
- Gong, L.; Ren, C.; Xu, Y. Deciphering the crucial roles of transcriptional regulator GadR on gamma-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb. Cell Fact. 2019, 18, 108. [Google Scholar] [CrossRef]
- Yogeswara, I.B.A.; Maneerat, S.; Haltrich, D. Glutamate Decarboxylase from Lactic Acid Bacteria—A Key Enzyme in GABA Synthesis. Microorganisms 2020, 8, 1923. [Google Scholar] [CrossRef]
- Lyu, C.; Zhao, W.; Peng, C.; Hu, S.; Fang, H.; Hua, Y.; Yao, S.; Huang, J.; Mei, L. Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production. Microb. Cell Fact. 2018, 17, 180. [Google Scholar] [CrossRef]
- Ma, D.; Lu, P.; Shi, Y. Substrate selectivity of the acid-activated glutamate/γ-aminobutyric acid (GABA) antiporter GadC from Escherichia coli. J. Biol. Chem. 2013, 288, 15148–15153. [Google Scholar] [CrossRef]

| Number | Strain | Medium | Temperature of Cultivation, °C | Source | References |
|---|---|---|---|---|---|
| Bulgarian National Bank for Industrial Microorganisms and Cell Cultures (NBIMCC) | |||||
| 1 | Lactobacillus delbrueckii subsp. bulgaricus NBIMCC 1132 | MRS | 37 | Collection | [23] |
| 2 | Streptococcus thermophilus NBIMCC 3916 | MRS | 37 | Collection | [23] |
| 3 | Lacticaseibacillus rhamnosus NBIMCC 1013 | MRS | 37 | Collection | [23] |
| 4 | Limosilactobacillus helveticus NBIMCC 275 | MRS | 37 | Collection | [23] |
| 5 | Lentilactobacillus kefiri NBIMCC 3446 | MRS | 30 | Collection | [23] |
| 6 | Levilactobacillus brevis NBIMCC 8426 | MRS | 30 | Collection | [23] |
| 7 | Liquorilactobacillus mali NBIMCC 3316 | MRS | 30 | Collection | [23] |
| 8 | Lactococcus lactis subsp. lactis NBIMCC 350 | MRS | 30 | Collection | [23] |
| 9 | Lactococcus lactis subsp. lactis NBIMCC 355 | MRS | 30 | Collection | [23] |
| Cryobiotica Ltd. | |||||
| 10 | Lactobacillus helveticus 13 | MRS | 37 | Cheese | |
| 11 | Lactobacillus helveticus 14 | MRS | 37 | Cheese | |
| 12 | Lactobacillus rhamnosus 1 | MRS | 37 | Cheese | |
| 13 | Lactobacillus rhamnosus 2 | MRS | 37 | Cheese | |
| 14 | Lactobacillus paracasei 8 | MRS | 37 | Cheese | |
| 15 | Lactobacillus paracasei | MRS | 37 | Cheese | |
| 16 | Lactobacillus plantarum Y6 | MRS | 37 | Cheese | |
| 17 | Levilactobacillus brevis NCTC 13768 | MRS | 37 | Collection | [24] |
| 18 | Lactobacillus delbrueckii subsp. delbrueckii NBIMCC 2449 | MRS | 37 | Collection | [23] |
| 19 | Lactobacillus delbrueckii subsp. bulgaricus LB6 | MRS | 37 | Yogurt | |
| 20 | Lactobacillus delbrueckii subsp. bulgaricus S10 | MRS | 37 | Yogurt | |
| 21 | Lactobacillus rhamnosus S25-1 | MRS | 37 | Cheese | |
| Laboratory of Lactic Acid Bacteria and Probiotics, Institute of Microbiology—BAS | |||||
| 22 | Lactiplantibacillus plantarum AC131 | MRS | 37 | Artisanal white brined cheese | [25] |
| 23 | Lactiplantibacillus plantarum LC2 | MRS | 37 | Artisanal white brined cheese | |
| 24 | Lactiplantibacillus plantarum L3 | MRS | 37 | Home-made katak | |
| 25 | Limosilactobacillus fermentum LB4 | MRS | 37 | Vaginal sample | |
| 26 | Limosilactobacillus helveticus 611 | MRS | 37 | Baby feces | [26] |
| Laboratory of Cell Biosystems, Institute of Microbiology—BAS | |||||
| 27 | Lacticaseibacillus rhamnosus LR2 | MRS | 37 | Home-made yogurt | |
| 28 | Lacticaseibacillus rhamnosus PglGER32 | MRS | 37 | Isolated from Panax ginseng | [27] |
| 29 | Lacticaseibacillus rhamnosus PglGER1 | MRS | 37 | Isolated from Panax ginseng | [27] |
| 30 | Lacticaseibacillus rhamnosus PglGER26 | MRS | 37 | Isolated from Panax ginseng | [27] |
| 31 | Lacticaseibacillus rhamnosus PglGER3 | MRS | 37 | Isolated from Panax ginseng | [27] |
| Gene | Forward Primer 5′-3′ | Reverse Primer 5′-3′ | Reference |
