Revealing the Complexity of Polysaccharides: Advances in NMR Spectroscopy for Structural Elucidation and Functional Characterization
Abstract
1. Introduction
2. Evolution of NMR Methodologies
2.1. Solution-State NMR for Polysaccharides
2.1.1. Structural Elucidation
2.1.2. Conformational Dynamics
2.1.3. Applications in Complex Mixtures
2.2. Solid-State NMR for Polysaccharides
2.2.1. Crystalline and Non-Crystalline Polysaccharides
2.2.2. Interactions with Water and Small Molecules
2.3. Multidimensional and Advanced NMR Techniques
2.3.1. Multidimensional NMR
2.3.2. Advanced ssNMR for Dynamics and Interactions
3. Applications of NMR in Polysaccharide Research
3.1. Biomedical Applications
3.2. Food Science and Nutrition
3.3. Environmental and Industrial Applications
4. Challenges and Future Directions
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NMR | Nuclear magnetic resonance spectroscopy |
| DNP | Dynamic nuclear polarization |
| CP/MAS | Cross-polarization magic-angle spinning |
| RDC | Residual dipolar coupling |
| DOSY | Diffusion-ordered spectroscopy |
References
- Lacelle, S.; Gerstein, B.C. Nmr studies on the presence of domain structures in polysaccharides. Biopolymers 1987, 26, 849–861. [Google Scholar] [CrossRef]
- Zujovic, Z.; Chen, D.; Melton, L.D. Comparison of celery (Apium graveolens L.) collenchyma and parenchyma cell wall polysaccharides enabled by solid-state 13C NMR. Carbohydr. Res. 2015, 420, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Debnath, D.; Gautam, I.; Fernando, L.D.; Wang, T. Charting the solid-state NMR signals of polysaccharides: A database-driven roadmap. Magn. Reson. Chem. 2024, 62, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Uhliariková, I.; Vršanská, M.; McCleary, B.V.; Biely, P. Positional specifity of acetylxylan esterases on natural polysaccharide: An NMR study. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3365–3372. [Google Scholar] [CrossRef] [PubMed]
- Fernando, L.D.; Zhao, W.; Gautam, I.; Ankur, A.; Wang, T. Polysaccharide assemblies in fungal and plant cell walls explored by solid-state NMR. Structure 2023, 31, 1375–1385. [Google Scholar] [CrossRef]
- Gautam, I.; Yarava, J.R.; Xu, Y.; Reina Li, R.; Scott, F.J.; Mentink-Vigier, F.; Momany, M.; Latge, J.; Wang, J. Comparative analysis of polysaccharide and cell wall structure in Aspergillus nidulans and Aspergillus fumigatus by solid-state NMR. Carbohydr. Polym. 2025, 348, 122907. [Google Scholar] [CrossRef]
- Huckerby, T.N. Accurate referencing of 13C NMR spectra from glycosaminoglycan and other polysaccharides in aqueous medium. Org. Magn. Reson. 1983, 21, 67–70. [Google Scholar] [CrossRef]
- Yao, H.Y.Y.; Wang, J.Q.; Yin, J.Y.; Nie, S.P.; Xie, M.Y. A review of NMR analysis in polysaccharide structure and conformation: Progress, challenge and perspective. Food Res. Int. 2021, 143, 110290. [Google Scholar] [CrossRef]
- Rodríguez Sánchez, R.A.; Matulewicz, M.C.; Ciancia, M. NMR spectroscopy for structural elucidation of sulfated polysaccharides from red seaweeds. Int. J. Biol. Macromol. 2022, 199, 386–400. [Google Scholar] [CrossRef]
- Xue, Y.; Li, H.; Kang, X. Molecular unraveling of polysaccharide digestion in wood-feeding termites: A solid-state NMR perspective. Carbohydr. Polym. 2024, 331, 121843. [Google Scholar] [CrossRef]
