Artificial Intelligence in Microbiome Research and Beyond: Connecting Human Health, Animal Husbandry, and Aquaculture
Abstract
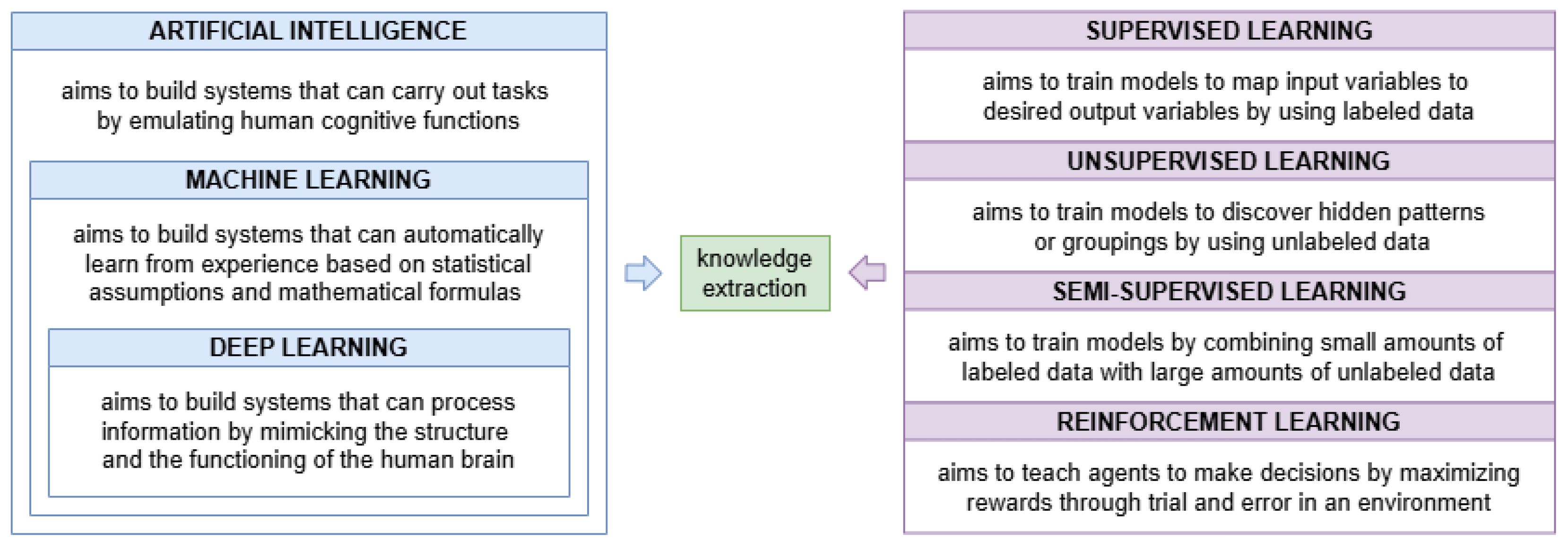
1. Introduction
2. Harnessing Artificial Intelligence for Human Health Insights
2.1. Connection Between Intestinal Microbiome and Diseases
2.2. Interrelationship Between Humans and Fish in Microbiome Research
3. Enhancing Animal Health Through Artificial Intelligence
Intestinal Microorganisms for Improved Animal Husbandry
4. Trends in Artificial Intelligence Application to Aquaculture
4.1. Microbial Diversity in Fish Microbiota
4.2. Benefits of Artificial Intelligence in Fish Welfare and Farming
5. Overview of Current Limitations and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | AdaBoost |
| ADG | average daily gain |
| AEs | autoencoders |
| AI | artificial intelligence |
| AMPs | antimicrobial peptides |
| AMR | antimicrobial resistance |
| ANFIS | adaptive neuro-fuzzy inference system |
| ANs | attention networks |
| ARG | antibiotic-resistant gene |
| AUC | area under the receiver-operating characteristic curve |
| AutoML | automatic machine learning |
| BERT | bidirectional encoder representations from transformers |
| BMI | body mass index |
| BNs | Bayesian networks |
| CAEs | compromised aquatic environments |
| CB | CatBoost |
| CFUs | colony forming units |
| CNNs | convolutional neural networks |
| CV | computer vision |
| DeepLIFT | deep learning important features |
| DES | discrete event system |
| DL | deep learning |
| DRT | dimensionality reduction techniques |
| DT | decision tree |
| EBM | ensemble-based models |
| ET | extra trees |
| FCR | feed conversion ratio |
| FE | feeding efficiency |
| FNNs | feedforward neural networks |
| GANs | generative adversarial networks |
| GB | gradient boosting |
| GNB | Gaussian Naïve Bayes |
| GNNs | graph neural networks |
| HAC | hierarchical agglomerative clustering |
| IBM | instance-based models |
| ICE | individual conditional expectation |
| IoE | Internet of Everything |
| IoT | Internet of Things |
| k-NN | k-nearest neighbor |
| LIME | local interpretable model-agnostic explanations |
| LR | logistic regression |
| LSTM | long short-term memory |
| MAE | mean absolute error |
| mAP | mean average precision |
| MBM | margin-based models |
| MDS | multidimensional scaling |
| ML | machine learning |
| MUN | milk urea nitrogen |
| NIR | near-infrared |
| NMDS | non-metric multidimensional scaling |
| NMF | non-negative matrix factorization |
| NNs | neural networks |
| NNM | neural network models |
| NUE | nitrogen utilization efficiency |
| PAM | partitioning around medoids |
| PCA | principal component analysis |
| PCoA | principal coordinate analysis |
| PLS-DA | partial least squares discriminant analysis |
| TBM | tree-based models |
| RB | random block |
| RF | random forest |
| RiPPs | ribosomally synthetized and post-translationally modified peptides |
| RNNs | recurrent neural networks |
| SHAP | Shapely additive explanations |
| SPM | statistical and probabilistic models |
| SVM | support vector machine |
| vOTUs | viral operational taxonomic units |
| XAI | explainable artificial intelligence |
| XGB | extreme gradient boosting |
| YOLO | You Only Look Once |
References
- Cullen, C.M.; Aneja, K.K.; Beyhan, S.; Cho, C.E.; Woloszynek, S.; Convertino, M.; McCoy, S.J.; Zhang, Y.; Anderson, M.Z.; Alvarez-Ponce, D.; et al. Emerging priorities for microbiome research. Front. Microbiol. 2020, 11, 136. [Google Scholar] [CrossRef]
- Trinh, P.; Zaneveld, J.R.; Safranek, S.; Rabinowitz, P.M. One health relationships between human, animal, and environmental microbiomes: A mini-review. Front. Public Health 2018, 6, 235. [Google Scholar] [CrossRef]
- Foo, J.L.; Ling, H.; Lee, Y.S.; Chang, M.W. Microbiome engineering: Current applications and its future. Biotechnol. J. 2017, 12, 1600099. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Theis, K.R. Host biology in light of the microbiome: Ten principles of holobionts and hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef] [PubMed]
- Bosch, T.C.; McFall-Ngai, M.J. Metaorganisms as the new frontier. Zoology 2011, 114, 185–190. [Google Scholar] [CrossRef]
- Iebba, V.; Totino, V.; Gagliardi, A.; Santangelo, F.; Cacciotti, F.; Trancassini, M.; Mancini, C.; Cicerone, C.; Corazziari, E.; Pantanella, F.; et al. Eubiosis and dysbiosis: The two sides of the microbiota. New Microbiol. 2016, 39, 1–12. [Google Scholar]
- Vayssier-Taussat, M.; Albina, E.; Citti, C.; Cosson, J.F.; Jacques, M.A.; Lebrun, M.H.; Le Loir, Y.; Ogliastro, M.; Petit, M.A.; Roumagnac, P.; et al. Shifting the paradigm from pathogens to pathobiome: New concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 2014, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Hooks, K.B.; O’Malley, M.A. Dysbiosis and its discontents. mBio 2017, 8, e01492-17. [Google Scholar] [CrossRef] [PubMed]
- Proctor, L. Priorities for the next 10 years of human microbiome research. Nature 2019, 569, 623–625. [Google Scholar] [CrossRef]
- Sudhakar, P.; Machiels, K.; Verstockt, B.; Korcsmaros, T.; Vermeire, S. Computational biology and machine learning approaches to understand mechanistic microbiome-host interactions. Front. Microbiol. 2021, 12, 618856. [Google Scholar] [CrossRef]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-generation machine learning for biological networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- McCoubrey, L.E.; Elbadawi, M.; Orlu, M.; Gaisford, S.; Basit, A.W. Harnessing machine learning for development of microbiome therapeutics. Gut Microbes 2021, 13, 1872323. [Google Scholar] [CrossRef]
- Chen, V.; Yang, M.; Cui, W.; Kim, J.S.; Talwalkar, A.; Ma, J. Applying interpretable machine learning in computational biology-pitfalls, recommendations and opportunities for new developments. Nat. Methods 2024, 21, 1454–1461. [Google Scholar] [CrossRef]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef]
