PneumoNet: Artificial Intelligence Assistance for Pneumonia Detection on X-Rays
Abstract
1. Introduction
2. Background and Related Work
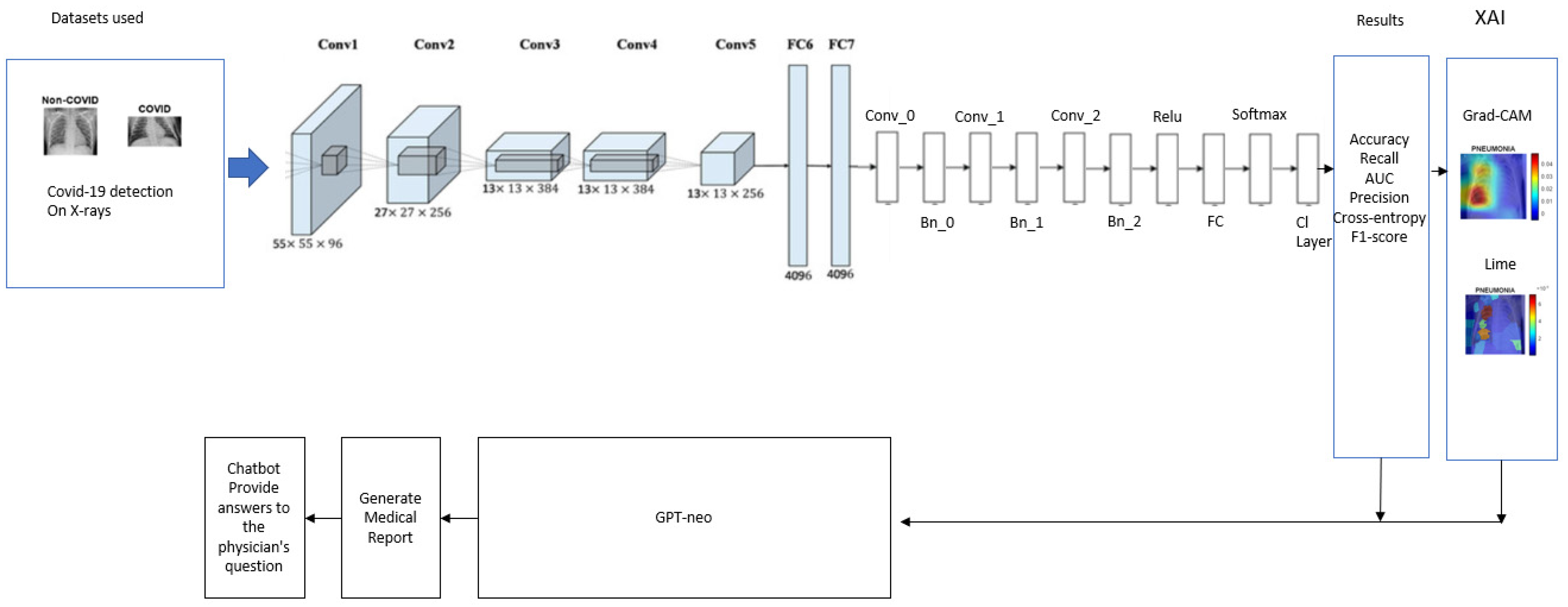
3. Pneumonet Framework
3.1. Pneumonia Classification Module
Tests, Validations, and Results Discussion
| Study | Method | Dataset Train and Test | Results |
|---|---|---|---|
| [26] | AlexNet, GoogLeNet and ResNet | Chest X-ray14 | ACC = 90.70% |
| Dataset #1 [8] | |||
| [19] | Deep learning model using VGG16 | Dataset #1 [8] | ACC = 95.40% |
| [27] | HOG + CNN | Dataset #1 [8] | ACC = 96.70% |
| Dataset #2 [9] | |||
| [25] | VGG-16 | Dataset #2 [9] | ACC = 87.50% |
| [28] | Custom trained Sequential CNN Arch. | Dataset #2 [9] | ACC = 90.20% |
| [29] | Generated Models | Dataset #2 [9] | ACC = 83.30% |
| [30] | VGG-16 | Dataset #2 [9] | ACC = 96.40% |
| [20] | MobileNet | Dataset #2 [9] | ACC = 94.23% |
| [31] | Comb. Inceptionv3 and Logistic Regression | Dataset #2 [9] | ACC = 79.32% |
| Dataset #3 [10] | |||
| [32] | EL Approach | Dataset #3 [10] | ACC = 93.91% |
| [33] | Quaternion-customised DNN Architecture | Dataset #3 [10] | ACC = 94.53% |
| [34] | DenseNet169 | Dataset #3 [10] | ACC = 95.72% |
| [35] | CNN + Modified Dropout Model | Dataset #3 [10] | ACC = 97.20% |
| [36] | Layer-wise Relevance Propagation (LRP) | Dataset #3 [10] | ACC = 91.00% |
| Datasest #1, 2, and 3 [8,9,10] | |||
| [37] | Modified AlexNet | Dataset #1, 2, and 3 [8,9,10] | ACC = 93.42% |
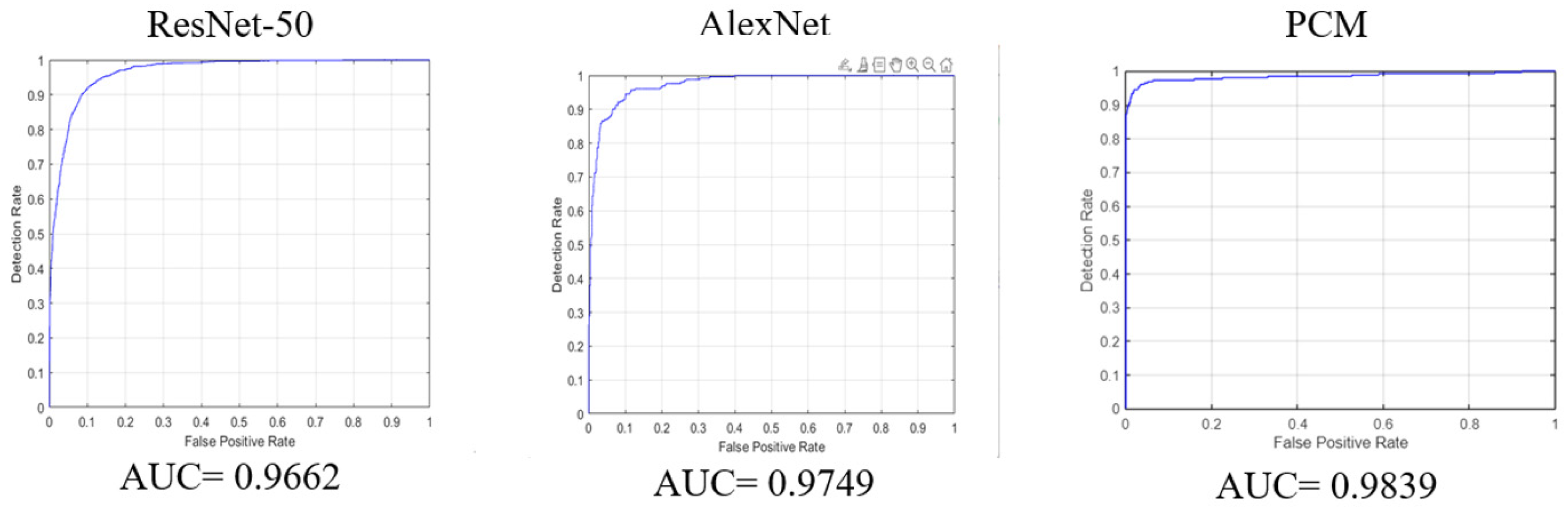
| PCM | PCM: AlexNet backbone + CNN + BN + FC | Dataset #1 [8] | ACC = 96.70% AUC = 98.39% |
| Dataset #2 [9] | ACC = 98.70% AUC = 99.70% | ||
| Dataset #3 [10] | ACC = 97.70% AUC = 98.04% | ||
| Dataset #1, 2, and 3 [8,9,10] | ACC = 98.70% AUC = 99.70% | ||
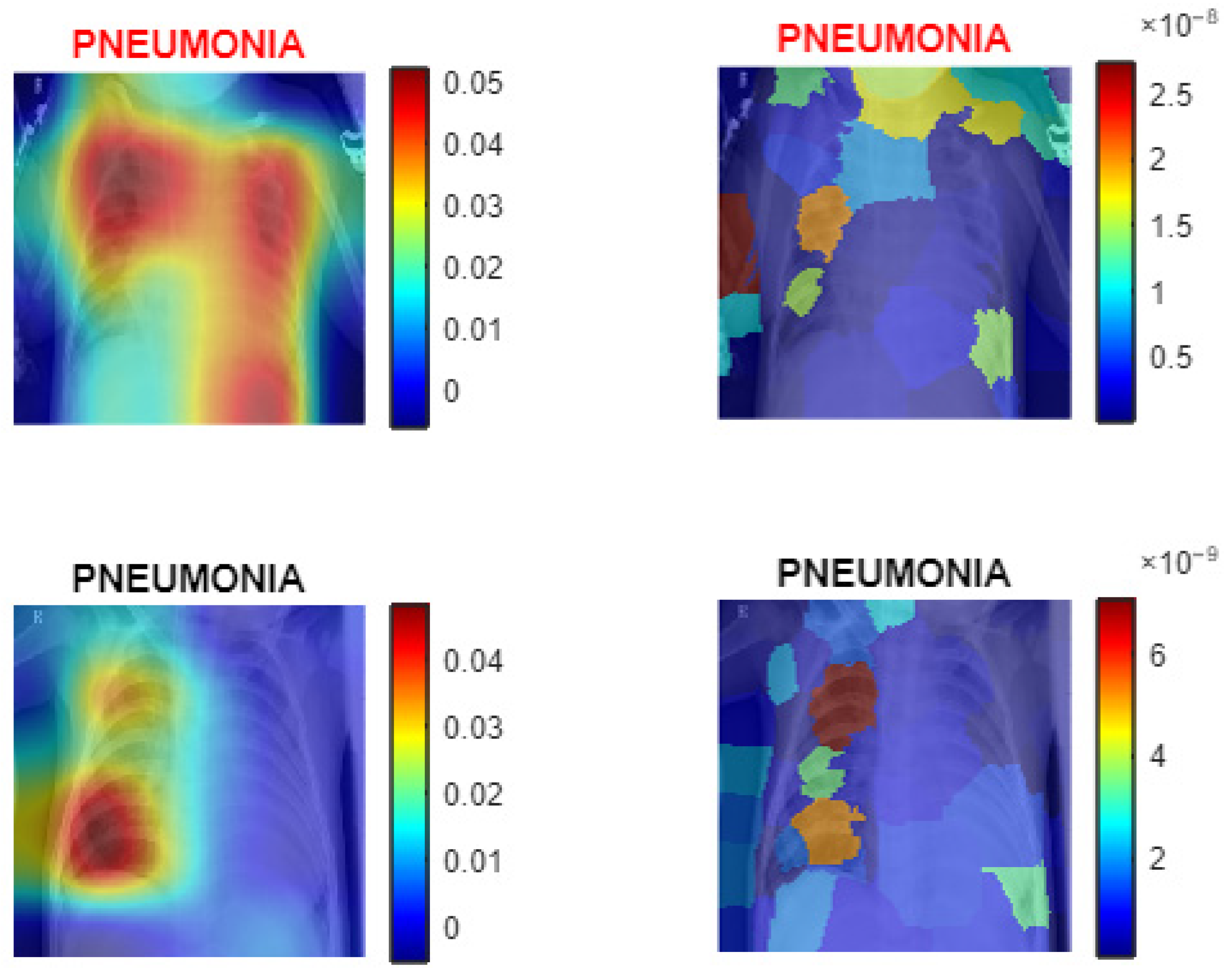
3.2. XAI Module
| Algorithm 1: PneumoNet—image classification with modified AlexNet and explainable AI |
