Featured Application
Endophytic bacteria identified in this research may find potential applications as biofertilizers to improve nutrient utilization in plants.
Abstract
Endophytic bacteria, especially those that participate in nitrogen fixation, play critical roles in supplying essential nutrients for legume plant growth. Despite that there have been numerous investigations targeting bacterial microbiomes in legume roots and nodules, little is known about embryonic bacteria that facilitate plant nutrient utilization after seed germination. Here, we collected and investigated endophytic bacterial microbiome in edible pea (Pisum sativum) embryos using five representative cultivars and a pea sprout (shoot of pea [SHP]) control. Twenty-six nitrogen-fixing bacteria (NFB) were isolated from pea embryos, with three strains found in fresh grain pea (FGP) and snow pea (SP) exhibiting the strongest nitrogenase activity of above 85 nmol C2H4/mL/h. Some NFB isolates are also potassium-solubilizing bacteria (KSB) or phosphorus-solubilizing bacteria (PSB) utilizing inorganic and/or organic phosphorus. All 26 NFB showed variable levels (0.41 to 7.10 μg/mL) of indole-3-acetic acid (IAA) secretion. The nutrient-solubilizing NFB identified in our research are potential targets for biofertilizer development. They could be useful in converting nitrogen, potassium, and/or phosphorus into usable forms for the plants. At the microbiome level, high-throughput 16S ribosomal RNA (rRNA) sequencing of 40 bacterial collections from pea embryos generated 4234 operational taxonomic units (OTUs) using 97% identity as the threshold for clustering high-quality effective reads (valid tags). Analysis of OTU annotation results revealed similar species community structures, abundance, and diversity in most samples. Our embryo-derived endophytic bacterial pool provides a microbiome platform for seed dormancy and germination research of edible peas, as well as for digging new biofertilizer resources in general.
1. Introduction
The edible pea (Pisum sativum) is a major grain legume crop with about 7.5 million hectares of planting area worldwide [1]. Major edible pea cultivars in the market include dry grain pea (DGP), fresh grain pea (FGP), snow pea (SP), sweet crispy pea (SCP), and pea tip (seedling of pea [SEP]). On an average, the pea grain consists of 55–60% starch, 20–25% protein, and 6–10% crude fiber, making it a healthy food resource of plant proteins, dietary fiber, vitamins, and essential minerals [2]. Like soybeans [3] and peanut [4,5], pea growth and nutrient uptake require growth-promoting endophytic bacteria [6], especially those involved in nitrogen fixation [7].
Endophytic bacteria thrive inside plant tissues and organs with no immediate impact on normal plant growth [8]. Over time, they may either cause damage to plants or promote plant growth under normal and/or stressful conditions [9]. Growth-promoting endophytic bacteria can enhance plant nutrient utilization, growth, and health via multiple approaches, including but not limited to enhancing plant resistance against phytopathogens, production of enzymes for nutrition solubilization, uptake and absorption, canonical phytohormone biosynthesis and modulation of their homeostasis, production and secretions of volatile organic acids and secondary metabolites, and heavy metal phytoremediation [3,10,11]. Endophytic bacteria-assisted acquisitions of macro- and micronutrients in plants especially involve nitrogen (N), phosphorus (P), and potassium (K) [3]. Legume-symbiotic nitrogen-fixing bacteria (NFB) have nitrogenase activity capable of transforming atmospheric nitrogen into plant-usable inorganic compounds [12]. Phosphorus-solubilizing bacteria (PSB) [13] and potassium-solubilizing bacteria (KSB) [14] can solubilize soil inorganic/organic phosphorus and potassium, respectively. These nutrients can be easily utilized by plants in their soluble forms. Endophytic bacteria may also produce auxins [15], cytokinins [16], and/or gibberellins [17] to modulate plant hormonal homeostasis, which may have significant impacts on plant growth and development. Nitrogen fixation, nutrient solubilization, and phytohormone production activities are not mutually exclusive in growth-promoting endophytic bacteria, with some strains playing two or more roles in promoting plant nutrient uptake and utilization [9].
Cultivable endophytic bacteria only account for 0.01–10% of the total bacterial species. Their species/genera can be determined by morphology observation, Gram staining, biochemical/physiological assays, and 16S ribosomal RNA (rRNA) sequencing [18,19,20]. High-throughput 16S rRNA sequencing enables an accurate and complete investigation of endophytic bacterial community structures, species diversity, and abundance at the microbiome level, with no need for individual strain isolation and culture [21,22,23].
Considered to originate from the external environment, endophytic bacteria can be found in plant seeds, leaves, flowers, roots, and root nodules [24]. Seed endophytic bacteria can be horizontally transferred through multiple generations of plants and help to keep seed dormancy status, protect the seed from pathogen infection, promote seed germination in the soil, and improve seedling tolerance to abiotic stress [25,26,27,28,29,30]. Leaf endophytic bacterial microbiomes have high variation associated with plant species, leaf sub-locations, and growth stages. They can suppress pathogen attachment on leaves and enhance plant growth and biomass accumulation [31,32,33,34]. Flower endophytic bacteria reside in stigmas, pollens, petals, and nectars to function in pollination [35]. Roots harbor far more endophytic bacterial species and abundance than any other organs. These bacteria are highly diverse in both community structure and functions [36]. Both rhizobia and nonrhizobial endophytic bacteria in root nodules are critical for legume growth and resilience [37].
Most plant endophytic bacterial studies, especially those that target peas, focus on roots and root nodules due to their legume and symbiotic nitrogen fixation nature [38,39,40,41]. In contrast, little is known about endophytic bacteria in plant embryos, which may harbor bacteria transmitted from flowers and seeds and facilitate plant nutrient utilization after seed germination. The embryo bacterial microbiome also shapes the blueprint of bacterial communities in the next generation of plants. For example, the high bacterial diversity in watermelon embryos mainly includes spore-forming Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and other minor phyla that are vertically transmittable to seedlings [42]. In addition, embryonic microbiota may be involved in plant disease resistance. For example, bacterial communities in maize embryos are associated with their susceptibility to fusarium ear rot [43].
We previously reported the community structure and associated networks of endophytic bacteria in pea roots throughout the plant life cycle [44]. Here, we collected and investigated the endophytic bacterial microbiomes in edible pea embryos using five representative cultivars and a pea sprout control. Twenty-six NFB were isolated from pea embryos. Some of the NFB isolates are also KSB or PSB. All 26 NFB showed variable levels of indole-3-acetic acid (IAA) secretion. High-throughput 16S RNA sequencing of 40 bacterial collections from pea embryos revealed similar community structures, species abundance, and diversity in most samples. Our work identified nutrient-solubilizing NFB with biofertilizer potential and established an embryo-derived endophytic bacterial pool for seed dormancy and germination research of edible peas.