- Xu, C.; Jackson, S.A. Machine learning and complex biological data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- D’Urso, F.; Broccolo, F. Applications of artificial intelligence in microbiome analysis and probiotic interventions—An overview and perspective based on the current state of the art. Appl. Sci. 2024, 14, 8627. [Google Scholar] [CrossRef]
- Hernández Medina, R.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine learning and deep learning applications in microbiome research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef]
- Rosenberg, E. Diversity of bacteria within the human gut and its contribution to the functional unity of holobionts. NPJ Biofilms Microbiomes 2024, 10, 134. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Li, P.; Luo, H.; Ji, B.; Nielsen, J. Machine learning for data integration in human gut microbiome. Microb. Cell Fact. 2022, 21, 241. [Google Scholar] [CrossRef]
- Visconti, A.; Le Roy, C.I.; Rosa, F.; Rossi, N.; Martin, T.C.; Mohney, R.P.; Li, W.; de Rinaldis, E.; Bell, J.T.; Venter, J.C.; et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019, 10, 4505. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Abavisani, M.; Foroushan, S.K.; Ebadpour, N.; Sahebkar, A. Deciphering the gut microbiome: The revolution of artificial intelligence in microbiota analysis and intervention. Curr. Res. Biotechnol. 2024, 7, 100211. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Kibria, M.K.; Sifat, I.K.; Hossen, M.B.; Hasan, F.; Mosharaf, M.P.; Hassan, M.Z. Identification of bacterial key genera associated with breast cancer using machine learning techniques. Microbe 2025, 6, 100228. [Google Scholar] [CrossRef]
- Yu, B.; Zhang, H.; Zhang, M. Deep learning-based differential gut flora for prediction of Parkinson’s. PLoS ONE 2025, 20, e0310005. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Duan, J.; Wang, R.; Ying, H.; Feng, Q.; Zhu, B.; Yang, C.; Yang, L. Landscape of gut microbiota and metabolites and their interaction in comorbid heart failure and depressive symptoms: A random forest analysis study. mSystems 2023, 8, e0051523. [Google Scholar] [CrossRef]
- Liu, W.; Fang, X.; Zhou, Y.; Dou, L.; Dou, T. Machine learning-based investigation of the relationship between gut microbiome and obesity status. Microbes Infect. 2022, 24, 104892. [Google Scholar] [CrossRef]
- Park, I.G.; Yoon, S.J.; Won, S.M.; Oh, K.K.; Hyun, J.Y.; Suk, K.T.; Lee, U. Gut microbiota-based machine-learning signature for the diagnosis of alcohol-associated and metabolic dysfunction-associated steatotic liver disease. Sci. Rep. 2024, 14, 16122. [Google Scholar] [CrossRef] [PubMed]
- Asher, E.E.; Bashan, A. Model-free prediction of microbiome compositions. Microbiome 2024, 12, 17. [Google Scholar] [CrossRef]
- Komaki, S.; Sahoyama, Y.; Hachiya, T.; Koseki, K.; Ogata, Y.; Hamazato, F.; Shiozawa, M.; Nakagawa, T.; Suda, W.; Hattori, M.; et al. Dimension reduction of microbiome data linked Bifidobacterium and Prevotella to allergic rhinitis. Sci. Rep. 2024, 14, 7983. [Google Scholar] [CrossRef]
- Chen, G.; Wang, X.; Sun, Q.; Tang, Z.Z. Multidimensional scaling improves distance-based clustering for microbiome data. Bioinformatics 2025, 41, btaf042. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.; Wong, R.J.; Shaw, G.; et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef]
- Yan, H.; Liang, X.; Luo, H.; Tang, X.; Xiao, X. Association between gut microbiota, microbial network, and immunity in pregnancy with a focus on specific bacterial clusters. Front. Microbiol. 2023, 14, 1314257. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of machine learning in human microbiome studies: A review on feature selection, biomarker identification, disease prediction and treatment. Front. Microbiol. 2021, 12, 634511. [Google Scholar] [CrossRef]
- Armstrong, G.; Rahman, G.; Martino, C.; McDonald, D.; Gonzalez, A.; Mishne, G.; Knight, R. Applications and comparison of dimensionality reduction methods for microbiome data. Front. Bioinform. 2022, 2, 821861. [Google Scholar] [CrossRef]
- Lu, Y.; Phillips, C.A.; Langston, M.A. A robustness metric for biological data clustering algorithms. BMC Bioinform. 2019, 20, 503. [Google Scholar] [CrossRef]
- Mayer, E.A.; Nance, K.; Chen, S. The gut-brain axis. Annu. Rev. Med. 2022, 73, 439–453. [Google Scholar] [CrossRef]
- Mayer, E.A.; Knight, R.; Mazmanian, S.K.; Cryan, J.F.; Tillisch, K. Gut microbes and the brain: Paradigm shift in neuroscience. J. Neurosci. 2014, 34, 15490–15496. [Google Scholar] [CrossRef]
- Przymus, P.; Rykaczewski, K.; Martín-Segura, A.; Truu, J.; Carrillo De Santa Pau, E.; Kolev, M.; Naskinova, I.; Gruca, A.; Sampri, A.; Frohme, M.; et al. Deep learning in microbiome analysis: A comprehensive review of neural network models. Front. Microbiol. 2025, 15, 1516667. [Google Scholar] [CrossRef] [PubMed]
- Lawson, P.A.; Saavedra Perez, L.; Sankaranarayanan, K. Reclassification of Clostridium cocleatum, Clostridium ramosum, Clostridium spiroforme and Clostridium saccharogumia as Thomasclavelia cocleata gen. nov., comb. nov., Thomasclavelia ramose comb. nov., gen. nov., Thomasclavelia spiroformis comb. nov. and Thomasclavelia saccharogumia comb. nov. Int. J. Syst. Evol. Microbiol. 2023, 73, 005694. [Google Scholar] [CrossRef]
- Duyar, C.; Senica, S.O.; Kalkan, H. Detection of cardiovascular disease using explainable artificial intelligence and gut microbiota data. Intell. Based Med. 2024, 10, 100180. [Google Scholar] [CrossRef]
- Gou, W.; Ling, C.W.; He, Y.; Jiang, Z.; Fu, Y.; Xu, F.; Miao, Z.; Sun, T.Y.; Lin, J.S.; Zhu, H.L.; et al. Interpretable machine learning framework reveals robust gut microbiome features associated with type 2 diabetes. Diabetes Care 2021, 44, 358–366. [Google Scholar] [CrossRef]