| Input: Image Data Path: Directory containing X-ray images. Pretrained AlexNet model. Training dataset percentage = 80%. Test dataset percentage = 20%. Validation dataset percentage = 10% of the Test dataset. Learning_rate: = MiniBatchSize: = 128. NumberOfEpochs: = 10. Output: Trained Deep Learning Model: . Predicted Labels: Performance Metrics: Accuracy: , Precision: = Recall = F1-Score: = Confusion Matrix: where represents the count of true class classified as ROC curve: AUC value calculated from true positive rate (TPR) and false positive rate (FPR). XAI Visualizations: Explanatory visuals for model predictions using Grad-CAM and LIME. Steps of the Algorithm: Load the dataset , assign labels based on folder structure and divide into subsets: ← Split (, 0.8, 0.2) ← 0.1 × If total images are sufficient: Assign 80% to training, 10% of training for validation, and 20% to testing. End If For each image in the dataset: If is grayscale: Convert to RGB by replicating channels: . End If Resize to the required input size: . End For Apply transformations to enhance the training data and retain AlexNet layers up to the penultimate layers. For each new layer: Add convolutional layers, batch normalisation, and ReLU activation. Add fully connected, SoftMax, and classification layers. End For Set learning rate α, mini-batch size , epochs , and validation parameters and Train the modified network. For each epoch : Update model weights using Adam optimizer: Perform validation checks periodically. End For Test the trained model on . For each class : Calculate precision: , Calculate recall: , Calculate F1-score: End For Predict the class label for each image . If use_gradcam: For each selected image : Generate a class activation map and overlay it on . End For End If If use_ LIME: For each selected image : Compute feature importance and overlay it on . End For End If Provide the trained model, evaluation metrics, and visualisations. |
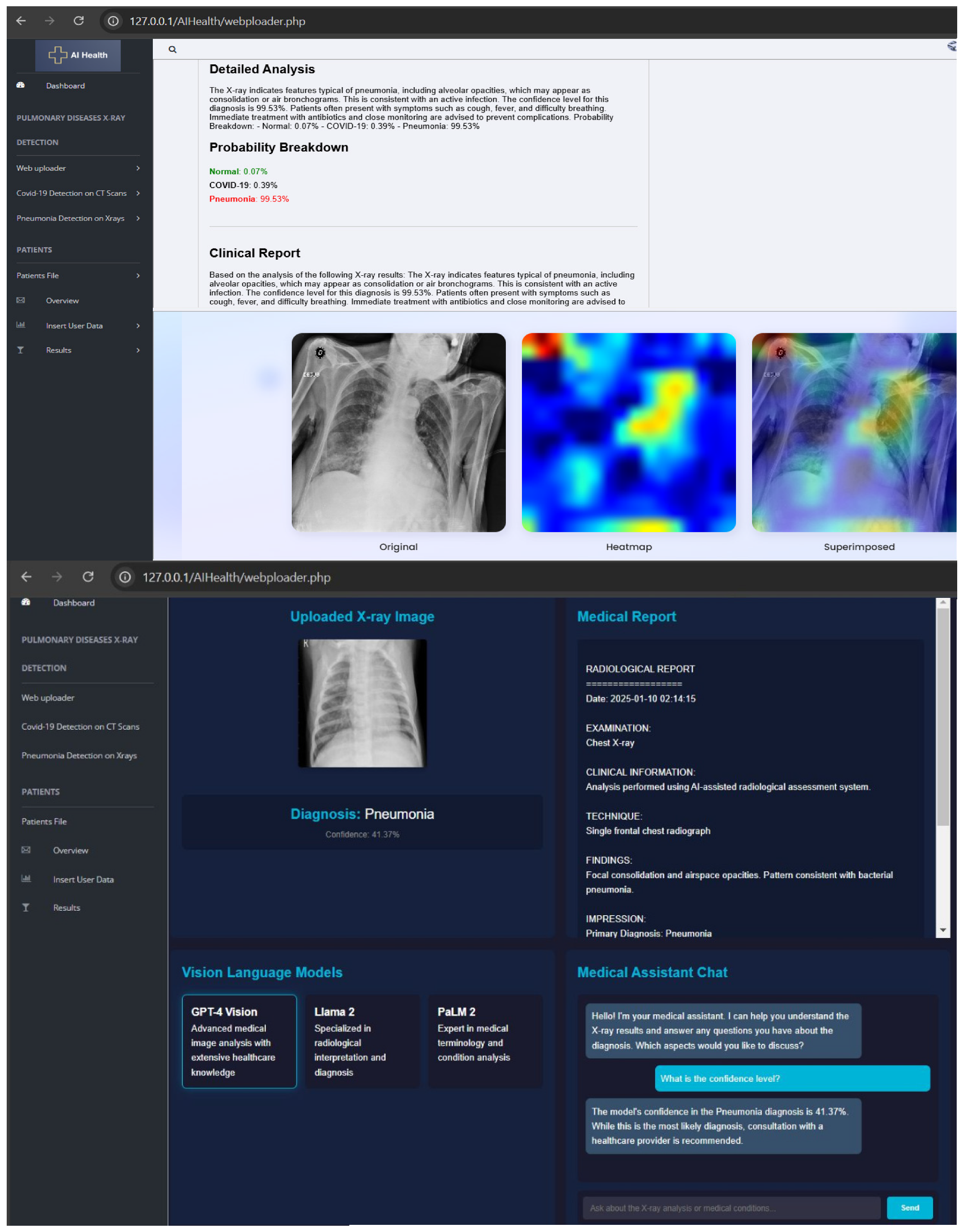