2. Materials and Methods
2.1. Plant Materials and Sampling
Edible pea seeds were stocks kept at Qingdao Academy of Agricultural Sciences. Five pea cultivars with different fiber and sugar contents were used in this research, including dry grain pea (DGP), fresh grain pea (FGP), snow pea (SP), sweet crispy pea (SCP), and pea tip (seedling of pea [SEP]). These five cultivars are the most common edible peas found in the market, representing wide ranges of nutritional and chemical compositions (proteins, dietary fibers, starch/sugar/carbohydrates, vitamins, polyphenolic compounds, antioxidants/bioactive compounds, etc.) [45,46,47]. Bacterial microbiome analysis on these cultivars would generate a comprehensive blueprint of endophytic bacteria distributions in pea embryos. Twenty pea accessions were sown with three to four accessions included in each cultivar and a pea sprout (shoot of pea [SHP]) accession used as the control. Pea plants were grown from March to June 2018. Samples were collected in the seedling, flowering, grain-filling, and seed maturity stages. All collected samples were stored at −20 °C.
2.2. Isolation and Purification of Cultivable Endophytic Bacteria
Pea embryos were isolated from de-coated seeds, sterilized with 70% ethanol for 3 min, and then rinsed/washed with sterilized water. Adequate sterilization was tested by spreading 100 µL water from the last rinse to Trypticase Soy Agar (TSA) plates (15 g/L tryptone, 5.0 g/L soybean peptone, 30 g/L NaCl, 15 g/L agar). Embryo sterilization was considered successful if no microbe grew after 72 h incubation of the TSA plates at 28 °C. Plastic-wrapped sterilized embryos were crushed and transferred to 1.5 mL Eppendorf tubes. After adding 1 mL saline solution to each tube, embryo samples were sonicated for 1 min. After 10-fold dilution, 100 µL suspension was spread on either TSA or Ashby’s nitrogen-free (20 g/L mannitol, 0.2 g/L K2HPO4, 0.2 g/L MgSO₄, 0.2 g/L NaCl, 0.1 g/L K2SO4, 5.0 g/L CaCO3, 15 g/L agar, pH 7.4 ± 0.2) plate. After incubation at 28 °C for 72 h, single bacterial colonies were picked for multiple rounds of streak cultivation. Purified isolates were cultured and stored at −80 °C. Bacteria strains isolated from the last round of streak were classified based on colony sizes, colors, transparency, and morphology.
2.3. Nitrogenase Activity and Nutrient Solubilization Assays of Plant Growth-Promoting Bacteria
Nitrogenase activity was measured by acetylene reduction assay (ARA). Ethylene absorption in ARA was determined spectrophotometrically since the change in the absorbance over time is linear with nitrogenase activity [48,49,50]. A total of 1 mL of NFB culture was added to a 13 mL glass vial with 1 mL of air replaced with the same volume of acetylene (99.99% purity). After culturing at 28 °C for 24 h, the bacterial sample was mixed with 2 mL of saturated NaCl solution. A total of 200 μL of the mixture (10% gas concentration) was then extracted and added to the absorption buffer (5.0 g/L CuSO4, 5.3 g/L ammonia, 23 g/L NH2OH·HCl, 0.9 g/L gelatin, 31.6% ethanol). The optical density (OD) was measured at 548 nm after shaking the mixture for 5 min. Blank medium without NFB inoculation was used as the mock control.
To measure inorganic phosphorus solubilization, bacterial strains were inoculated to inorganic phosphorus screening medium (10 g/L glucose, 0.5 g/L (NH4)2SO4, 0.2 g/L NaCl, 0.2 g/L KCl, 0.03 g/L MgSO4·7H2O, 0.03 g/L MnSO4, 0.003 g/L FeSO4, 0.5 g/L yeast extract, 5.0 g/L Ca3(PO4)2, 15 g/L agar, pH 6.8–7.0) and cultured at 30 °C for 3 d. Inorganic phosphorus solubilization was indicated by the emergence of clear halos on the plates, with halo sizes positively correlated to solubilization activity. Similarly, organic phosphorus solubilization was tested by inoculating the bacterium to egg yolk medium (10 g/L tryptone, 3.0 g/L beef extract, 5.0 g/L NaCl, 15 g/L agar, pH 7.0–7.5; add 3 mL of fresh egg yolk to a 50 mL medium plate before use) and checking the formation of turbid halos after incubation at 30 °C for 3 d.
To measure potassium solubilization, bacterial strains were inoculated to silicate medium (10 g/L sucrose, 0.5 g/L yeast extract, 1.0 g/L (NH4)2SO4, 2.0 g/L Na2HPO4, 0.5 g/L MgSO4·7H2O, 1.0 g/L CaCO3, 1.0 g/L potash feldspar powder, 15 g/L agar) and incubated at 30 °C for 3 d. The emergence of clear halos on silicate plates indicated successful solubilization of inorganic potassium, with halo sizes positively correlated to bacterial potassium solubilization activity.
Bacterial IAA production was quantified by using the Salkowski reagent (a fresh mixture solution of 0.5 M FeCl3 and 35% HClO4) to generate a standard curve [51]. A total of 50 μL of bacterial culture at the logarithmic growth phase was inoculated to the beef extract liquid medium containing 0.5 g/L tryptophan. After shaking at 140 rpm and 28 °C for 36 h, 50 μL of activated bacterial culture at the logarithmic growth phase were mixed with the Salkowski reagent and incubated (avoid light) at 25 °C for 30 min. Bacterial IAA production was indicated by the emergence of pink color in the solution. IAA-producing bacterial cultures were then centrifugated at 8000× g for 5 min. A total of 2 mL of supernatant from each strain was mixed with 4 mL of the Salkowski reagent and incubated (avoid light) at 25 °C for 30 min. IAA production was quantified by measuring ODs at 530 nm with three biological replicates performed for each sample. The standard curve was generated by using a concentration gradient of eight IAA standards for Salkowski reactions and subsequent OD measurements. In all inorganic/organic phosphorus and potassium solubilization and IAA production assays, all the bacterial samples were tested in the same batch to facilitate side-by-side activity comparisons.