- Nguyen, T.K.N.; Tran, V.M.; Victor, X.V.; Skalicky, J.J.; Kuberan, B. Characterization of uniformly and atom-specifically 13C-labeled heparin and heparan sulfate polysaccharide precursors using 13C NMR spectroscopy and ESI mass spectrometry. Carbohydr. Res. 2010, 345, 2228–2232. [Google Scholar] [CrossRef] [PubMed]
- Kokoulin, M.S.; Belova, V.S.; Romanenko, L.A. Effect of bacterial dissociation on lipopolysaccharide structure: A study of O-polysaccharide from the marine bacterium Pseudoalteromonas agarivorans KMM 232 (O-form). Carbohydr. Res. 2024, 545, 109300. [Google Scholar] [CrossRef]
- Kou, Y.; Guo, R.; Sun, X.; Ma, X.; Chen, Y.; Song, L.; Song, L.; Yuan, C.; Huang, S.; Tang, J.; et al. An investigation of the mechanism of emulsion stabilization in octenyl succinic anhydride-modified tamarind seed polysaccharide. Food Hydrocoll. 2025, 158, 110590. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, W.; Li, J.; Tang, S.; Wang, M.; Huang, W.; Yao, W.; Gao, X. A polysaccharide extracted from Astragalus membranaceus residue improves cognitive dysfunction by altering gut microbiota in diabetic mice. Carbohydr. Polym. 2019, 205, 500–512. [Google Scholar] [CrossRef]
- Mulloy, B.; Mourão, P.A.S.; Gray, E. Structure/function studies of anticoagulant sulphated polysaccharides using NMR. J. Biotechnol. 2000, 77, 123–135. [Google Scholar] [CrossRef]
- Marvelys, L.; Maritza, M.; Lilian, S.; de Pinto Gladys, L.; Julio, H. Structural elucidation of the polysaccharide from Sterculia apetala gum by a combination of chemical methods and NMR spectroscopy. Food Hydrocoll. 2006, 20, 908–913. [Google Scholar] [CrossRef]
- Cai, P.; Moran, J.; Pavliak, V.; Deng, C.; Khoury, N.; Marcq, O.; Ruppen, M.E. NMR structural analysis of the capsular polysaccharide from Streptococcus pneumoniae serotype 6C. Carbohydr. Res. 2012, 351, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Naumenko, O.I.; Zheng, H.; Wang, J.; Senchenkova, S.N.; Wang, H.; Shashkov, A.S.; Chizhov, A.O.; Li, Q.; Knirel, Y.A.; Xiong, Y. Structure elucidation of the O-specific polysaccharide by NMR spectroscopy and selective cleavage and genetic characterization of the O-antigen of Escherichia albertii O5. Carbohydr. Res. 2018, 457, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Merkx, D.W.H.; Westphala, Y.; van Velzena, E.J.J.; Thakoera, K.V.; de Rooa, N.; van Duynhovena, J.P.M. Quantification of food polysaccharide mixtures by 1H NMR. Carbohydr. Polym. 2018, 179, 379–385. [Google Scholar] [CrossRef]
- Li, T.; Xu, S.; Bi, J.; Huang, S.; Fan, B.; Qian, C. Metabolomics study of polysaccharide extracts from Polygonatum sibiricum in mice based on 1H NMR technology. J. Sci. Food Agric. 2020, 100, 4627–4635. [Google Scholar] [CrossRef]
- Cao, L.; Zhao, J.; Wang, M.; Khan, I.A.; Li, X. Rapid preparation and proton NMR fingerprinting of polysaccharides from Radix Astragali. Carbohydr. Res. 2024, 536, 109053. [Google Scholar] [CrossRef]
- Poveda, A.; Santamaria, M.; Bernabe, M.; Rivera, A.; Corzo, J.; Jimenez-Barbero, J. Solution conformation and dynamics of an extracellular polysaccharide isolated from Bradyrhyzobium as deduced from 1H-NMR off resonance ROESY and 13C-NMR relaxation measurements. Carbohydr. Res. 1997, 304, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Vlachou, S.; Politou, A.; Dais, P.; Mazeau, K.; Taravel, F.R. Structure and dynamics of the branched polysaccharide scleroglucan in dilute solutions studied by 1D and 2D NMR spectroscopy. Carbohydr. Polym. 2001, 46, 349–363. [Google Scholar] [CrossRef]