3.3. Medical Report Module and User Interface
| Algorithm 2: Application with artificial intelligence assistance |
| Input: An uploaded chest X-ray image in JPG/PNG format via a web interface. User text query (optional) for chatbot interaction. Output: A classification result: “Normal” or “Pneumonia”. A detailed medical report explaining the implications of the classification. Optional chatbot-generated medical responses based on user input. Steps of the Algorithm: Initialize the Flask application and configure the upload folder. Load the ONNX model using the ONNX Runtime. Load the GPT-Neo language model and tokenizer. Define the image preprocessing function: If the image is provided: Convert it to RGB format. Resize to (227, 227). Normalize to the range (0, 1). Convert to NCHW format and add a batch dimension. End If Define the prediction function: If a pre-processed image is given: Pass the image to the ONNX model. Get the predicted class with the highest probability. If the class is 1: Return “Pneumonia.” Else: Return “Normal.” End If End If Define the report generation function: If a prediction result is available: Create a prompt based on the prediction. Generate a medical explanation using GPT-Neo. Postprocess the response to ensure clarity. End If Create the home route: If the method is GET: Render the index.html page. Else If the method is POST: Save the uploaded image. Preprocess the image. Predict the result using the model. Generate the report. Render the result.html page with the result, report, and image. End Else If End If Create the upload route: If an image is uploaded: Save the image. Preprocess and predict. Generate the medical report. Render the result.html page. End If Create the chatbot route: If a user query is received: Generate a coherent response using GPT-Neo. Postprocess the response. Return the response as a JSON object. |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krajcik, S.; Haniskova, T.; Mikus, P. Pneumonia in Older People. Rev. Clin. Gerontol. 2010, 21, 16–27. [Google Scholar] [CrossRef]