2.4. Bacterial DNA Extraction and 16S Ribosomal DNA Sequencing
Genomic DNA samples were extracted from endophytic bacterial collections derived from pea embryos. Their 16S rDNA V3/V4-specific fragments were amplified using the high-fidelity TaKaRa Ex Taq DNA Polymerase (TakaRa, Dalian, China) and two specific barcoded PCR primers named 343F (5′-TACGGRAGGCAGCAG-3′) and 798R (5′-AGGGTATCTAATCCT-3′). PCR products were sequenced by OE Biotech (Shanghai, China) using the Illumina PE250 platform.
2.5. Bioinformatic Analysis of High-Throughput 16S rDNA Sequencing Data
The raw bacterial 16S rDNA V3/V4-specific fragment sequences in FASTQ format were processed by Trimmomatic v0.22 [52] to remove low-quality reads. Optimized paired-end reads were assembled using FLASH v1.2.11 [53]. Shorter-than-normal reads and reads containing ambiguous or single repetitive bases were discarded to improve assembly accuracy. At the same time, UCHIME v4.1 [54] was used to detect and remove chimeric sequences. The high-quality 16S rDNA sequences generated afterwards were processed by VSEARCH v2.14.2 [55] to define operational taxonomic units (OTUs), with sequences sharing at least 97% identities being clustered into the same OTU. Representative 16S OTU sequences were selected by QIIME v1.9.1 [56] and used for BLAST searches against Greengenes v13.5 [57] and SILVA Release 138 [58] databases. BLAST results with confidence intervals larger than 0.7 were used for phylum/genus/species annotation using the Ribosomal Database Project (RDP) v11.4 Classifier [59].
3. Results
3.1. Morphological Identification of Cultivable Endophytic Bacteria in Pea Embryos with Putative Nitrogen Fixation Activity
Twenty pea accessions were divided into six cultivar groups, including DGPS (dry grain pea samples S18P01-04), FGPS (fresh grain pea samples S18P05-08), SPS (snow pea samples S18P09-12), SCPS (sweet crispy pea samples S18P13-16), SEPS (pea tip [seedling of pea] samples S18P17-19), and SHPS (pea sprout [shoot of pea] sample S18P20) which was used as a control. Twenty-seven putative NFB isolates were identified from pea embryos based on their sizes, shapes, texture, and colors (Table S1; Figure 1a). These strains were somewhat evenly distributed in DGPS (S18P01-1, S18P01-2, S18P02-1, S18P02-2, S18P02-3, S18P03-1, S18P03-2, S18P04-1, S18P04-2, and S18P04-3), FGPS (S18P05-1, S18P05-2, S18P06-1, S18P06-2, S18P06-3, S18P07-1, S18P07-2, S18P07-3, S18P08-1, S18P08-2, and S18P08-3), and SPS (S18P09, S18P10, S18P11-1, S18P11-2, S18P12-1, and S18P12-2), with no putative NFB isolates being identified from SCPS, SEPS, or SHPS.
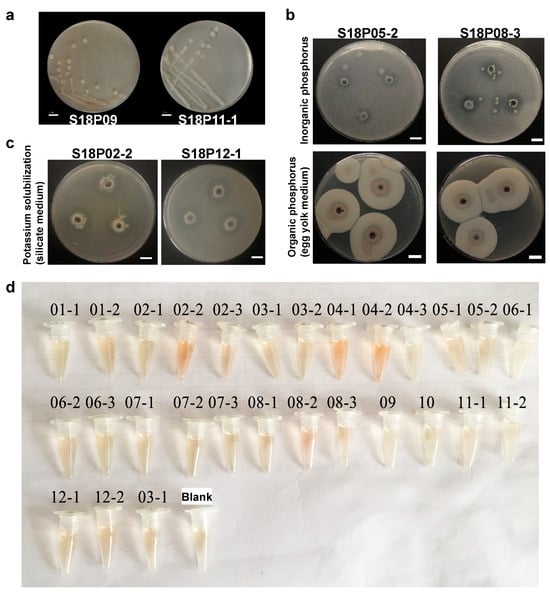
Figure 1.
Isolation of nitrogen-fixing bacteria (NFB) from pea embryos and their nutrient solubilization activities. (a) Colonial morphology of two (S18P11-1 and S18P09) representative endophytic bacterial strains. Twenty-seven culturable endophytic bacterial strains isolated in this research may possess putative nitrogen fixation capability. (b) Two NFB isolates S18P05-2 and S18P08-3 are also phosphorus-solubilizing bacteria (PSB) capable of solubilizing both inorganic and organic phosphorus. Inorganic phosphorus screen plates (top) and egg yolk plates (bottom) plates were used to test bacterial abilities to solubilize inorganic and organic phosphorus, respectively. The emergence of clear (top) and turbid (bottom) halos indicates bacterial solubilization of inorganic and organic phosphorus, respectively. (c) Among 13 isolated NFB that are also potassium-solubilizing bacteria (KSB), S18P02-2 (left) and S18P12-1 (right) have the highest potassium solubilization capacities. Bacterial potassium solubilization capability was measured by the emergence of clear halos on the silicate plates. Both S18P02-2 and S18P12-1 generated clear halos with diameters as large as 2.2 cm. (d) Quantification assays using the Salkowski reagent show that all isolated NFB secrete IAA, with the highest and the lowest levels of IAA production observed in S18P05-2 and S18P11-1, respectively.
3.2. Nutrient Solubilization and Utilization Activities of Nitrogen-Fixing Bacteria (NFB) Isolated from Pea Embryos
Nitrogenase activities of the 27 putative NFB cultures ranged from 10.67 to 89.45 and averaged at 71.95 nmol C2H4/mL/h (Table 1). They were classified into four nitrogenase activity categories of less than 40 (S18P02-1 [10.67]), 40–60 (S18P11-1 [54.37]), 60–80 (21 isolates), and above 80 (S18P05-1 [85.02], S18P08-2 [89.45], and S18P12-1 [85.86]). The three strongest NFB were isolated from FGPS (S18P05-1 and S18P08-02) and SPS (S18P12-1). Overall, 26 out of 27 isolates, excluding S18P02-1, were verified as NFB with considerable nitrogen fixation capability.

Table 1.
Nitrogen fixation (nitrogenase), nutrient (phosphorus and potassium) solubilization, and IAA production activities of the 27 putative nitrogen-fixing bacterial (NFB) strains identified from pea embryos. Solubilization of inorganic and organic phosphorus (P) and potassium (K) is indicated by the diameters of halos produced on the respective nutrient plates.