- Martin-Pastor, M.; Bush, C.A. Comparison of the conformation and dynamics of a polysaccharide and of its isolated heptasaccharide repeating unit on the basis of nuclear overhauser effect, long-range C-C and C-H coupling constants, and NMR relaxation data. Biopolymers 2000, 54, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.J.; Imberty, A.; Phalipon, A.; Perez, S.; Simenel, C.; Mulard, L.A.; Delepierre, M. Conformational Studies of the O-specific Polysaccharide of Shigella flexneri 5a and of Four Related Synthetic Pentasaccharide Fragments Using NMR and Molecular Modeling. J. Biol. Chem. 2003, 278, 47928–47936. [Google Scholar] [CrossRef] [PubMed]
- Coxon, B.; Sari, N.; Batta, G.; Pozsgay, V. NMR spectroscopy, molecular dynamics, and conformation of a synthetic octasaccharide fragment of the O-specific polysaccharide of Shigella dysenteriae type 1. Carbohydr. Res. 2000, 324, 53–65. [Google Scholar] [CrossRef]
- de Carvalho, M.M.; Ellefsen, C.F.; Eltvik, A.A.; Hiorth, M.; Samuelsen, A.B.C. Chemical structure characterization of polysaccharides using diffusion ordered NMR spectroscopy (DOSY). Carbohydr. Polym. 2025, 349, 123021. [Google Scholar] [CrossRef]
- Chen, J.; Wang, S.; Guo, F.; Gong, Y.; Chen, T.; Shaw, C.; Jiang, R.; Huang, F.; Lin, D. 1H-NMR-based metabolomics reveals the preventive effect of Enteromorpha prolifera polysaccharides on diabetes in Zucker diabetic fatty rats. Food Sci. Nutr. 2024, 12, 4049–4062. [Google Scholar] [CrossRef]
- De Souza, A.C.; Rietkerk, T.; Selin, C.G.M.; Lankhorst, P.P. A robust and universal NMR method for the compositional analysis of polysaccharides. Carbohydr. Polym. 2013, 95, 657–663. [Google Scholar] [CrossRef]
- Ye, L.; Li, J.; Zhang, J.; Pan, Y. NMR characterization for polysaccharide moiety of a glycopeptide. Fitoterapia 2010, 81, 93–96. [Google Scholar] [CrossRef]
- Wang, H.X.; Zhao, J.; Li, D.; Song, S.; Song, L.; Fu, Y.; Zhang, L. Structural investigation of a uronic acid-containing polysaccharide from abalone by graded acid hydrolysis followed by PMP-HPLC-MSn and NMR analysis. Carbohydr. Res. 2015, 402, 95–101. [Google Scholar] [CrossRef]
- Cordeiro, A.R.; Bezerra, I.L.; Santana-Filho, A.P.; Benedetti, P.R.; Ingberman, M.; Sassaki, G.L. Wine fermentation process evaluation through NMR analysis: Polysaccharides, ethanol quantification and biological activity. Food Chem. 2024, 451, 139531. [Google Scholar] [CrossRef] [PubMed]
- Falourd, X.; Rondeau-Mouro, C.; Cambert, M.; Lahaye, M.; Chabbert, B.; Aguié-Béghin, V. Polysaccharide-water interactions: NMR and DVS data. Data Br. 2024, 53, 110106. [Google Scholar] [CrossRef]
- Naumann, C.; Kuchel, P.W. NMR (pro)chiral discrimination using polysaccharide gels. Chem. A Eur. J. 2009, 15, 12189–12191. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; Idbella, M.; Zotti, M.; Alteriis, E.D.; Diano, M.; Lanzotti, V.; Spaccini, R.; Mazzoleni, S. Multi-kingdom characterization of living organisms by 13C CPMAS NMR spectroscopy reveals unique traits in bacteria, fungi, algae, and higher plants. Geoderma 2024, 448, 116978. [Google Scholar] [CrossRef]
- Delcourte, L.; Berbon, M.; Rodriguez, M.; Subban, K.; Lends, A.; Grelard, A.; Morvan, E.; Habenstein, B.; Saupe, S.J.; Delhaes, L.; et al. Magic-angle spinning NMR spectral editing of polysaccharides in whole cells using the DREAM scheme. Methods 2024, 230, 59–67. [Google Scholar] [CrossRef]