|---|---|---|---|
| gadB | TGGCTAAGTATGGTTGGCAAGTTCC | TCATCGGCAATCGTCATGGTCATG | [20] |
| gadC | TACCTCGTACAAGGAAACCCA | GATAAAC-GGAACAAATCCCACT | [20] |
| gadR | GTCGTCGATTCTCATGCTTATT | TGCCTGCTTCAGACTCTGTTT | [20] |
| 16S rRNA | CACATTGGGACTGAGACACG | AGCCGAAACCCTTCTTCACT | [20] |
| № | Strain | GABA, mg/L |
|---|---|---|
| 1 | Lactobacillus delbrueckii subsp. bulgaricus NBIMCC 1132 | 236.4 ± 2.6 |
| 2 | Streptococcus thermophilus NBIMCC 3916 | 225.4 ± 4.9 |
| 3 | Lacticaseibacillus rhamnosus NBIMCC 1013 | 269.3 ± 1.7 |
| 4 | Limosilactobacillus helveticus NBIMCC 275 | 344.1 ± 10.3 |
| 5 | Lentilactobacillus kefiri NBIMCC 3446 | 239.4 ± 2.6 |
| 6 | Levilactobacillus brevis NBIMCC 8426 | 196.8 ± 2.2 |
| 7 | Liquorilactobacillus mali NBIMCC 3316 | 328.9 ± 4.4 |
| 8 | Lactococcus lactis subsp. lactis NBIMCC 350 | 259.6 ± 6.6 |
| 9 | Lactococcus lactis subsp. lactis NBIMCC 355 | 315.2 ± 3.8 |
| 10 | Lactobacillus helveticus 13 | 269.2 ± 2.3 |
| 11 | Lactobacillus helveticus 14 | 293.9 ± 7.5 |
| 12 | Lactobacillus rhamnosus 1 | 232.6 ± 4.7 |
| 13 | Lactobacillus rhamnosus 2 | 256.6 ± 1.7 |
| 14 | Lactobacillus paracasei 8 | 257.3 ± 3.2 |
| 15 | Lactobacillus paracasei | 269.4 ± 6.8 |
| 16 | Lactobacillus plantarum Y6 | 343.4 ± 5.7 |
| 17 | Levilactobacillus brevis NCTC 13768 | 3830.7 ± 12.1 |
| 18 | Lactobacillus delbrueckii subsp. delbrueckii NBIMCC 2449 | 304.6 ± 8.9 |
| 19 | Lactobacillus delbrueckii subsp. bulgaricus LB6 | 337.5 ± 0.5 |
| 20 | Lactobacillus delbrueckii subsp. bulgaricus S10 | 286.7 ± 1.6 |
| 21 | Lactobacillus rhamnosus S25-1 | 418.1 ± 1.2 |
| 22 | Lactiplantibacillus plantarum AC131 | n.d. |
| 23 | Lactiplantibacillus plantarum LC2 | n.d. |
| 24 | Lactiplantibacillus plantarum L3 | 27.9 ± 0.2 |
| 25 | Limosilactobacillus fermentum LB4 | 27.7 ± 0.2 |
| 26 | Limosilactobacillus helveticus 611 | n.d. |
| 27 | Lacticaseibacillus rhamnosus LR2 | 165.5 ± 0.5 |
| 28 | Lacticaseibacillus rhamnosus PglGER32 | 234.0 ± 3.2 |
| 29 | Lacticaseibacillus rhamnosus PglGER1 | 246.4 ± 0.5 |
| 30 | Lacticaseibacillus rhamnosus PglGER26 | 245.5 ± 0.80 |
| 31 | Lacticaseibacillus rhamnosus PglGER3 | 247.8 ± 0.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teneva, D.; Pencheva, D.; Teneva-Angelova, T.; Danova, S.; Atanasova, N.; Dobreva, L.; Ognyanov, M.; Petrova, A.; Slavchev, A.; Georgiev, V.; et al. Screening of 31 Lactic Acid Bacteria Strains Identified Levilactobacillus brevis NCTC 13768 as a High-Yield GABA Producer. Appl. Sci. 2025, 15, 10670. https://doi.org/10.3390/app151910670
Teneva D, Pencheva D, Teneva-Angelova T, Danova S, Atanasova N, Dobreva L, Ognyanov M, Petrova A, Slavchev A, Georgiev V, et al. Screening of 31 Lactic Acid Bacteria Strains Identified Levilactobacillus brevis NCTC 13768 as a High-Yield GABA Producer. Applied Sciences. 2025; 15(19):10670. https://doi.org/10.3390/app151910670
Chicago/Turabian StyleTeneva, Desislava, Daniela Pencheva, Tsvetanka Teneva-Angelova, Svetla Danova, Nikoleta Atanasova, Lili Dobreva, Manol Ognyanov, Ani Petrova, Aleksandar Slavchev, Vasil Georgiev, and et al. 2025. "Screening of 31 Lactic Acid Bacteria Strains Identified Levilactobacillus brevis NCTC 13768 as a High-Yield GABA Producer" Applied Sciences 15, no. 19: 10670. https://doi.org/10.3390/app151910670
APA StyleTeneva, D., Pencheva, D., Teneva-Angelova, T., Danova, S., Atanasova, N., Dobreva, L., Ognyanov, M., Petrova, A., Slavchev, A., Georgiev, V., & Denev, P. (2025). Screening of 31 Lactic Acid Bacteria Strains Identified Levilactobacillus brevis NCTC 13768 as a High-Yield GABA Producer. Applied Sciences, 15(19), 10670. https://doi.org/10.3390/app151910670