PSB and KSB solubilize soil phosphorus and potassium, respectively, for their easier absorption by plants [60,61]. We screened our NFB collection for PSB and KSB by measuring the halos they generated on certain nutrient plates, which has been demonstrated to be a convenient and reliable approach [62]. Eleven out of twenty-six NFB isolates generated clear halos on inorganic phosphorus screen plates (Table 1), demonstrating their ability to solubilize inorganic phosphorus. On the other hand, only 3 out of 26 isolates were organic phosphorus solubilizers, as revealed by their ability to generate turbid halos on egg yolk plates (Table 1). Of particular interest, S18P05-2 and S18P08-3 are the only two NFB that are also dual-role PSB capable of solubilizing both inorganic and organic phosphorus (Figure 1b).
KSB catabolize potash feldspar powder in the silicate medium. They can also degrade apatite and aluminosilicate minerals in the soil [63]. These solubilized nutrients may then be utilized by plants. Thirteen out of twenty-six NFB isolates generated clear halos on the silicate plates (Table 1), demonstrating that they are also KSB. Among them, S18P02-2 and S18P12-1 are the two strongest potassium solubilizers which generated clear halos as large as 2.2 cm in diameter (Figure 1c).
Tests using the Salkowski reagent showed that all NFB isolates secreted IAA (Figure 1d). They were classified into four categories based on their IAA production capacity (μg/mL) [64], including “below 2” (S18P11-1 [0.41] and S18P12-2 [1.97]), “2–4” (18 isolates), “4–6” (5 isolates), and “above 6” (S18P05-2 [7.10]) (Table 1). Isolates with the highest (S18P05-2) and the lowest (S18P11-1) IAA production levels were isolated from FGPS and SPS, respectively.
3.3. Quality Control of Bacterial 16S rDNA Sequencing Data and Operational Taxonomic Unit (OTU) Clustering
A range of 23449 to 37104 (33793 on average) effective DNA sequencing reads were obtained from each replicate of the 20 pea embryo bacterial collections (two replicates per collection, 40 samples total) after removing low-quality sequences (Table S2). Most of these high-quality reads are 410–450 base pairs (bp) in lengths (Figure S1). A total of 4234 OTUs were obtained using 97% identity as the threshold for clustering high-quality effective reads (valid tags). Clustered OTUs per pea embryo sample varied from 271 (S18P02.1) to 690 (S18P01.1), with 396 OTUs being annotated on average.
3.4. The Community Structure of Endophytic Bacteria in Pea Embryos
Based on OTU annotation results, endophytic bacteria isolated from the embryos of 20 pea accessions were clustered into 28 phyla, with Bacteroidetes (27.82%), Firmicutes (26.60%), Proteobacteria (17.92%), and Actinobacteria (4.10) exhibiting the highest abundance on average (Figure 2a). These OTUs were further clustered into 517 genera (Figure 2b). Overall, dominant genera include Bacteroides, Enterococcus, Bacillus, Lactobacillus, Lachnospiraceae_NK4A136_group, Alloprevotella, Prevotellaceae_UCG_001, Streptococcus, Neisseria, and Fusobacterium. In contrast to most pea embryo samples, S18P07.2 and S18P10.2/S18P20.2/S18P11.2 harbored the most dominant bacterial genera of Streptococcus and Enterococcus, respectively. In the heat map of bacterial genera abundance (Figure 3), OTUs were somewhat evenly distributed across different genera in most samples except for S18P03.2 (low abundance of Lactobacillus and Bacteroides and high abundance of Prevotella_7, Streptococcus, and Rothia), S18P01.1 (high abundance of Bacillus, Blautia, Prevotellaceae_NK3B31_group, Neisseria, and Fusobacterium), S18P011.2 (very high abundance of Enterococcus and low abundance of all other genera), and S18P14.2 (high abundance of Bacteroides and Lachnospiraceae_NK4A136_group).
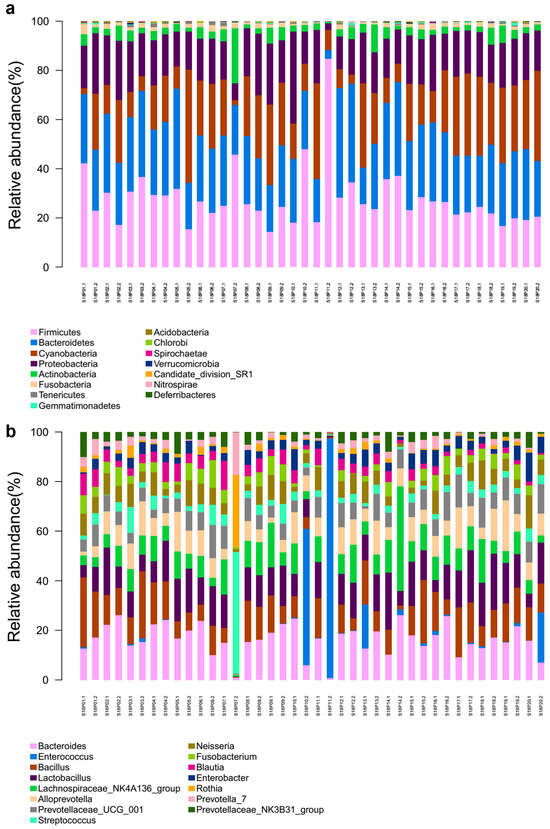
Figure 2.
Horizontal community structure of endogenous bacteria in pea embryos at the phylum (a) and genus (b) levels.
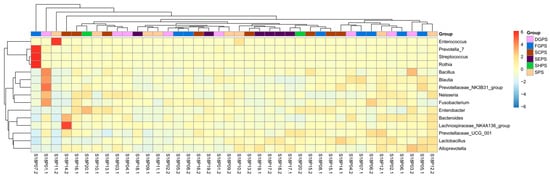
Figure 3.
The heat map of the horizontal abundance of bacterial genera in pea embryos. Red/orange and blue colors indicate relatively high and low abundance, respectively. DGPS: dry grain pea samples; FGPS: fresh grain pea samples; SPS; snow pea samples; SCPS: sweet crispy pea samples; SEPS: pea tip (seedling of pea) samples; SHPS: pea sprout (shoot of pea) samples.