- Ng, J.K.T.; Zujovic, Z.D.; Smith, B.G.; Johnston, J.W.; Schroder, R.; Melton, L.D. Solid-state 13C NMR study of the mobility of polysaccharides in the cell walls of two apple cultivars of different firmness. Carbohydr. Res. 2014, 386, 1–6. [Google Scholar] [CrossRef]
- Davies, L.M.; Harris, P.J.; Newman, R.H. Molecular ordering of cellulose after extraction of polysaccharides from primary cell walls of Arabidopsis thaliana: A solid-state CP/MAS 13C NMR study. Carbohydr. Res. 2002, 337, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Widanage, M.C.D.; Yarava, J.R.; Ankur, A.; Wang, T. Molecular architecture of chitin and chitosan-dominated cell walls in zygomycetous fungal pathogens by solid-state NMR. Nat. Commun. 2024, 15, 8295. [Google Scholar] [CrossRef]
- Adam, O.; Rivière, M.; Vercellone, A.; Monsan, P.F.; Puzo, G. Complete NMR structural elucidation of the capsular polysaccharide adjuvant from klebsiella I-, 714. Eur. J. Biochem. 1996, 241, 602–610. [Google Scholar] [CrossRef]
- Falourd, X.; Rondeau-Mouro, C.; Cambert, M.; Lahaye, M.; Chabbert, B.; Aguie-Beghin, V. Assessing the complementarity of time domain NMR, solid-state NMR and dynamic vapor sorption in the characterization of polysaccharide-water interactions. Carbohydr. Polym. 2024, 326, 121579. [Google Scholar] [CrossRef] [PubMed]
- Bellich, B.; Cacioppo, M.; Zorzi, R.D.; Rizzo, R.; Brady, J.W.; Cescutti, P. Interactions of biofilm polysaccharides produced by human infective bacteria with molecules of the quorum sensing system. A microscopy and NMR study. Int. J. Biol. Macromol. 2024, 281, 136222. [Google Scholar] [CrossRef] [PubMed]
- Agles, A.A.; Bourg, I.C. Structure and Dynamics of Water in Polysaccharide (Alginate) Solutions and Gels Explained by the Core-Shell Model. Biomacromolecules 2024, 25, 6403–6415. [Google Scholar] [CrossRef]
- Baranowska, H.M.; Sikora, M.; Kowalski, S.; Tomasik, P. Interactions of potato starch with selected polysaccharide hydrocolloids as measured by low-field NMR. Food Hydrocoll. 2008, 22, 336–345. [Google Scholar] [CrossRef]
- Yuris, A.; Hindmarsh, J.; Hardacre, A.K.; Goh, K.K.T.; Matia-Merino, L. The interactions between wheat starch and Mesona chinensis polysaccharide: A study using solid-state NMR. Food Chem. 2019, 284, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Savary, G.; Moreau, C.; Cayot, N. Impact of the composition of polysaccharide composite gels on small molecules diffusion: A rheological and NMR study. Food Res. Int. 2010, 43, 364–368. [Google Scholar] [CrossRef]
- Leeflang, B.R.; Faber, E.J.; Erbel, P.; Vliegenthart, J.F.G. Structure elucidation of glycoprotein glycans and of polysaccharides by NMR spectroscopy. J. Biotechnol. 2000, 77, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Kulik, A.S.; Haverkamp, J. Molecular mobility of polysaccharide chains in starch investigated by two-dimensional solid-state NMR spectroscopy. Carbohydr. Polym. 1997, 34, 49–54. [Google Scholar] [CrossRef]
- Fontana, C.; Weintraub, A.; Widmalm, G. Facile structural elucidation of glycans using NMR spectroscopy data and the program CASPER: Application to the O-antigen polysaccharide of Escherichia coli O155. Chempluschem 2013, 78, 1327–1329. [Google Scholar] [CrossRef]