- Papoutsoglou, G.; Tarazona, S.; Lopes, M.B.; Klammsteiner, T.; Ibrahimi, E.; Eckenberger, J.; Novielli, P.; Tonda, A.; Simeon, A.; Shigdel, R.; et al. Machine learning approaches in microbiome research: Challenges and best practices. Front. Microbiol. 2023, 14, 1261889. [Google Scholar] [CrossRef]
- Brugman, S.; Ikeda-Ohtsubo, W.; Braber, S.; Folkerts, G.; Pieterse, C.M.J.; Bakker, P.A.H.M. A comparative review on microbiota manipulation: Lessons from fish, plants, livestock, and human research. Front. Nutr. 2018, 5, 80. [Google Scholar] [CrossRef]
- Shima, H.; Sato, Y.; Sakata, K.; Asakura, T.; Kikuchi, J. Identifying a correlation among qualitative non-numeric parameters in natural fish microbe dataset using machine learning. Appl. Sci. 2022, 12, 5927. [Google Scholar] [CrossRef]
- King, C.H.; Desai, H.; Sylvetsky, A.C.; LoTempio, J.; Ayanyan, S.; Carrie, J.; Crandall, K.A.; Fochtman, B.C.; Gasparyan, L.; Gulzar, N.; et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS ONE 2019, 14, e0206484. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.R.; Lee, J.B.; Kim, M.S.; Whon, T.W.; Hyun, D.W.; Yun, J.H.; Jung, M.J.; Kim, J.Y.; Bae, J.W. Host habitat is the major determinant of the gut microbiome of fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef]
- Rimoldi, S.; Quiroz, K.F.; Kalemi, V.; McMillan, S.; Stubhaug, I.; Martinez-Rubio, L.; Betancor, M.B.; Terova, G. Interactions between nutritional programming, genotype, and gut microbiota in Atlantic salmon: Long-term effects on gut microbiota, fish growth and feed efficiency. Aquaculture 2025, 596, 741813. [Google Scholar] [CrossRef]
- Rimoldi, S.; Montero, D.; Torrecillas, S.; Serradell, A.; Acosta, F.; Haffray, P.; Hostins, B.; Fontanillas, R.; Allal, F.; Bajek, A.; et al. Genetically superior European sea bass (Dicentrarchus labrax) and nutritional innovations: Effects of functional feeds on fish immune response, disease resistance, and gut microbiota. Aquac. Rep. 2023, 33, 101747. [Google Scholar] [CrossRef]
- Ofek, T.; Lalzar, M.; Laviad-Shitrit, S.; Izhaki, I.; Halpern, M. Comparative study of intestinal microbiota composition of six edible fish species. Front. Microbiol. 2021, 12, 760266. [Google Scholar] [CrossRef]
- Degregori, S.; Schiettekatte, N.M.; Casey, J.M.; Brandl, S.J.; Mercière, A.; Amato, K.R.; Mazel, F.; Parravicini, V.; Barber, P.H. Host diet drives gut microbiome convergence between coral reef fishes and mammals. Mol. Ecol. 2024, 33, e17520. [Google Scholar] [CrossRef] [PubMed]
- Uniacke-Lowe, S.; Stanton, C.; Hill, C.; Ross, R.P. The marine fish gut microbiome as a source of novel bacteriocins. Microorganisms 2024, 12, 1346. [Google Scholar] [CrossRef] [PubMed]
- Adhish, M.; Manjubala, I. Effectiveness of zebrafish models in understanding human diseases—A review of models. Heliyon 2023, 9, e14557. [Google Scholar] [CrossRef]
- Sree Kumar, H.; Wisner, A.S.; Refsnider, J.M.; Martyniuk, C.J.; Zubcevic, J. Small fish, big discoveries: Zebrafish shed light on microbial biomarkers for neuro-immune-cardiovascular health. Front. Physiol. 2023, 14, 1186645. [Google Scholar] [CrossRef]
- Tonon, F.; Grassi, G. Zebrafish as an experimental model for human disease. Int. J. Mol. Sci. 2023, 24, 8771. [Google Scholar] [CrossRef]
- Soussi-Yanicostas, N. Zebrafish as a model for neurological disorders. Int. J. Mol. Sci. 2022, 23, 4321. [Google Scholar] [CrossRef]
- Basheer, F.; Sertori, R.; Liongue, C.; Ward, A.C. Zebrafish: A relevant genetic model for human primary immunodeficiency (PID) disorders? Int. J. Mol. Sci. 2023, 24, 6468. [Google Scholar] [CrossRef]
- Bowley, G.; Kugler, E.; Wilkinson, R.; Lawrie, A.; van Eeden, F.; Chico, T.J.; Evans, P.C.; Noël, E.S.; Serbanovic-Canic, J. Zebrafish as a tractable model of human cardiovascular disease. Br. J. Pharmacol. 2022, 179, 900–917. [Google Scholar] [CrossRef] [PubMed]
- Hason, M.; Bartůněk, P. Zebrafish models of cancer—New insights on modeling human cancer in a non-mammalian vertebrate. Genes 2019, 10, 935. [Google Scholar] [CrossRef] [PubMed]
- Mani, A.; Henn, C.; Couch, C.; Patel, S.; Lieke, T.; Chan, J.T.H.; Korytar, T.; Salinas, I. A brain microbiome in salmonids at homeostasis. Sci. Adv. 2024, 10, eado0277. [Google Scholar] [CrossRef]
- Link, C.D. Is there a brain microbiome? Neurosci. Insights 2021, 16, 26331055211018709. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.S.; Harkins, D.M.; Nelson, K.E. Advances in microbiome research for animal health. Annu. Rev. Anim. Biosci. 2021, 9, 289–311. [Google Scholar] [CrossRef]
- Ezenwa, V.O.; Gerardo, N.M.; Inouye, D.W.; Medina, M.; Xavier, J.B. Animal behavior and the microbiome. Science 2012, 338, 198–199. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Voolstra, C.R.; Sweet, M.; Duarte, C.M.; Carvalho, S.; Villela, H.; Lunshof, J.E.; Gram, L.; Woodhams, D.C.; Walter, J.; et al. Harnessing the microbiome to prevent global biodiversity loss. Nat. Microbiol. 2022, 7, 1726–1735. [Google Scholar] [CrossRef]
- Yan, M.; Andersen, T.O.; Pope, P.B.; Yu, Z. Probing the eukaryotic microbes of ruminants with a deep-learning classifier and comprehensive protein databases. Genome Res. 2025, 35, 368–378. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, H.; Qin, L.; Wang, T.; Zhang, Y.; Sun, Y. Interpretable machine learning reveals microbiome signatures strongly associated with dairy cow milk urea nitrogen. iScience 2024, 27, 109955. [Google Scholar] [CrossRef]
- Wang, D.; Tang, G.; Wang, Y.; Yu, J.; Chen, L.; Chen, J.; Wu, Y.; Zhang, Y.; Cao, Y.; Yao, J. Rumen bacterial cluster identification and its influence on rumen metabolites and growth performance of young goats. Anim. Nutr. 2023, 15, 34–44. [Google Scholar] [CrossRef]