3.5. Alpha Diversity Analysis of Endophytic Bacteria in Pea Embryos
Meaningful microbial community diversity and abundance analysis requires sufficient sequencing depth to cover most bacterial species in the samples. Constructed from the diversity indexes of different sequencing volumes, the Shannon–Wiener dilution curves [65] all went flat (Figure 4) with sequencing depths above 99% (Table S3), indicating that our sequencing data sizes are large enough to accurately reflect the endophytic bacterial microbiome in pea embryos.
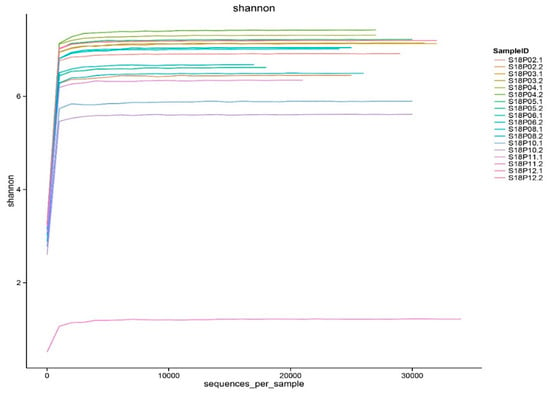
Figure 4.
The dilution curves of bacterial diversity indexes in pea embryos.
Chao1, Shannon, and Simpson indexes [66] were calculated (Table S3) to estimate the Alpha (within-sample) diversity [67] of endophytic bacteria in pea embryos. The Chao1 index is the estimation of total OTUs in a sample by counting OTUs that appear only one to two times. High Shannon index values indicate high diversity and a relatively even distribution of bacterial species. The Simpson index represents the possibility that two randomly selected bacterial isolates are different species. Simpson values are positively correlated to species diversity in samples. To facilitate diversity comparison among samples, an OTU matrix was constructed by randomly retrieving sequences from all samples with an identical sampling depth. This new matrix was used to normalize the deviation caused by sequencing depth differences so that all diversity indexes of different samples can be calculated with a uniform depth. Sample Chao1 values of our samples varied from 226.26 to 394.01 (Table S3), which are comparable to the actual annotated OTU numbers (Table S2). Shannon and Simpson index values are also consistent in each sample (Table S3). Unlike most samples, endophytic bacteria from S18P05.2, S18P11.1, and S18P11.2 exhibited much lower Alpha diversity and uneven distributions (Shannon index < 5; Simpson index < 0.9), with S18P11.2 showing the lowest values for both indexes (Table S3).
3.6. Principal Component Analysis (PCA) of Endophytic Bacteria in Pea Embryos
Principal component analysis (PCA) was carried out on endophytic bacterial samples from the embryos of all six pea cultivar groups, including DGPS, FGPS, SPS, SCPS, SEPS, and SHPS. PCA is an unsupervised learning algorithm used to essentially cluster unlabeled bacterial microbiome data through dimensionality reduction, which ends up with principal components that explain as much variation as possible [68]. Principal components 1, 2, and 3 (PC1, PC2, and PC3) contributed 51.9%, 29.7%, and 6.5% of the total variation, respectively (Figure 5). S18P07.2 in the fresh grain pea sample (FGPS) group, as well as S18P11.2 and S18P10.1 in the snow pea sample (SPS) group, showed relatively long distances and low similarities to all the other samples in the PCA map (Figure 5). Except for these two outliers, all bacterial collections, especially those from SEPS, exhibited short distances, good sample parallelism, and similar components and community structures (Figure 5). Most samples aggregated in both PC1 and PC2 (Figure 5), indicating that there is no significant structural difference in endophytic bacterial communities from pea embryos of different cultivars/accessions. In conclusion, the PCA results implicate that most endophytic bacteria from all six pea embryo cultivar groups can all be clustered together with relatively short distances (Figure 5).
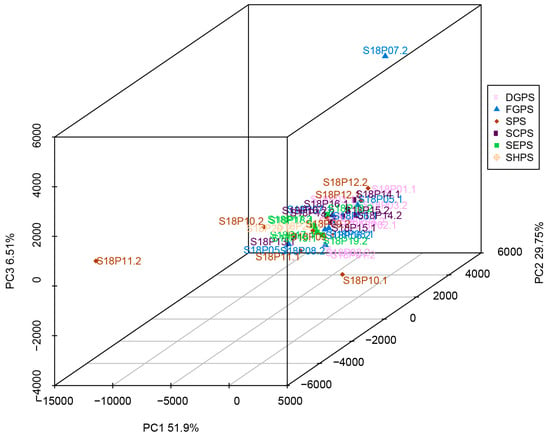
Figure 5.
Principal component analysis (PCA) of endophytic bacteria in all six pea embryo cultivar groups. DGPS: dry grain pea samples; FGPS: fresh grain pea samples; SPS; snow pea samples; SCPS: sweet crispy pea samples; SEPS: pea tip (seedling of pea) samples; SHPS: pea sprout (shoot of pea) samples.
3.7. Linear Discriminant Analysis (LDA) Coupled with Effect Size Measurement (LEfSe) Analysis of Endophytic Bacteria in Pea Embryos
Linear discriminant analysis (LDA) coupled with effect size measurement (LEfSe) analysis was carried out to analyze endophytic bacterial difference in pea embryos. LEfSe is used for high-dimensional identification and characterization of two or more significant groups in bacterial microbiome data [69,70]. LEfSe scores represent the effective sizes of abundant species and indicate species enriched in the groups. Relatively abundant endophytic bacterial species (LDA score > 3, which is more stringent than the default LDA score threshold of 2) were identified from all six cultivar groups of DGPS (Bifidobacterium, Bifidobacteriales, Bifidobacteriaceae, and Blautia), FGPS (Prevotella_7, Eubacterium_coprostanoligenes_group, Entomoplasmatales, Entomoplasmatales_Incertae_Sedis, Candidatus_Hepatoplasma, Spirochaetae, Spirochaetaceae, Spirochaetales, Spirochaetes, and Treponema_2), SPS (Geodermatophilaceae), SCPS (Marvinbryantia), SEPS (Ruminococcus_2, Incertae_Sedis, and Peptoclostridium), and SHPS (Peptostreptococcaceae, Romboutsia, Flavisolibacter, Ruminococcaceae_UCG_013, Deferribacteraceae, Deferribacteres, uncultured_bacterium, Deferribacterales, Mucispirillum, BIrii41, Flammeovirgaceae, Ambiguous_taxa, Phaselicystidaceae, Phaselicystis, Candidatus_Nitrosoarchaeum_limnia_SFB1, and SAR324_clade_Marine_group_B) (Figure 6).