- Kupce, E.; Nishida, T.; Widmalm, G.; Freeman, R. Resolving overlap in two-dimensional NMR spectra: Nuclear Overhauser effects in a polysaccharide. Magn. Reson. Chem. 2005, 43, 791–794. [Google Scholar] [CrossRef]
- Kolz, J.; Yarovoy, Y.; Mitchell, J.; Johns, M.L.; Gladden, L.F. Interactions of binary liquid mixtures with polysaccharides studied using multi-dimensional NMR relaxation time measurements. Polymer 2010, 51, 4103–4109. [Google Scholar] [CrossRef]
- Kirui, A.; Du, J.; Zhao, W.; Barnes, W.; Kang, X.; Anderson, C.T.; Xiao, C.; Wang, T. A pectin methyltransferase modulates polysaccharide dynamics and interactions in Arabidopsis primary cell walls: Evidence from solid-state NMR. Carbohydr. Polym. 2021, 270, 118370. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ren, P.; Sun, Y.; Li, J.; Zhou, X.; Zhang, H.; He, C.; Dai, H.; Guan, L. Structure elucidation and molecular mechanism of an immunomodulatory polysaccharide from Nostoc commune. Int. J. Biol. Macromol. 2024, 283, 137435. [Google Scholar] [CrossRef]
- Li, W.Q.; Liu, H.; Guo, Y.R.; Wang, X.Y.; Zhao, P.T. Investigating the matrix effect of high/low methoxyl pectins on typical aroma of apple juice using HS-SPME-GC–MS and NMR analysis. Food Chem. 2025, 463, 141372. [Google Scholar] [CrossRef]
- Qin, C.J.; Hu, J.; Tong, W.; Zhang, T.; Tian, G.; Zou, X.; Liu, J.; Yin, J. Determination of ribose and phosphorus contents in Haemophilus influenzae type b capsular polysaccharide by a quantitative NMR method using a single internal standard. Chin. J. Nat. Med. 2022, 20, 633–640. [Google Scholar] [CrossRef]
- Hoang, T.V.; Alshiekheid, M.A. A study on anticancer and antioxidant ability of selected brown algae biomass yielded polysaccharide and their chemical and structural properties analysis by FT-IR and NMR analyses. Environ. Res. 2024, 260, 119567. [Google Scholar] [CrossRef]
- Davidson, J.; Gauthier-Signore, C.; Bishop, K.P.; Wicks, C.; Monteiro, M.A.; Roy, P.; Auzanneau, F. ROESY and 13C NMR to distinguish between d- and l-rhamnose in the α-d-Manp-(1 → 4)-β-Rhap-(1 → 3) repeating motif. Org. Biomol. Chem. 2022, 20, 2964–2980. [Google Scholar] [CrossRef] [PubMed]
- Tajima, K.; Uenishi, N.; Fujiwara, M.; Erata, T.; Munekata, M.; Takai, M. The production of a new water-soluble polysaccharide by Acetobacter xylinum NCI 1005 and its structural analysis by NMR spectroscopy. Carbohydr. Res. 1997, 305, 117–122. [Google Scholar] [CrossRef]
- Rustandi, R.R.; Xu, Q. Reactivity characterization of bromoacetyl derivatized polyribosylribitol polysaccharide in Haemophilus influenzae type b for PedvaxHIB® by NMR spectroscopy. Vaccine 2022, 40, 187–191. [Google Scholar] [CrossRef]
- Petersen, B.O.; Hindsgaul, O.; Paulsen, B.S.; Redondo, A.R.; Skovsted, I.C. Structural elucidation of the capsular polysaccharide from Streptococcus pneumoniae serotype 47A by NMR spectroscopy. Carbohydr. Res. 2014, 386, 62–67. [Google Scholar] [CrossRef]
- Li, C.; Andersen, K.B.; Elverdal, P.L.; Skovsted, I.C.; Duus, J.; Kjeldsen, C. Full NMR assignment, revised structure and biosynthetic analysis for the capsular polysaccharide from Streptococcus Pneumoniae serotype 15F. Carbohydr. Res. 2021, 508, 108418. [Google Scholar] [CrossRef]
- Jones, C.; Lemercinier, X. Use and validation of NMR assays for the identity and O-acetyl content of capsular polysaccharides from Neisseria meningitidis used in vaccine manufacture. J. Pharm. Biomed. Anal. 2002, 30, 1233–1247. [Google Scholar] [CrossRef] [PubMed]