- Stephen, O.; Sain, M.; Maduh, U.J.; Jeong, D.-U. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J. Healthc. Eng. 2019, 2019, 4180949. [Google Scholar] [CrossRef] [PubMed]
- Shuja, J.; Alanazi, E.; Alasmary, W.; Alashaikh, A. COVID-19 Open Source Data Sets: A Comprehensive Survey. Appl. Intell. 2021, 51, 1296–1325. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Plebani, M. Laboratory Abnormalities in Patients with COVID-2019 Infection. Clin. Chem. Lab. Med. 2020, 58, 1131–1134. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Gupta, K.D.; Islam, M.M.; Sen, S.; Rahman, M.L.; Shakhawat Hossain, M. COVID-19 Symptoms Detection Based on NasNetMobile with Explainable AI Using Various Imaging Modalities. Mach. Learn. Knowl. Extr. 2020, 2, 490–504. [Google Scholar] [CrossRef]
- Chhajed, G.; Surpur, S.; Suryawanshi, A.; Sherekar, H. A Review on Key Algorithms for Pneumonia Detection in X-ray Images. AIP Conf. Proc. 2024, 3156, 070007. [Google Scholar]
- Antunes, C.; Rodrigues, J.; Cunha, A. CTCovid19: Automatic Covid-19 Model for Computed Tomography Scans Using Deep Learning. Intell. Based Med. 2024, 11, 100190. [Google Scholar] [CrossRef]
- Chest X-Ray (COVID-19 & Pneumonia). Available online: https://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumonia (accessed on 2 May 2025).
- Kermany, D. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Mendeley Dataset 2018. Available online: https://data.mendeley.com/datasets/rscbjbr9sj/2 (accessed on 2 May 2025).
- Mooney, P. Chest X-Ray Images (Pneumonia). Available online: https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia (accessed on 2 May 2025).
- Xu, X. A Systematic Review: Deep Learning-Based Methods for Pneumonia Region Detection. Appl. Comput. Eng. 2023, 22, 210–217. [Google Scholar] [CrossRef]