- Azouggagh, L.; Ibáñez-Escriche, N.; Martínez-Álvaro, M.; Varona, L.; Casellas, J.; Negro, S.; Casto-Rebollo, C. Characterization of microbiota signatures in Iberian pig strains using machine learning algorithms. Anim. Microbiome 2025, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Jing, X.; Ma, C.; Yang, Y.; Li, Y.; Zhang, Y.; Long, R.; Zheng, H. Massive expansion of the pig gut virome based on global metagenomic mining. NPJ Biofilms Microbiomes 2024, 10, 76. [Google Scholar] [CrossRef]
- Sarpong, N.; Seifert, J.; Bennewitz, J.; Rodehutscord, M.; Camarinha-Silva, A. Microbial signatures and enterotype clusters in fattening pigs: Implications for nitrogen utilization efficiency. Front. Microbiol. 2024, 15, 1354537. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Prenafeta-Boldú, F.; Zingaretti, L.M.; Gonzalez-Rodriguez, O.; Dalmau, A.; Quintanilla, R.; Ballester, M. Gut eukaryotic communities in pigs: Diversity, composition and host genetics contribution. Anim. Microbiome 2020, 2, 18. [Google Scholar] [CrossRef]
- Ram Das, A.; Pillai, N.; Nanduri, B.; Rothrock, M.J., Jr.; Ramkumar, M. Exploring pathogen presence prediction in pastured poultry farms through transformer-based models and attention mechanism explainability. Microorganisms 2024, 12, 1274. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Zhang, X.; Maciel-Guerra, A.; Dong, Y.; Wang, W.; Hu, Y.; Renney, D.; Hu, Y.; Liu, L.; Li, H.; et al. Machine learning and metagenomics reveal shared antimicrobial resistance profiles across multiple chicken farms and abattoirs in China. Nat. Food 2023, 4, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Keum, G.B.; Pandey, S.; Kim, E.S.; Doo, H.; Kwak, J.; Ryu, S.; Choi, Y.; Kang, J.; Kim, S.; Kim, H.B. Understanding the diversity and roles of the ruminal microbiome. J. Microbiol. 2024, 62, 217–230. [Google Scholar] [CrossRef]
- Sanjorjo, R.A.; Tseten, T.; Kang, M.K.; Kwon, M.; Kim, S.W. In pursuit of understanding the rumen microbiome. Fermentation 2023, 9, 114. [Google Scholar] [CrossRef]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef]
- Moraïs, S.; Mizrahi, I. The road not taken: The rumen microbiome, functional groups, and community states. Trends Microbiol. 2019, 27, 538–549. [Google Scholar] [CrossRef]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Tröscher-Mußotter, J.; Saenz, J.S.; Grindler, S.; Meyer, J.; Kononov, S.U.; Mezger, B.; Borda-Molina, D.; Frahm, J.; Dänicke, S.; Camarinha-Silva, A.; et al. Microbiome clusters disclose physiologic variances in dairy cows challenged by calving and lipopolysaccharides. mSystems 2021, 6, e0085621. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Noverr, M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013, 21, 334–341. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, H.; Zhang, C.; Wang, G.; Shi, C.; Li, Z.; Gao, F.; Cui, Y.; Li, M.; Yang, G. Composition and evolutionary characterization of the gut microbiota in pigs. Int. Microbiol. 2024, 27, 993–1008. [Google Scholar] [CrossRef]
- Yang, J.; Chen, R.; Peng, Y.; Chai, J.; Li, Y.; Deng, F. The role of gut archaea in the pig gut microbiome: A mini-review. Front. Microbiol. 2023, 14, 1284603. [Google Scholar] [CrossRef]
- Luo, Y.; Ren, W.; Smidt, H.; Wright, A.G.; Yu, B.; Schyns, G.; McCormack, U.M.; Cowieson, A.J.; Yu, J.; He, J.; et al. Dynamic distribution of gut microbiota in pigs at different growth stages: Composition and contribution. Microbiol. Spectr. 2022, 10, e0068821. [Google Scholar] [CrossRef]
- Wang, C.; Wei, S.; Chen, N.; Xiang, Y.; Wang, Y.; Jin, M. Characteristics of gut microbiota in pigs with different breeds, growth periods and genders. Microb. Biotechnol. 2022, 15, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, Y.; Fu, H.; Xiong, X.; Fang, S.; Jiang, H.; Wu, J.; Yang, H.; Gao, J.; Huang, L. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat. Commun. 2021, 12, 1106. [Google Scholar] [CrossRef]
- Patil, Y.; Gooneratne, R.; Ju, X.H. Interactions between host and gut microbiota in domestic pigs: A review. Gut Microbes 2020, 11, 310–334. [Google Scholar] [CrossRef]
- Pena, R.N.; Noguera, J.L.; García-Santana, M.J.; González, E.; Tejeda, J.F.; Ros-Freixedes, R.; Ibáñez-Escriche, N. Five genomic regions have a major impact on fat composition in Iberian pigs. Sci. Rep. 2019, 9, 2031. [Google Scholar] [CrossRef]
- Urubschurov, V.; Janczyk, P.; Pieper, R.; Souffrant, W.B. Biological diversity of yeasts in the gastrointestinal tract of weaned piglets kept under different farm conditions. FEMS Yeast Res. 2008, 8, 1349–1356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wylezich, C.; Belka, A.; Hanke, D.; Beer, M.; Blome, S.; Höper, D. Metagenomics for broad and improved parasite detection: A proof-of-concept study using swine faecal samples. Int. J. Parasitol. 2019, 49, 769–777. [Google Scholar] [CrossRef]
- Mach, N.; Berri, M.; Estellé, J.; Levenez, F.; Lemonnier, G.; Denis, C.; Leplat, J.J.; Chevaleyre, C.; Billon, Y.; Doré, J.; et al. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015, 7, 554–569. [Google Scholar] [CrossRef]
- Lu, D.; Tiezzi, F.; Schillebeeckx, C.; McNulty, N.P.; Schwab, C.; Shull, C.; Maltecca, C. Host contributes to longitudinal diversity of fecal microbiota in swine selected for lean growth. Microbiome 2018, 6, 4. [Google Scholar] [CrossRef]
- Le Sciellour, M.; Renaudeau, D.; Zemb, O. Longitudinal analysis of the microbiota composition and enterotypes of pigs from post-weaning to finishing. Microorganisms 2019, 7, 622. [Google Scholar] [CrossRef]
- Shang, J.; Tang, X.; Sun, Y. PhaTYP: Predicting the lifestyle for bacteriophages using BERT. Brief. Bioinform. 2023, 24, bbac487. [Google Scholar] [CrossRef] [PubMed]
- Heinritz, S.N.; Mosenthin, R.; Weiss, E. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr. Res. Rev. 2013, 26, 191–209. [Google Scholar] [CrossRef]
- Fathima, S.; Shanmugasundaram, R.; Adams, D.; Selvaraj, R.K. Gastrointestinal microbiota and their manipulation for improved growth and performance in chickens. Foods 2022, 11, 1401. [Google Scholar] [CrossRef]