- Beeren, S.R.; Meier, S.; Hindsgaul, O. Probing helical hydrophobic binding sites in branched starch polysaccharides using NMR spectroscopy. Chem. A Eur. J. 2013, 19, 16314–16320. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, K.I.; Inukai, K.; Suzuki, M.; Kuga, H.; Korenaga, H. Structural studies on a sulfated polysaccharide from an Arthrobacter sp. by NMR spectroscopy and methylation analysis. Carbohydr. Res. 1997, 305, 253–260. [Google Scholar] [CrossRef]
- Kang, J.; Cui, S.W.; Phillips, G.O.; Chen, J.; Guo, Q.; Wang, Q. New studies on gum ghatti (Anogeissus latifolia) Part III: Structure characterization of a globular polysaccharide fraction by 1D, 2D NMR spectroscopy and methylation analysis. Food Hydrocoll. 2011, 25, 1999–2007. [Google Scholar] [CrossRef]
- Dourado, F.; Cardoso, S.M.; Silva, A.M.S.; Gama, F.M.; Coimbra, M.A. NMR structural elucidation of the arabinan from Prunus dulcis immunobiological active pectic polysaccharides. Carbohydr. Polym. 2006, 66, 27–33. [Google Scholar] [CrossRef]
- Beri, S.; Gandhi, D. Quantification of residual cetyltrimethylammonium bromide (CTAB) and sodium deoxycholate (DOC) in Haemophilus influenzae type b (Hib) polysaccharide using NMR. Biologicals 2021, 70, 22–27. [Google Scholar] [CrossRef]
- Li, H.; Chen, H.; Shi, Z.; Yi, Z.; Hu, W.; Zhou, S.; Yang, X.; Kan, J.; Awad, S.; Hegyi, F.; et al. Structure and physicochemical properties of rice starch modified with dodecenyl succinic anhydride and its use for microencapsulating Pediococcus acidilactici probiotic. Food Chem. 2025, 463, 141276. [Google Scholar] [CrossRef]
- Gamian, A.; Romanowska, E.; Romanowska, A.; Lugowski, C.; Dabrowski, J.; Trauner, K. Citrobacter lipopolysaccharides: Structure elucidation of the O-specific polysaccharide from strain PCM 1487 by mass spectrometry, one-dimensional and two-dimensional 1H-NMR spectroscopy and methylation analysis. Eur. J. Biochem. 1985, 146, 641–647. [Google Scholar] [CrossRef]
- Spevacek, J.; Brus, J. Solid-state NMR studies of polysaccharide systems. Macromol. Symp. 2008, 265, 69–76. [Google Scholar] [CrossRef]
- Sheng, S.; Cherniak, R. Structure of the capsular polysaccharide of Clostridium perfringens Hobbs 10 determined by NMR spectroscopy. Carbohydr. Res. 1997, 305, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Junker, F.; Michalski, K.; Guthausen, G.; Bunzel, M. Characterization of covalent, feruloylated polysaccharide gels by pulsed field gradient-stimulated echo (PFG-STE)-NMR. Carbohydr. Polym. 2021, 267, 118232. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Reid, D.G.; Bazin, D.; Daudon, M.; Duer, M.J. Solid state NMR of salivary calculi: Proline-rich salivary proteins, citrate, polysaccharides, lipids, and organic–mineral interactions. Comptes Rendus Chim. 2016, 19, 1665–1671. [Google Scholar] [CrossRef]
- Neiss, T.G.; Cheng, H.N.; Daas, P.J.H.; Schols, H.A. Compositional heterogeneity in pectic polysaccharides: NMR studies and statistical analysis. Macromol. Symp. 1999, 140, 165–178. [Google Scholar] [CrossRef]
- El Hariri El Nokab, M.; van der Wel, P.C.A. Use of solid-state NMR spectroscopy for investigating polysaccharide-based hydrogels: A review. Carbohydr. Polym. 2020, 240, 116276. [Google Scholar] [CrossRef]