- Yaraghi, S.; Khosravi, F. Diagnosis of Pneumonia from Chest X-Ray Images Using Transfer Learning and Generative Adversarial Network. Int. J. Innov. Sci. Res. Technol. 2024, 9, 7. [Google Scholar] [CrossRef]
- Waterer, G. What Is Pneumonia? Eur. Respir. Soc. 2021, 17, 210087. [Google Scholar] [CrossRef]
- Sharma, S.; Guleria, K. A Systematic Literature Review on Deep Learning Approaches for Pneumonia Detection Using Chest X-ray Images. Multimed. Tools Appl. 2023, 83, 24101–24151. [Google Scholar] [CrossRef]
- Siddiqi, R.; Javaid, S. Deep Learning for Pneumonia Detection in Chest X-ray Images: A Comprehensive Survey. J. Imaging 2024, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Eido, W.M.; Yasin, H.M. Pneumonia and COVID-19 Classification and Detection Based on Convolutional Neural Network: A Review. Asian J. Res. Comput. Sci. 2025, 18, 174–183. [Google Scholar] [CrossRef]
- Abueed, M.A.M.; Nor, D.M.; Ibrahim, N.; Ogier, J.-M. Pneumonia Detection Using Transfer Learning: A Systematic Literature Review. Int. J. Adv. Comput. Sci. Appl. 2025, 16, 2. [Google Scholar] [CrossRef]
- Bhalke, D.G.; Shaikh, A.S. Classification of Pneumonia Subtypes in Chest X-Rays Using a Custom CNN. In Proceedings of the 1st International Conference on AIML-Applications for Engineering & Technology, Pune, India, 16–17 January 2025; IEEE: Piscataway, NJ, USA, 2025; pp. 1–6. [Google Scholar]
- Sharma, S.; Guleria, K. A Deep Learning Based Model for the Detection of Pneumonia from Chest X-Ray Images Using VGG-16 and Neural Networks. Procedia Comput. Sci. 2023, 218, 357–366. [Google Scholar] [CrossRef]
- Reshan, M.S.A.; Gill, K.S.; Anand, V.; Gupta, S.; Alshahrani, H.; Sulaiman, A.; Shaikh, A. Detection of Pneumonia from Chest X-ray Images Utilizing MobileNet Model. Healthcare 2023, 11, 1561. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 3462–3471. [Google Scholar]