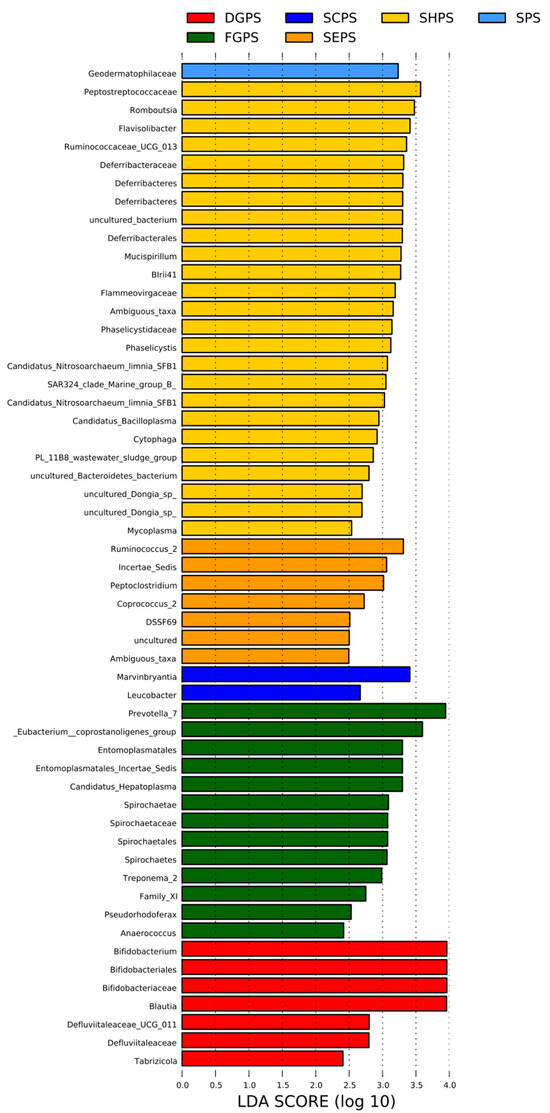
Figure 6.
Linear discriminant analysis (LDA) coupled with effect size measurement (LEfSe) analysis of endophytic bacteria in all six pea embryo cultivar groups. DGPS: dry grain pea samples; FGPS: fresh grain pea samples; SPS; snow pea samples; SCPS: sweet crispy pea samples; SEPS: pea tip (seedling of pea) samples; SHPS: pea sprout (shoot of pea) samples.
3.8. The Species of Four NFB Strains of Interest Are Determined by Long 16S rDNA Sequencing
We then sequenced long (nearly full-length) 16S rDNA fragments of S18P02-2 (a strong KSB), S18P08-2 (a strong NFB), S18P08-3 (a dual-role PSB), and S18P12-1 (a strong NFB and KSB) to determine their species (Table 2). Detailed sequencing results can be found in Table S4. Both S18P08-2 and S18P12-1 are Enterobacter roggenkampii with strong nitrogen fixation ability. However, S18P12-1 is a KSB but S18P08-2 cannot solubilize potassium, which indicates that they are probably different strains.

Table 2.
The species of four selected nitrogen-fixing bacterial (NFB) strains are determined by long 16S rDNA sequencing.
4. Discussion
Endophytic bacteria have previously been shown to benefit pea growth and health. For example, Bacillus pumilus strain SE34 in pea root tissues can induce ultrastructural modifications to enhance defense [71]. Enterobacter sp. MN17 in pea seeds enhances plant health and tolerance to cadmium [72]. In our research, the three NFB isolates with the strongest nitrogenase activity are obtained from FGPS (S18P05-1 and S18P08-02) and SPS (S18P12-1). Although they are embryo-localized and may not actually participate in nitrogen fixation at the current generation of peas, their transmission and subsequent symbiosis with the root nodules of offspring pea plants can contribute to nitrogen fixation. Pea lines with improved nitrogen fixation capacity can often be selected from nodulation mutants [73]. Crossing of the selected mutant lines with popular pea cultivars generate hybrid lines pertaining to the trait of higher nitrogen fixation potential [74]. Alternatively, the strong NFB isolates isolated in this research can potentially be inoculated to pea root nodules as biofertilizer. Their high nitrogenase activity is expected to improve the nitrogen fixation ability of host pea plants. Notably, no NFB isolate in our collection is from SCPS, SEPS, or SHPS (Table 1), indicating that the enrichment of NFB in pea embryos may be associated with pea cultivars and genotypes [75,76,77,78,79].
In legumes, endophytic bacteria are known to help plants directly and indirectly in acquiring nutrients beyond nodulation and nitrogen fixation, such as phosphate solubilization, iron chelation, phytohormone production, secreting antibacterial agents to prevent pathogen infection, outcompeting pathogens for nutrients by siderophore production, and facilitating the establishment of systemic resistance in plants [80]. For plants in general, mineral solubilizing bacteria may directly or indirectly stimulate plant growth and development by releasing plant growth regulators, solubilizing phosphorus, potassium, zinc, selenium, and silicon, producing siderophores, ammonia, hydrogen cyanide, hydrolytic enzymes (e.g., cellulases, pectinases, xylanases, amylases, and gelatinases), and bioactive compound/secondary metabolites [81,82]. At the microbiome level, nutrient-solubilizing endophytic bacteria have been identified in diverse phyla such as Actinobacteria, Bacteroidetes, Chlorobi, Cyanobacteria, Firmicutes, Gemmatimonadetes, Proteobacteria, and Tenericutes [81]. α-, β-, γ-, and δ-Proteobacteria constitute a dominant bacterial group in many plants [82]. In contrast, endophytic Bacteroidetes, Deinococcus-Thermus, and Acidobacteria exhibit the least diversity [82]. Achromobacter, Burkholderia, Bacillus, Enterobacter, Herbaspirillum, Pseudomonas, Pantoea, Rhizobium, and Streptomyces are dominant bacterial genera in most host plants [82].
In our NFB collection, the number of inorganic PSB isolates is almost three folds more than that of organic PSB. This observation is consistent with the proposed soil origination of most endophytic bacteria in embryos. The majority of soil PSB tend to solubilize and utilize inorganic phosphorus [83,84]. Since organic PSB are scarce, they can be valuable biofertilizer resources functionally supplementing with the more common inorganic PSB that have already been possessed by plants. Particularly, we obtained two dual-role PSB isolates (S18P05-2 and S18P08-3) capable of solubilizing both inorganic and organic phosphorus. When used as biofertilizer, they may help plants utilize broader types of soil phosphorus. Their potential to solubilize additional forms of inorganic and organic phosphorus should be tested in the future.