- Byeon, C.H.; Kinney, T.; Saricayir, H.; Hansen, K.H.; Scott, F.J.; Srinivasa, S.; Wells, M.K.; Mentink-Vigier, F.; Kim, W.; Akbey, Ü. Ultrasensitive characterization of native bacterial biofilms via dynamic nuclear polarization-enhanced solid-state NMR. Angew. Chem. Int. Ed. 2025, 64, e202418146. [Google Scholar] [CrossRef]
- Garrido, R.; Soubal, J.P.; Torres, L.; Ramírez, U.; Vérez, V. NMR line-fitting quantification of polysaccharide N-acylurea-based modification in glycoconjugates of Salmonella Typhi Vi polysaccharide. Magn. Reson. Chem. 2017, 55, 720–723. [Google Scholar] [CrossRef]
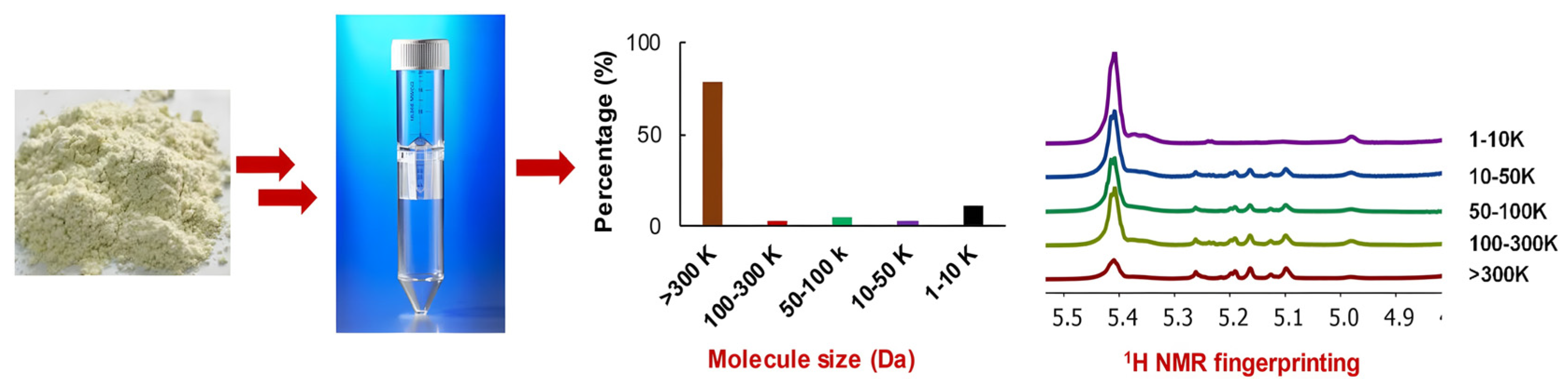

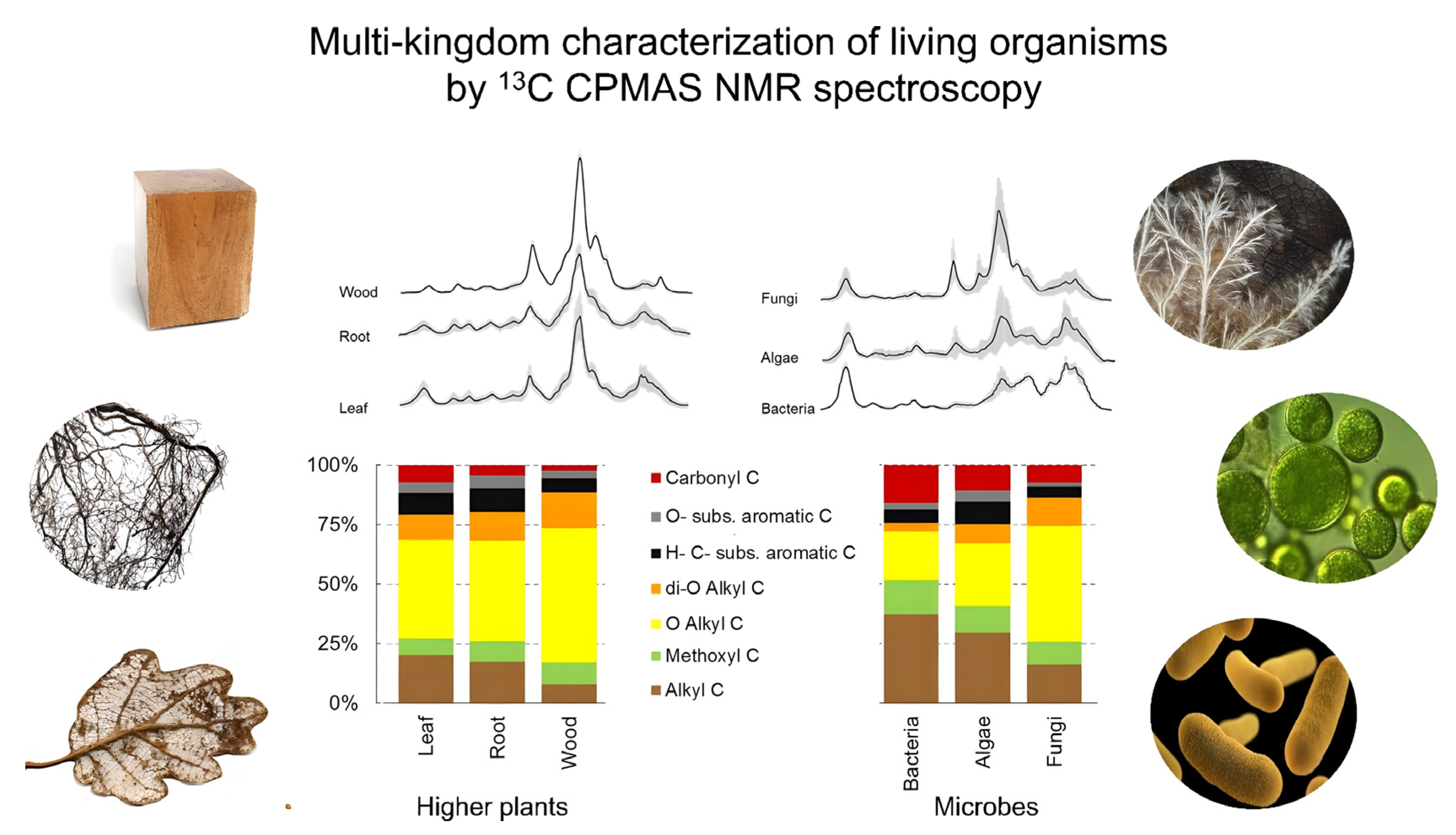
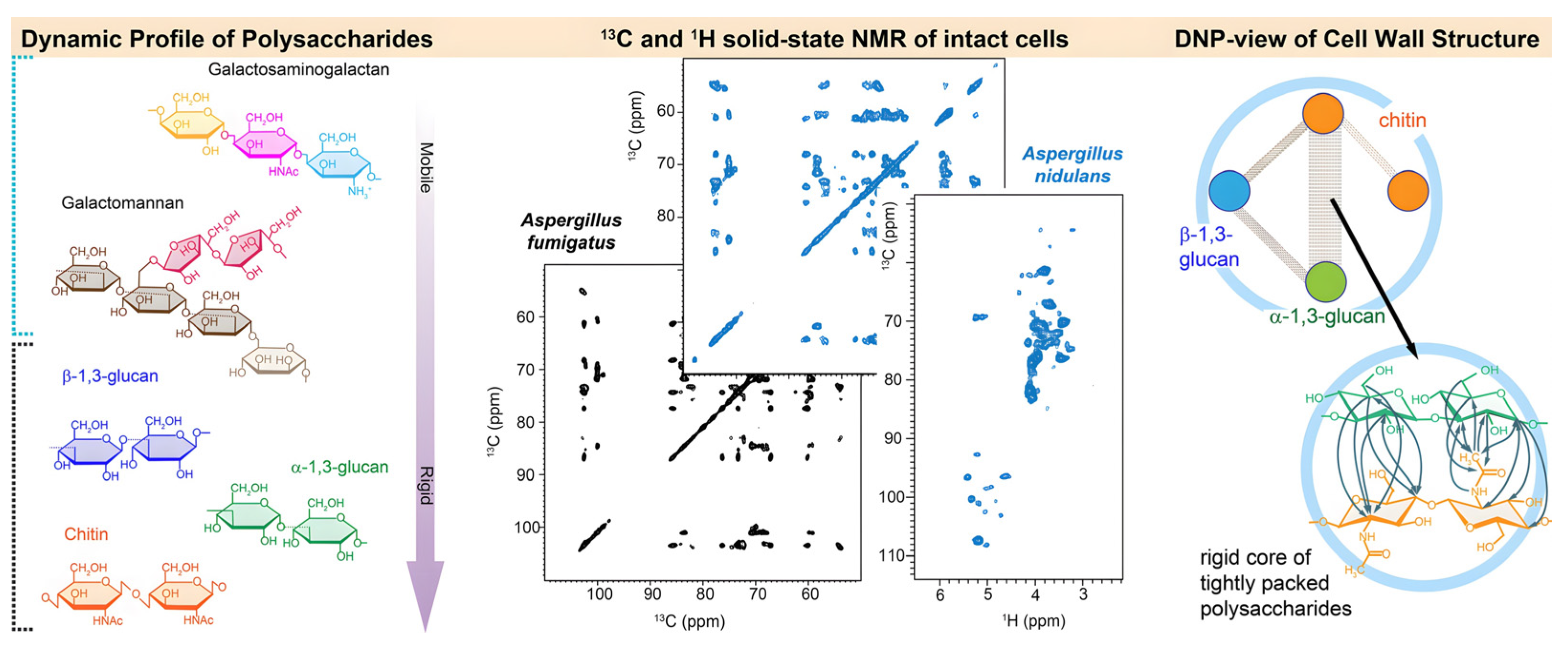
| Sample State | Techniques | Key Features | Optimal Applications |
|---|---|---|---|
| Solution-state | 1D 1H/13C | Rapid chemical shift assignment; simple operation | Purity assessment, monosaccharide fingerprinting (e.g., herbal polysaccharides) |
| 2D COSY/TOCSY | 1H-1H coupling networks; intra-ring connectivity | Homopolysaccharide validation (e.g., starch, cellulose) | |
| 2D HSQC/HMBC | 1H-13C correlations; glycosidic linkage mapping | Heteropolysaccharide structure (e.g., microbial EPS) | |
| DOSY | Diffusion-based component separation; Hydrodynamic radius | interactions in mixtures (e.g., pectin–protein complexes) | |
| Solid-state | CP/MAS | Resolves rigid molecular structures; crystallinity analysis | Plant cell wall polysaccharides (cellulose–lignin interactions) |
| DNP-NMR | 10–100× sensitivity enhancement; low-abundance components | Biofilm matrix, fungal cell walls | |
| HETCOR | Heteronuclear correlations; hydration studies | Food gel hydration mechanisms |
| Technique | Advantages | Limitations | Typical Applications |
|---|---|---|---|
| HSQC | 1H-13C direct correlation; ideal for glycosidic bond identification and monosaccharide composition | Lower sensitivity for low-abundance samples | Primary structure determination (branching patterns, anomeric configurations) |
| HMBC | Detects 1H-13C long-range couplings (2–4 bonds); reveals cross-glycosidic linkages | Weak signal intensity; requires longer acquisition time; challenging for overlapping signals | Sequence determination in complex polysaccharides (e.g., bacterial O-antigens) |
| NOESY | Provides spatial proximity information (<5 Å); reveals conformation and intermolecular interactions | Sensitive to molecular motion; difficult to quantify | Conformational studies (e.g., food gels, biofilms) |
| Field | Key Findings | Methodology | Potential Impact |
|---|---|---|---|
| Biomedical | Elucidation of immunomodulatory and anticoagulant polysaccharides | Solution-state NMR, HSQC, NOESY | Development of therapeutics for immune modulation and clotting |
| Structural characterization of bacterial capsular polysaccharides for vaccines | Multidimensional NMR, isotopic labeling | Improved vaccine design and efficacy | |
| Antioxidant activity of polysaccharides | 1H NMR, MD | Reduced oxidative stress; therapeutic innovation | |
| Food Science | Analysis of polysaccharide interactions in food matrices | 1H NMR, DOSY | Optimization of texture, stability, and sensory properties |
| Study of gelation and hydration dynamics | Solid-state NMR, relaxation measurements | Enhanced food product formulation and quality control | |
| Environmental | Investigation of microbial exopolysaccharides in biofilms | Solid-state NMR, DNP-enhanced NMR | Development of sustainable biofilm management technologies |