- Wang, T.; Nie, Z.; Wang, R.; Xu, Q.; Huang, H.; Xu, H.; Liu, X.-J. PneuNet: Deep Learning for COVID-19 Pneumonia Diagnosis on Chest X-ray Image Analysis Using Vision Transformer. Med. Biol. Eng. Comput. 2023, 61, 1395–1408. [Google Scholar] [CrossRef]
- Puspita, R.; Rahayu, C. Pneumonia Prediction on Chest X-ray Images Using Deep Learning Approach. IAES Int. J. Artif. Intell. 2024, 13, 467–474. [Google Scholar] [CrossRef]
- Black, S.; Leo, G.; Wang, P.; Leahy, C.; Biderman, S. GPT-Neo: Large Scale Autoregressive Language Modeling with Mesh-Tensorflow. Zenodo 2021. Available online: https://www.semanticscholar.org/paper/GPT-Neo%3A-Large-Scale-Autoregressive-Language-with-Black-Gao/7e5008713c404445dd8786753526f1a45b93de12 (accessed on 2 May 2025).
- Saboo, Y.S.; Kapse, S.; Prasanna, P. Convolutional Neural Networks (CNNs) for Pneumonia Classification on Pediatric Chest Radiographs. Cureus 2023, 15, 8. [Google Scholar] [CrossRef]
- Varshni, D.; Thakral, K.; Agarwal, L.; Nijhawan, R.; Mittal, A. Pneumonia Detection Using CNN Based Feature Extraction. In Proceedings of the IEEE International Conference on Electrical, Computer and Communication Technologies, Coimbatore, India, 20–22 February 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–7. [Google Scholar]
- Rahman, M.M.; Nooruddin, S.; Hasan, K.M.A.; Dey, N.K. HOG + CNN Net: Diagnosing COVID-19 and Pneumonia by Deep Neural Network from Chest X-Ray Images. SN Comput. Sci. 2021, 2, 371. [Google Scholar] [CrossRef]
- Kusk, M.W.; Lysdahlgaard, S. The Effect of Gaussian Noise on Pneumonia Detection on Chest Radiographs, Using Convolutional Neural Networks. Radiography 2023, 29, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Toro, C.; García-Pedrero, A.; Lillo-Saavedra, M.; Gonzalo-Martín, C. Automatic Detection of Pneumonia in Chest X-ray Images Using Textural Features. Comput. Biol. Med. 2022, 145, 105466. [Google Scholar] [CrossRef]