- Wickramasuriya, S.S.; Park, I.; Lee, K.; Lee, Y.; Kim, W.H.; Nam, H.; Lillehoj, H.S. Role of physiology, immunity, microbiota, and infectious diseases in the gut health of poultry. Vaccines 2022, 10, 172. [Google Scholar] [CrossRef]
- Rychlik, I. Composition and function of chicken gut microbiota. Animals 2020, 10, 103. [Google Scholar] [CrossRef]
- Sood, U.; Gupta, V.; Kumar, R.; Lal, S.; Fawcett, D.; Rattan, S.; Poinern, G.E.J.; Lal, R. Chicken gut microbiome and human health: Past scenarios, current perspectives, and futuristic applications. Indian J. Microbiol. 2020, 60, 2–11. [Google Scholar] [CrossRef]
- Maki, J.J.; Klima, C.L.; Sylte, M.J.; Looft, T. The microbial pecking order: Utilization of intestinal microbiota for poultry health. Microorganisms 2019, 7, 376. [Google Scholar] [CrossRef]
- Kers, J.G.; Velkers, F.C.; Fischer, E.A.J.; Hermes, G.D.A.; Stegeman, J.A.; Smidt, H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018, 9, 235. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Chia, N.; Jeraldo, P.; Sipos, M.; Goldenfeld, N.D.; White, B.A. The microbiome of the chicken gastrointestinal tract. Anim. Health Res. Rev. 2012, 13, 89–99. [Google Scholar] [CrossRef]
- Hwang, D.; Rothrock, M.J., Jr.; Pang, H.; Kumar, G.D.; Mishra, A. Farm management practices that affect the prevalence of Salmonella in pastured poultry farms. LWT 2020, 127, 109423. [Google Scholar] [CrossRef]
- Rothrock, M.J., Jr.; Locatelli, A.; Feye, K.M.; Caudill, A.J.; Guard, J.; Hiett, K.; Ricke, S.C. A microbiomic analysis of a pasture-raised broiler flock elucidates foodborne pathogen ecology along the farm-to-fork continuum. Front. Vet. Sci. 2019, 6, 260. [Google Scholar] [CrossRef]
- Gilroy, R. Spotlight on the avian gut microbiome: Fresh opportunities in discovery. Avian Pathol. 2021, 50, 291–294. [Google Scholar] [CrossRef]
- Luan, Y.; Li, M.; Zhou, W.; Yao, Y.; Yang, Y.; Zhang, Z.; Ringø, E.; Olsen, R.E.; Clarke, J.L.; Xie, S.; et al. The fish microbiota: Research progress and potential applications. Engineering 2023, 29, 137–146. [Google Scholar] [CrossRef]
- Medina-Félix, D.; Garibay-Valdez, E.; Vargas-Albores, F.; Martínez-Porchas, M. Fish disease and intestinal microbiota: A close and indivisible relationship. Rev. Aquac. 2023, 15, 820–839. [Google Scholar] [CrossRef]
- Diwan, A.D.; Harke, S.N.; Gopalkrishna; Panche, A.N. Aquaculture industry prospective from gut microbiome of fish and shellfish: An overview. J. Anim. Physiol. Anim. Nutr. 2022, 106, 441–469. [Google Scholar] [CrossRef]
- Legrand, T.P.; Wynne, J.W.; Weyrich, L.S.; Oxley, A.P. A microbial sea of possibilities: Current knowledge and prospects for an improved understanding of the fish microbiome. Rev. Aquac. 2020, 12, 1101–1134. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Ringø, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 626–640. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The gut microbiota of marine fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- Daly, K.; Kelly, J.; Moran, A.W.; Bristow, R.; Young, I.S.; Cossins, A.R.; Bravo, D.; Shirazi-Beechey, S.P. Host selectively contributes to shaping intestinal microbiota of carnivorous and omnivorous fish. J. Gen. Appl. Microbiol. 2019, 65, 129–136. [Google Scholar] [CrossRef]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595–1608. [Google Scholar] [CrossRef]
- Kokou, F.; Sasson, G.; Friedman, J.; Eyal, S.; Ovadia, O.; Harpaz, S.; Cnaani, A.; Mizrahi, I. Core gut microbial communities are maintained by beneficial interactions and strain variability in fish. Nat. Microbiol. 2019, 4, 2456–2465. [Google Scholar] [CrossRef]
- Zarkasi, K.Z.; Abell, G.C.; Taylor, R.S.; Neuman, C.; Hatje, E.; Tamplin, M.L.; Katouli, M.; Bowman, J.P. Pyrosequencing-based characterization of gastrointestinal bacteria of Atlantic salmon (Salmo salar L.) within a commercial mariculture system. J. Appl. Microbiol. 2014, 117, 18–27. [Google Scholar] [CrossRef]
- Yoshimizu, M.; Kimura, T. Study on the intestinal microflora of salmonids. Fish Pathol. 1976, 10, 243–259. [Google Scholar] [CrossRef]
- Domingo-Bretón, R.; Cools, S.; Moroni, F.; Belenguer, A.; Calduch-Giner, J.A.; Croes, E.; Holhorea, P.G.; Naya-Català, F.; Boon, H.; Pérez-Sánchez, J. Intestinal microbiota shifts by dietary intervention during extreme heat summer episodes in farmed gilthead sea bream (Sparus aurata). Aquac. Rep. 2025, 40, 102566. [Google Scholar] [CrossRef]
- Hasan, I.; Rimoldi, S.; Saroglia, G.; Terova, G. Sustainable fish feeds with insects and probiotics positively affect freshwater and marine fish gut microbiota. Animals 2023, 13, 1633. [Google Scholar] [CrossRef]
- Torrecillas, S.; Rimoldi, S.; Montero, D.; Serradell, A.; Acosta, F.; Fontanillas, R.; Allal, F.; Haffray, P.; Bajek, A.; Terova, G. Genotype x nutrition interactions in European sea bass (Dicentrarchus labrax): Effects on gut health and intestinal microbiota. Aquaculture 2023, 574, 739639. [Google Scholar] [CrossRef]
- Bondad-Reantaso, M.G.; MacKinnon, B.; Karunasagar, I.; Fridman, S.; Alday-Sanz, V.; Brun, E.; Le Groumellec, M.; Li, A.; Surachetpong, W.; Karunasagar, I.; et al. Review of alternatives to antibiotic use in aquaculture. Rev. Aquac. 2023, 15, 1421–1451. [Google Scholar] [CrossRef]
- Wong, S.; Rawls, J.F. Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol. Ecol. 2012, 21, 3100–3102. [Google Scholar] [CrossRef]
- Butt, R.L.; Volkoff, H. Gut microbiota and energy homeostasis in fish. Front. Endocrinol. 2019, 10, 9. [Google Scholar] [CrossRef]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Clements, K.D.; Angert, E.R.; Montgomery, W.L.; Choat, J.H. Intestinal microbiota in fishes: What’s known and what’s not. Mol. Ecol. 2014, 23, 1891–1898. [Google Scholar] [CrossRef]