Most soil potassium exists in mineral forms such as aluminosilicate, which cannot be directly utilized by plants. Endophytic KSB play a critical role in the generation of soluble potassium for plant utilization [85]. Bacteria from the Klebsiella genus have been previously reported to be KSB [86]. Consistently, one of the strongest KSB we obtained, S18P02-2, is also a Klebsiella bacterium.
Members from the Pseudomonas genus are well known for their growth promotion and IAA production activities [87,88]. It is no surprise that a Pseudomonas isolate (S18P05-2) is the strongest IAA producer of all 26 NFB in our collection.
Data analysis of high-throughput 16S rDNA sequencing results found little variation in endophytic bacterial abundance and diversity in most embryo samples, indicating that bacterial community structures are not significantly affected by pea cultivars or accessions. The abundant endophytic bacteria identified in this research will be the targets for further biochemical, physiological, and functional investigations due to their strong presence in the pea embryo bacterial microbiome.
Endophytic bacteria support their plant hosts through vegetative and embryonic/reproductive growth promotion, pathogen reduction/protection, and abiotic stress alleviation [89]. They often employ similar colonization strategies as their pathogenic counterparts, but instead differentially and beneficially fine-tune host plant immune response, motility, and metabolic crosstalk [89]. Overall, our endophytic bacterial collection from pea embryos is a significant addition to known pea microbiomes, as endophytic bacteria in pea embryos haven’t been systematically investigated previously. NFB/PSB/KSB isolates obtained in our study can potentially be adopted as agri-environmental sustainable biofertilizers. Their efficient application in agriculture practice requires detailed understanding of the molecular mechanisms of nutrient solubilization and environmental conditions that facilitate in planta accommodation in peas and other crops [89]. In the future, their nitrogen fixation and nutrient solubilization activities may be further tested after being applied to the field. Additional challenges associated with developing these endophytic bacteria into commercial biofertilizer products include successful product refinement, the passing of toxicology analysis/evaluation, and prototype formulation [90].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app14020788/s1, Figure S1: Length–density distribution diagram of effective sequences from culturable endophytic bacteria in pea embryos; Table S1: Morphological description of the 27 putative nitrogen-fixing bacterial (NFB) strains identified from pea embryos; Table S2: Summary of effective DNA sequencing reads from endophytic bacteria in pea embryos and derived operational taxonomic units (OTUs); Table S3: Sequencing depths and diversity indexes of endophytic bacterial samples in pea embryos; Table S4: The long 16S rDNA sequences of four selected nitrogen-fixing bacterial (NFB) strains.
Author Contributions
Conceptualization, J.H., H.P. and Y.M.; methodology, X.Z., S.Q., F.S. and X.L.; validation, X.Z., S.Q., F.S. and X.L.; formal analysis, X.Z., S.Q., F.S. and X.L.; investigation, X.Z., S.Q., F.S. and X.L.; resources, J.H., H.P. and Y.M.; data curation, X.Z., S.Q., F.S. and X.L.; writing—original draft preparation, H.P., X.Z., Y.M. and J.H.; writing—review and editing, H.P., X.Z., Y.M. and J.H.; visualization, X.Z., S.Q., F.S. and X.L.; supervision, J.H., H.P. and Y.M.; project administration, J.H., H.P. and Y.M.; funding acquisition, J.H. and H.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the internal grant of Qingdao Academy of Agricultural Sciences (J.H.) and the USDA-ARS appropriated project 2034-22000-015-000D (H.P.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Acknowledgments
We thank Quanlan Liu (College of Marine Science and Biological Engineering, Qingdao University of Science and Technology) for assistance in the sequencing data analysis. Any mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not constitute endorsement by USDA. USDA is an equal opportunity provider and employer.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
DGP: Dry grain pea; DGPS: Dry grain pea samples; FGP: Fresh grain pea; FGPS: Fresh grain pea samples; IAA: Indole-3-acetic acid; KSB: Potassium-solubilizing bacteria; LDA: Linear discriminant analysis; LEfSe: Linear discriminant analysis (LDA) coupled with effect size measurement; NFB: Nitrogen-fixing bacteria; OTU: Operational taxonomic unit; PCA: Principal component analysis; PSB: Phosphorus-solubilizing bacteria; SCP: Sweet crispy pea; SCPS: Sweet crispy pea samples; SEP: Pea tip (seedling of pea); SEPS: Pea tip (seedling of pea) samples; SHP: Pea sprout (shoot of pea); SHPS: Pea sprout (shoot of pea) samples; SP: Snow pea; SPS: Snow pea samples; TSA: Trypticase Soy Agar.