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, D.; Tiwari, P.; Moreira, C.; de Albuquerque, V.H.C. A Novel Transfer Learning Based Approach for Pneumonia Detection in Chest X-ray Images. Appl. Sci. 2020, 10, 559. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, B.; Liu, J.; Dong, Z.; Zhou, Y.; Meng, X.; Toe, T.T. Pneumonia Image Classification: Deep Learning and Machine Learning Fusion. In Proceedings of the 7th International Conference on Artificial Intelligence and Big Data, Beijing, China, 5–7 July 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 440–447. [Google Scholar]
- Mabrouk, A.; Díaz Redondo, R.P.; Dahou, A.; Abd Elaziz, M.; Kayed, M. Pneumonia Detection on Chest X-ray Images Using Ensemble of Deep Convolutional Neural Networks. Appl. Sci. 2022, 12, 6448. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, M.; Kumar, A.; Verma, B.K.; Shitharth, S. Pneumonia Detection with QCSA Network on Chest X-ray. Sci. Rep. 2023, 13, 9025. [Google Scholar] [CrossRef]
- Hammoudi, K.; Benhabiles, H.; Melkemi, M.; Dornaika, F.; Arganda-Carreras, I.; Collard, D.; Scherpereel, A. Deep Learning on Chest X-ray Images to Detect and Evaluate Pneumonia Cases at the Era of COVID-19. J. Med. Syst. 2021, 45, 75. [Google Scholar] [CrossRef] [PubMed]
- Szepesi, P.; Szilágyi, L. Detection of Pneumonia Using Convolutional Neural Networks and Deep Learning. Biocybern. Biomed. Eng. 2022, 42, 1012–1022. [Google Scholar] [CrossRef]
- Colin, J.; Surantha, N. Interpretable Deep Learning for Pneumonia Detection Using Chest X-Ray Images. Information 2025, 16, 53. [Google Scholar] [CrossRef]
- Antunes, C.; Rodrigues, J.M.F.; Cunha, A. Web Diagnosis for COVID-19 and Pneumonia Based on Computed Tomography Scans and X-rays. In Universal Access in Human-Computer Interaction; HCII 2024; Antona, M., Stephanidis, C., Eds.; Springer: Cham, Switzerland, 2024; Volume LNCS14698. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2019, 128, 336–359. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 1135–1144. [Google Scholar]