- Monteiro, M.; Rimoldi, S.; Costa, R.S.; Kousoulaki, K.; Hasan, I.; Valente, L.M.P.; Terova, G. Polychaete (Alitta virens) meal inclusion as a dietary strategy for modulating gut microbiota of European seabass (Dicentrarchus labrax). Front. Immunol. 2023, 14, 1266947. [Google Scholar] [CrossRef]
- Banerjee, G.; Ray, A.K. Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 2017, 72, 1–11. [Google Scholar] [CrossRef]
- Turner, J.W., Jr.; Cheng, X.; Saferin, N.; Yeo, J.Y.; Yang, T.; Joe, B. Gut microbiota of wild fish as reporters of compromised aquatic environments sleuthed through machine learning. Physiol. Genomics 2022, 54, 177–185. [Google Scholar] [CrossRef]
- Zhang, B.; Xiao, J.; Liu, H.; Zhai, D.; Wang, Y.; Liu, S.; Xiong, F.; Xia, M. Vertical habitat preferences shape the fish gut microbiota in a shallow lake. Front. Microbiol. 2024, 15, 1341303. [Google Scholar] [CrossRef]
- Soriano, B.; Hafez, A.I.; Naya-Català, F.; Moroni, F.; Moldovan, R.A.; Toxqui-Rodríguez, S.; Piazzon, M.C.; Arnau, V.; Llorens, C.; Pérez-Sánchez, J. SAMBA: Structure-learning of aquaculture microbiomes using a Bayesian approach. Genes 2023, 14, 1650. [Google Scholar] [CrossRef]
- Yuniarti, I.; Glenk, K.; McVittie, A.; Nomosatryo, S.; Triwisesa, E.; Suryono, T.; Santoso, A.B.; Ridwansyah, I. An application of Bayesian Belief Networks to assess management scenarios for aquaculture in a complex tropical lake system in Indonesia. PLoS ONE 2021, 16, e0250365. [Google Scholar] [CrossRef]
- Barbedo, J.G. A review on the use of computer vision and artificial intelligence for fish recognition, monitoring, and management. Fishes 2022, 7, 335. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, S.; Liu, J.; Gao, Q.; Dong, S.; Zhou, C. Deep learning for smart fish farming: Applications, opportunities and challenges. Rev. Aquac. 2021, 13, 66–90. [Google Scholar] [CrossRef]
- Li, J.; Lian, Z.; Wu, Z.; Zeng, L.; Mu, L.; Yuan, Y.; Bai, H.; Guo, Z.; Mai, K.; Tu, X.; et al. Artificial intelligence-based method for the rapid detection of fish parasites (Ichthyophthirius multifiliis, Gyrodactylus kobayashii, and Argulus japonicus). Aquaculture 2023, 563, 738790. [Google Scholar] [CrossRef]
- Iqbal, U.; Li, D.; Akhter, M. Intelligent diagnosis of fish behavior using deep learning method. Fishes 2022, 7, 201. [Google Scholar] [CrossRef]
- Zhou, C.; Lin, K.; Xu, D.; Chen, L.; Guo, Q.; Sun, C.; Yang, X. Near infrared computer vision and neuro-fuzzy model-based feeding decision system for fish in aquaculture. Comput. Electron. Agric. 2018, 146, 114–124. [Google Scholar] [CrossRef]
- De Verdal, H.; Komen, H.; Quillet, E.; Chatain, B.; Allal, F.; Benzie, J.A.; Vandeputte, M. Improving feed efficiency in fish using selective breeding: A review. Rev. Aquac. 2018, 10, 833–851. [Google Scholar] [CrossRef]
- Pěnka, T.; Malinovskyi, O.; Imentai, A.; Kolářová, J.; Kučera, V.; Policar, T. Evaluation of different feeding frequencies in RAS-based juvenile pikeperch (Sander lucioperca) aquaculture. Aquaculture 2023, 562, 738815. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, Y.G.; Yang, X.G.; Gao, Q.F.; Chen, Y.N.; Ren, Y.C.; Dong, S.L. Optimizing feeding frequencies in fish: A meta-analysis and machine learning approach. Aquaculture 2025, 595, 741678. [Google Scholar] [CrossRef]
- Young, T.; Laroche, O.; Walker, S.P.; Miller, M.R.; Casanovas, P.; Steiner, K.; Esmaeili, N.; Zhao, R.; Bowman, J.P.; Wilson, R.; et al. Prediction of feed efficiency and performance-based traits in fish via integration of multiple omics and clinical covariates. Biology 2023, 12, 1135. [Google Scholar] [CrossRef]
- Hornung, R.; Wright, M.N. Block Forests: Random forests for blocks of clinical and omics covariate data. BMC Bioinform. 2019, 20, 358. [Google Scholar] [CrossRef]
- Navarro, L.C.; Azevedo, A.; Matos, A.; Rocha, A.; Ozório, R. Predicting weight dispersion in seabass aquaculture using discrete event system simulation and machine learning modeling. Aquac. Rep. 2024, 38, 102315. [Google Scholar] [CrossRef]
- Slette, H.T.; Asbjørnslett, B.E.; Pettersen, S.S.; Erikstad, S.O. Simulating emergency response for large-scale fish welfare emergencies in sea-based salmon farming. Aquac. Eng. 2022, 97, 102243. [Google Scholar] [CrossRef]
- Humer, C.; Hinterreiter, A.; Leichtmann, B.; Mara, M.; Streit, M. Reassuring, misleading, debunking: Comparing effects of XAI methods on human decisions. ACM Trans. Interact. Intell. Syst. 2024, 14, 16. [Google Scholar] [CrossRef]
- Tourab, H.; Pérez, L.L.; Arroyo-Gallego, P.; Georga, E.; Rujas, M.; Ponziani, F.R.; Torrego-Ellacuría, M.; Merino-Barbancho, B.; Ciuti, G.; Fotiadis, D.; et al. The use of machine learning and explainable artificial intelligence in gut microbiome research: A scoping review. TechRxiv 2024. [Google Scholar] [CrossRef]
- Boge, F.; Mosig, A. Causality and scientific explanation of artificial intelligence systems in biomedicine. Pflugers Arch.-Eur. J. Physiol. 2025, 477, 543–554. [Google Scholar] [CrossRef]
- Odriozola, I.; Rasmussen, J.A.; Gilbert, M.T.P.; Limborg, M.T.; Alberdi, A. A practical introduction to holo-omics. Cell Rep. Methods 2024, 4, 100820. [Google Scholar] [CrossRef]
- Alberdi, A.; Andersen, S.B.; Limborg, M.T.; Dunn, R.R.; Gilbert, M.T.P. Disentangling host-microbiota complexity through hologenomics. Nat. Rev. Genet. 2022, 23, 281–297. [Google Scholar] [CrossRef]
- Intelligence, N.M. The rewards of reusable machine learning code. Nat. Mach. Intell. 2024, 6, 369. [Google Scholar] [CrossRef]
- Haibe-Kains, B.; Adam, G.A.; Hosny, A.; Khodakarami, F.; Massive Analysis Quality Control (MAQC) Society Board of Directors; Waldron, L.; Wang, B.; McIntosh, C.; Goldenberg, A.; Kundaje, A.; et al. Transparency and reproducibility in artificial intelligence. Nature 2020, 586, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Reynoso-García, J.; Miranda-Santiago, A.E.; Meléndez-Vázquez, N.M.; Acosta-Pagán, K.; Sánchez-Rosado, M.; Díaz-Rivera, J.; Rosado-Quiñones, A.M.; Acevedo-Márquez, L.; Cruz-Roldán, L.; Tosado-Rodríguez, E.L.; et al. A complete guide to human microbiomes: Body niches, transmission, development, dysbiosis, and restoration. Front. Syst. Biol. 2022, 2, 951403. [Google Scholar] [CrossRef] [PubMed]