| Analysis of lignocellulose degradation in biofuel production | CP/MAS NMR | Advancement in renewable energy technologies |
| Challenge | Current Limitations | Emerging Solutions | Future Directions |
|---|---|---|---|
| Signal Overlap | Overlapping signals in complex mixtures | Multidimensional NMR, computational modeling, isotopic labeling | Development of advanced spectral deconvolution algorithms |
| Sensitivity Limitations | Low sensitivity for dilute samples or small amounts of material | DNP-enhanced NMR, isotopic labeling (13C, 15N) | Integration of DNP with ultra-high-field NMR |
| Dynamic Processes | Difficulty in studying dynamic polysaccharide–protein interactions in real time | Time-resolved NMR, in situ solid-state NMR | Development of portable NMR systems for real-time monitoring |
| Sample Heterogeneity | Heterogeneous samples (e.g., plant cell walls) complicate structural analysis | Integration with complementary techniques (e.g., mass spectrometry, X-ray) | Holistic approaches combining NMR with other methods |
| Accessibility of NMR Instruments | High cost and limited access | Cost-effective NMR systems, open-access NMR facilities | Democratization of NMR technology |
| Polysaccharide Type | Structural Features | Recommended Techniques | Rationale |
|---|---|---|---|
| Linear homopolysaccharides (e.g., cellulose) | Simple repeats, rigid chains | Solution: 1D 13C NMR; solid: CP/MAS | 1D NMR confirms composition; CP/MAS distinguishes crystalline domains |
| Branched heteropolysaccharides (e.g., pectin) | Complex side chains, acidic groups | HSQC (linkages) + HMBC (sequence) + NOESY (conformation) | Multidimensional approach needed for branching and charge effects |
| Microbial EPS | High heterogeneity, modified groups (e.g., sulfation) | HSQC/TOCSY + DOSY; solid: DNP-NMR (low-abundance) | Combination addresses structural diversity; DNP enhances sensitivity |
| Food matrix polysaccharides | Multi-component, dynamic interactions | DOSY + 1H NMR metabolomics; Solid: HETCOR | DOSY separates components; HETCOR reveals hydration/protein interactions |
| Biofilm polysaccharides | Semi-rigid, environment-sensitive | Solution: NOESY; solid: DNP-NMR + 1H-detection | DNP solves sensitivity issues; NOESY elucidates biofilm formation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Gao, L.; Yu, Z. Revealing the Complexity of Polysaccharides: Advances in NMR Spectroscopy for Structural Elucidation and Functional Characterization. Appl. Sci. 2025, 15, 5246. https://doi.org/10.3390/app15105246
Liu Y, Gao L, Yu Z. Revealing the Complexity of Polysaccharides: Advances in NMR Spectroscopy for Structural Elucidation and Functional Characterization. Applied Sciences. 2025; 15(10):5246. https://doi.org/10.3390/app15105246
Chicago/Turabian StyleLiu, Yaqin, Lina Gao, and Zeling Yu. 2025. "Revealing the Complexity of Polysaccharides: Advances in NMR Spectroscopy for Structural Elucidation and Functional Characterization" Applied Sciences 15, no. 10: 5246. https://doi.org/10.3390/app15105246
APA StyleLiu, Y., Gao, L., & Yu, Z. (2025). Revealing the Complexity of Polysaccharides: Advances in NMR Spectroscopy for Structural Elucidation and Functional Characterization. Applied Sciences, 15(10), 5246. https://doi.org/10.3390/app15105246