- ISO 13485; Medical Devices—Quality Management Systems—Requirements for Regulatory Purposes. ISO: Geneva, Switzerland, 2016.

| Dataset | X-Ray Normal | X-Ray Pneumonia |
|---|---|---|
| Dataset #1 [8] Chest X-ray (COVID-19 and Pneumonia) | Train: 1266 Test: 317 | Train: 3418 Test: 855 |
| Dataset #2 [9] Optical Coherence Tomography and Chest X-ray | Train: 1349 Test: 234 | Train: 3883 Test: 390 |
| (Dataset #3) [10] Chest X-ray Images (Pneumonia) | Train: 1341 Val: 8; Test: 234 | Train: 3875 Val: 8; Test: 390 |
| Model | Accuracy | Precision | Recall | AUC | Specificity | F1 Score |
|---|---|---|---|---|---|---|
| ResNet-50 trained and tested with [8] | 91.3% | 82.1% | 86.6% | 96.6% | 93% | 84.2% |
| AlexNet trained and tested with [8] | 91.1% | 78.3% | 92.9% | 97.4% | 90.5% | 84.9% |
| PCM trained and tested with [8] | 96.7% | 97.8% | 89.7% | 98.39% | 99.3% | 93.12% |
| PCM trained and tested with [9] | 98.7% | 98.9% | 95.9% | 99.77% | 99.6% | 98.35% |
| PCM trained and tested with [10] | 97.7% | 98.4% | 92.5% | 98.04% | 99.5% | 96.97% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antunes, C.; Rodrigues, J.M.F.; Cunha, A. PneumoNet: Artificial Intelligence Assistance for Pneumonia Detection on X-Rays. Appl. Sci. 2025, 15, 7605. https://doi.org/10.3390/app15137605
Antunes C, Rodrigues JMF, Cunha A. PneumoNet: Artificial Intelligence Assistance for Pneumonia Detection on X-Rays. Applied Sciences. 2025; 15(13):7605. https://doi.org/10.3390/app15137605
Chicago/Turabian StyleAntunes, Carlos, João M. F. Rodrigues, and António Cunha. 2025. "PneumoNet: Artificial Intelligence Assistance for Pneumonia Detection on X-Rays" Applied Sciences 15, no. 13: 7605. https://doi.org/10.3390/app15137605
APA StyleAntunes, C., Rodrigues, J. M. F., & Cunha, A. (2025). PneumoNet: Artificial Intelligence Assistance for Pneumonia Detection on X-Rays. Applied Sciences, 15(13), 7605. https://doi.org/10.3390/app15137605