- Probul, N.; Huang, Z.; Saak, C.C.; Baumbach, J.; List, M. AI in microbiome-related healthcare. Microb. Biotechnol. 2024, 17, e70027. [Google Scholar] [CrossRef]
- Ratiner, K.; Ciocan, D.; Abdeen, S.K.; Elinav, E. Utilization of the microbiome in personalized medicine. Nat. Rev. Microbiol. 2024, 22, 291–308. [Google Scholar] [CrossRef] [PubMed]
- Kirk, D.; Kok, E.; Tufano, M.; Tekinerdogan, B.; Feskens, E.J.M.; Camps, G. Machine learning in nutrition research. Adv. Nutr. 2022, 13, 2573–2589. [Google Scholar] [CrossRef]
- Forcina, G.; Pérez-Pardal, L.; Carvalheira, J.; Beja-Pereira, A. Gut microbiome studies in livestock: Achievements, challenges, and perspectives. Animals 2022, 12, 3375. [Google Scholar] [CrossRef] [PubMed]
- Gerber, G.K. AI in microbiome research: Where have we been, where are we going? Cell Host Microbe 2024, 32, 1230–1234. [Google Scholar] [CrossRef]
- Akyildiz, I.F.; Chen, J.; Ghovanloo, M.; Guler, U.; Ozkaya-Ahmadov, T.; Pierobon, M.; Sarioglu, A.F.; Unluturk, B.D. Microbiome-gut-brain axis as a biomolecular communication network for the internet of bio-nanothings. IEEE Access 2019, 7, 136161–136175. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, T.; Zhang, Z.; Chen, J. An intelligent grazing development strategy for unmanned animal husbandry in China. Drones 2023, 7, 542. [Google Scholar] [CrossRef]
- Centelleghe, C.; Carraro, L.; Gonzalvo, J.; Rosso, M.; Esposti, E.; Gili, C.; Bonato, M.; Pedrotti, D.; Cardazzo, B.; Povinelli, M.; et al. The use of unmanned aerial vehicles (UAVs) to sample the blow microbiome of small cetaceans. PLoS ONE 2020, 15, e0235537. [Google Scholar] [CrossRef]
- Baindara, P.; Dinata, R.; Mandal, S.M. Marine bacteriocins: An evolutionary gold mine to payoff antibiotic resistance. Mar. Drugs 2024, 22, 388. [Google Scholar] [CrossRef] [PubMed]
- Merwin, N.J.; Mousa, W.K.; Dejong, C.A.; Skinnider, M.A.; Cannon, M.J.; Li, H.; Dial, K.; Gunabalasingam, M.; Johnston, C.; Magarvey, N.A. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products. Proc. Natl. Acad. Sci. USA 2020, 117, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Gayathri, K.V.; Aishwarya, S.; Kumar, P.S.; Rajendran, U.R.; Gunasekaran, K. Metabolic and molecular modelling of zebrafish gut biome to unravel antimicrobial peptides through metagenomics. Microb. Pathog. 2021, 154, 104862. [Google Scholar] [CrossRef]
- Xia, H.; Chen, H.; Cheng, X.; Yin, M.; Yao, X.; Ma, J.; Huang, M.; Chen, G.; Liu, H. Zebrafish: An efficient vertebrate model for understanding role of gut microbiota. Mol. Med. 2022, 28, 161. [Google Scholar] [CrossRef]
- Lu, H.; Li, P.; Huang, X.; Wang, C.H.; Li, M.; Xu, Z.Z. Zebrafish model for human gut microbiome-related studies: Advantages and limitations. Med. Microecol. 2021, 8, 100042. [Google Scholar] [CrossRef]
- Kanika, N.H.; Liaqat, N.; Chen, H.; Ke, J.; Lu, G.; Wang, J.; Wang, C. Fish gut microbiome and its application in aquaculture and biological conservation. Front. Microbiol. 2025, 15, 1521048. [Google Scholar] [CrossRef]
- Rimoldi, S.; Di Rosa, A.R.; Armone, R.; Chiofalo, B.; Hasan, I.; Saroglia, M.; Kalemi, V.; Terova, G. The replacement of fish meal with poultry by-product meal and insect exuviae: Effects on growth performance, gut health and microbiota of the European seabass, Dicentrarchus labrax. Microorganisms 2024, 12, 744. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.; Di Rosa, A.R.; Oteri, M.; Chiofalo, B.; Hasan, I.; Saroglia, M.; Terova, G. The impact of diets containing Hermetia illucens meal on the growth, intestinal health, and microbiota of gilthead seabream (Sparus aurata). Fish Physiol. Biochem. 2024, 50, 1003–1024. [Google Scholar] [CrossRef] [PubMed]
- Terova, G.; Gini, E.; Gasco, L.; Moroni, F.; Antonini, M.; Rimoldi, S. Effects of full replacement of dietary fishmeal with insect meal from Tenebrio molitor on rainbow trout gut and skin microbiota. J. Anim. Sci. Biotechnol. 2021, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Ghosh, A.R. Role of artificial intelligence (AI) in fish growth and health status monitoring: A review on sustainable aquaculture. Aquac. Int. 2024, 32, 2791–2820. [Google Scholar] [CrossRef]
- Wang, T.; Xu, X.; Wang, C.; Li, Z.; Li, D. From smart farming towards unmanned farms: A new mode of agricultural production. Agriculture 2021, 11, 145. [Google Scholar] [CrossRef]
- Wang, C.; Li, Z.; Wang, T.; Xu, X.; Zhang, X.; Li, D. Intelligent fish farm—The future of aquaculture. Aquac. Int. 2021, 29, 2681–2711. [Google Scholar] [CrossRef]
- Bradley, D.; Merrifield, M.; Miller, K.M.; Lomonico, S.; Wilson, J.R.; Gleason, M.G. Opportunities to improve fisheries management through innovative technology and advanced data systems. Fish Fish. 2019, 20, 564–583. [Google Scholar] [CrossRef]
- Ashraf Rather, M.; Ahmad, I.; Shah, A.; Ahmad Hajam, Y.; Amin, A.; Khursheed, S.; Ahmad, I.; Rasool, S. Exploring opportunities of artificial intelligence in aquaculture to meet increasing food demand. Food Chem. X 2024, 22, 101309. [Google Scholar] [CrossRef]
- Mustapha, U.F.; Alhassan, A.W.; Jiang, D.N.; Li, G.L. Sustainable aquaculture development: A review on the roles of cloud computing, internet of things and artificial intelligence (CIA). Rev. Aquac. 2021, 13, 2076–2091. [Google Scholar] [CrossRef]
- Vo, T.T.; Ko, H.; Huh, J.H.; Kim, Y. Overview of smart aquaculture system: Focusing on applications of machine learning and computer vision. Electronics 2021, 10, 2882. [Google Scholar] [CrossRef]
- Civas, M.; Cetinkaya, O.; Kuscu, M.; Akan, O.B. Universal transceivers: Opportunities and future directions for the Internet of Everything (IoE). Front. Commun. Netw. 2021, 2, 733664. [Google Scholar] [CrossRef]
- Babar, A.Z.; Akan, O.B. Sustainable and precision agriculture with the Internet of Everything (IoE). arXiv 2024, arXiv:2404.06341. [Google Scholar] [CrossRef]
- Tian, L.; Fang, G.; Li, G.; Li, L.; Zhang, T.; Mao, Y. Metagenomic approach revealed the mobility and co-occurrence of antibiotic resistomes between non-intensive aquaculture environment and human. Microbiome 2024, 12, 107. [Google Scholar] [CrossRef] [PubMed]
- Lorgen-Ritchie, M.; Uren Webster, T.; McMurtrie, J.; Bass, D.; Tyler, C.R.; Rowley, A.; Martin, S.A. Microbiomes in the context of developing sustainable intensified aquaculture. Front. Microbiol. 2023, 14, 1200997. [Google Scholar] [CrossRef]
- Thakur, I.S.; Roy, D. Environmental DNA and RNA as records of human exposome, including biotic/abiotic exposures and its implications in the assessment of the role of environment in chronic diseases. Int. J. Mol. Sci. 2020, 21, 4879. [Google Scholar] [CrossRef]
- Zhang, Y.; Thompson, K.N.; Branck, T.; Yan, Y.; Nguyen, L.H.; Franzosa, E.A.; Huttenhower, C. Metatranscriptomics for the human microbiome and microbial community functional profiling. Annu. Rev. Biomed. Data Sci. 2021, 4, 279–311. [Google Scholar] [CrossRef]
- Rieder, J.; Berezenko, A.; Meziti, A.; Adrian-Kalchhauser, I. The future of pathogen detection in aquaculture: Miniature labs, field-compatible assays, environmental DNA and RNA, CRISPR and metatranscriptomics. Aquacult. Fish Fish. 2025, 5, e70062. [Google Scholar] [CrossRef]
- Oh, M.; Zhang, L. DeepMicro: Deep representation learning for disease prediction based on microbiome data. Sci. Rep. 2020, 10, 6026. [Google Scholar] [CrossRef] [PubMed]
| Category | Health Condition | Algorithm | Data Type | Reference |
|---|---|---|---|---|
| Patient stratification | Breast cancer | XGB | 16S rRNA gene | [29] |
| Parkinson’s disease | LSTM + SVM | metagenomics | [30] | |
| Heart failure and depression | RF | metagenomics, metabolomics | [31] | |
| Obesity | SVM | metagenomics | [32] | |
| Liver disease | CNN | 16S rRNA gene | [33] | |
| Microbiota composition analysis | Microbial dysbiosis | k-NN | metagenomics | [34] |
| Dietary intake and allergic rhinitis | NMDS, NMF, PCA, PCoA | 16S rRNA gene | [35] | |
| Geography- and season-based variation | MDS + PAM | 16S rRNA gene | [36] | |
| Pregnancy | PAM | 16S rRNA gene | [37] | |
| Pregnancy | PAM | 16S rRNA gene | [38] |
| Livestock Group | Algorithm | Data Type | Purpose | Reference |
|---|---|---|---|---|
| Ruminants | FNN + CNN | metagenomics | identification of fungal and protozoan sequences | [71] |
| RF | 16S rRNA gene | stratification based on MUN values | [72] | |
| PAM | 16S rRNA gene | investigation of rumen bacterial clusters | [73] | |
| Swine | DT, RF, XGB, AB, CB, SVM, GNB + PLS-DA, LR + PLS-DA | 16S rRNA gene | stratification based on breeding status | [74] |
| BERT ⁕ | metagenomics | identification of viral sequences | [75] | |
| PAM | 16S rRNA gene | association between microbial clusters and NUE values | [76] | |
| GB | ITS and 18S rRNA gene | association between eukaryotic communities and body weight | [77] | |
| Poultry | transformer | 16S rRNA gene | pathogen prediction in microbial clusters | [78] |
| ET | metagenomics | prediction of antibiotic resistance/susceptibility | [79] |
| Algorithm | Data Type | Purpose | Reference |
|---|---|---|---|
| PAM | 16S rRNA gene | microbial clustering according to trophic levels | [135] |
| BN | 16S rRNA gene | network-based modeling of microbiome relationships with rearing conditions | [136] |
| RF | 16S rRNA gene | contamination monitoring in aquatic environments | [134] |
| Task Type | Algorithm | Purpose | Reference |
|---|---|---|---|
| Image-based | YOLO | automatic identification of parasite infection | [140] |
| CNN | automatic identification of feeding behavior | [141] | |
| NIR-CV + ANFIS | automatic feeding decision making | [142] | |
| Non-image-based | GB | quantification of feeding frequency impact on ADG and FCR | [134] |
| RF | automatic quantification of weight dispersion | [137] | |
| RB | prediction of growth performance metrics | [135] |
| Research Domain | Data Available | Data Unavailable |
|---|---|---|
| Human health | [29,30,31,34,36,37,38] | [32,33,35] |
| Animal husbandry | [71,72,73,74,75,76,77,79] | [78] |
| Aquaculture | [134,136] | [135,140,141,142,145,146,148] |
| Research Domain | Code Available | Code Unavailable |
|---|---|---|
| Human health | [34,37] | [29,30,31,32,33,35,36,38] |
| Animal husbandry | [71,72,75,79] | [73,74,76,77,78] |
| Aquaculture | [136] | [134,135,140,141,142,145,146,148] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizzi, S.; Saroglia, G.; Kalemi, V.; Rimoldi, S.; Terova, G. Artificial Intelligence in Microbiome Research and Beyond: Connecting Human Health, Animal Husbandry, and Aquaculture. Appl. Sci. 2025, 15, 9781. https://doi.org/10.3390/app15179781
Rizzi S, Saroglia G, Kalemi V, Rimoldi S, Terova G. Artificial Intelligence in Microbiome Research and Beyond: Connecting Human Health, Animal Husbandry, and Aquaculture. Applied Sciences. 2025; 15(17):9781. https://doi.org/10.3390/app15179781
Chicago/Turabian StyleRizzi, Silvio, Giulio Saroglia, Violeta Kalemi, Simona Rimoldi, and Genciana Terova. 2025. "Artificial Intelligence in Microbiome Research and Beyond: Connecting Human Health, Animal Husbandry, and Aquaculture" Applied Sciences 15, no. 17: 9781. https://doi.org/10.3390/app15179781
APA StyleRizzi, S., Saroglia, G., Kalemi, V., Rimoldi, S., & Terova, G. (2025). Artificial Intelligence in Microbiome Research and Beyond: Connecting Human Health, Animal Husbandry, and Aquaculture. Applied Sciences, 15(17), 9781. https://doi.org/10.3390/app15179781