References
- Powers, S.; Mirsky, E.; Bandaranayake, A.; Thavarajah, P.; Shipe, E.; Bridges, W.; Thavarajah, D. Field pea (Pisum sativum L.) shows genetic variation in phosphorus use efficiency in different P environments. Sci. Rep. 2020, 10, 18940. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Hong, S.; Li, Y. Pea protein composition, functionality, modification, and food applications: A review. Adv. Food. Nutr. Res. 2022, 101, 71–127. [Google Scholar] [PubMed]
- Ayilara, M.S.; Adeleke, B.S.; Babalola, O.O. Bioprospecting and Challenges of Plant Microbiome Research for Sustainable Agriculture, a Review on Soybean Endophytic Bacteria. Microb. Ecol. 2023, 85, 1113–1135. [Google Scholar] [CrossRef]
- Lucero, C.T.; Lorda, G.S.; Anzuay, M.S.; Ludueña, L.M.; Taurian, T. Peanut Endophytic Phosphate Solubilizing Bacteria Increase Growth and P Content of Soybean and Maize Plants. Curr. Microbiol. 2021, 8, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.K.; Dey, R.; Sherathia, D.N.; Devidayal; Mangalassery, S.; Kumar, A.; Rupapara, R.B.; Mandaliya, M.; Rawal, P.; Bhadania, R.A.; et al. Alleviation of Salinity Stress in Peanut by Application of Endophytic Bacteria. Front. Microbiol. 2021, 12, 650771. [Google Scholar] [CrossRef]
- Maheshwari, R.; Kumar, P.; Bhutani, N.; Suneja, P. Exploration of plant growth-promoting endophytic bacteria from Pisum sativum and Cicer arietinum from South-West Haryana. J. Basic Microbiol. 2022, 62, 857–874. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.; Hameed, S.; Yasmeen, T.; Zahid, M.; Zafar, M. Molecular characterization and identification of plant growth promoting endophytic bacteria isolated from the root nodules of pea (Pisum sativum L.). World J. Microbiol. Biotechnol. 2014, 30, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Freitas, H. Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 2009, 77, 153–160. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Zhang, C.; Freitas, H. Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manag. 2016, 174, 14–25. [Google Scholar] [CrossRef]
- Yang, J.; Lan, L.; Jin, Y.; Yu, N.; Wang, D.; Wang, E. Mechanisms underlying legume-rhizobium symbioses. J. Integr. Plant Biol. 2022, 64, 244–267. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Qin, Y.; Wu, H.; Zuo, W.; He, H.; Tan, J.; Wang, Y.; He, D. Isolation and Characterization of Phosphorus Solubilizing Bacteria with Multiple Phosphorus Sources Utilizing Capability and Their Potential for Lead Immobilization in Soil. Front. Microbiol. 2020, 11, 752. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, H.; Shen, Z.; Ye, J. Whole-Genome Sequencing and Potassium-Solubilizing Mechanism of Bacillus aryabhattai SK1-7. Front. Microbiol. 2022, 12, 722379. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.; Poulev, A.; Chrisler, W.; Acosta, K.; Orr, G.; Lebeis, S.; Lam, E. Auxin-Producing Bacteria from Duckweeds Have Different Colonization Patterns and Effects on Plant Morphology. Plants 2022, 11, 721. [Google Scholar] [CrossRef] [PubMed]
- Bhore, S.J.; Ravichantar, N.; Loh, C.Y. Screening of endophytic bacteria isolated from leaves of Sambung Nyawa [Gynura procumbens (Lour.) Merr.] for cytokinin-like compounds. Bioinformation 2010, 5, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Bottini, R.; Cassán, F.; Piccoli, P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 2004, 65, 497–503. [Google Scholar] [CrossRef]
- Ceballos, I.; Mosquera, S.; Angulo, M.; Mira, J.J.; Argel, L.E.; Uribe-Velez, D.; Romero-Tabarez, M.; Orduz-Peralta, S.; Villegas, V. Cultivable bacteria populations associated with leaves of banana and plantain plants and their antagonistic activity against Mycosphaerella fijiensis. Microb. Ecol. 2012, 64, 641–653. [Google Scholar] [CrossRef]
- Hazarika, S.N.; Saikia, K.; Borah, A.; Thakur, D. Prospecting Endophytic Bacteria Endowed with Plant Growth Promoting Potential Isolated from Camellia sinensis. Front. Microbiol. 2021, 12, 738058. [Google Scholar] [CrossRef]
- Yarte, M.E.; Gismondi, M.I.; Llorente, B.E.; Larraburu, E.E. Isolation of endophytic bacteria from the medicinal, forestal and ornamental tree Handroanthus impetiginosus. Environ. Technol. 2022, 43, 1129–1139. [Google Scholar] [CrossRef]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O. Metagenomics methods for the study of plant-associated microbial communities: A review. J. Microbiol. Methods 2020, 170, 105860. [Google Scholar] [CrossRef] [PubMed]
- Riva, V.; Mapelli, F.; Bagnasco, A.; Mengoni, A.; Borin, S. A Meta-Analysis Approach to Defining the Culturable Core of Plant Endophytic Bacterial Communities. Appl. Environ. Microbiol. 2022, 88, e0253721. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.; Irizarry, I.; Bergen, M.; Kharwar, R.N.; White, J.F., Jr. Seed-vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 2017, 122, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; White, J.F. Indigenous endophytic seed bacteria promote seedling development and defend against fungal disease in browntop millet (Urochloa ramosa L.). J. Appl. Microbiol. 2018, 124, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Z.; Pan, S.; Li, L.; Li, X. Characterization and Metabolism Effect of Seed Endophytic Bacteria Associated with Peanut Grown in South China. Front. Microbiol. 2019, 10, 2659. [Google Scholar] [CrossRef]
- Krishnamoorthy, A.; Agarwal, T.; Kotamreddy, J.N.R.; Bhattacharya, R.; Mitra, A.; Maiti, T.K.; Maiti, M.K. Impact of seed-transmitted endophytic bacteria on intra- and inter-cultivar plant growth promotion modulated by certain sets of metabolites in rice crop. Microbiol. Res. 2020, 241, 126582. [Google Scholar] [CrossRef]
- Dowarah, B.; Agarwal, H.; Krishnatreya, D.B.; Sharma, P.L.; Kalita, N.; Agarwala, N. Evaluation of seed associated endophytic bacteria from tolerant chilli cv. Firingi Jolokia for their biocontrol potential against bacterial wilt disease. Microbiol. Res. 2021, 248, 126751. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, S.; Liu, N. Diversity of Endophytic Bacteria in Cardamine hupingshanensis and Potential of Culturable Selenium-Resistant Endophytes to Enhance Seed Germination under Selenate Stress. Curr. Microbiol. 2021, 78, 2091–2103. [Google Scholar] [CrossRef]
- Romero, F.M.; Marina, M.; Pieckenstain, F.L. The communities of tomato (Solanum lycopersicum L.) leaf endophytic bacteria, analyzed by 16S-ribosomal RNA gene pyrosequencing. FEMS Microbiol. Lett. 2014, 351, 187–194. [Google Scholar] [CrossRef]
- Pinto-Carbó, M.; Gademann, K.; Eberl, L.; Carlier, A. Leaf nodule symbiosis: Function and transmission of obligate bacterial endophytes. Curr. Opin. Plant Biol. 2018, 44, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chang, M.; Li, H.; Zhang, Z.; Chen, Q.; Chen, Y.; Yao, Y.; Pan, A.; Shi, C.; Wang, C.; et al. Endophytic Bacteria as Contributors to Theanine Production in Camellia sinensis. J. Agric. Food Chem. 2019, 67, 10685–10693. [Google Scholar] [CrossRef] [PubMed]
- Acar, T.; Moreau, S.; Coen, O.; De Meyer, F.; Leroux, O.; Beaumel, M.; Wilkin, P.; Carlier, A. Motility-Independent Vertical Transmission of Bacteria in Leaf Symbiosis. mBio 2022, 13, e0103322. [Google Scholar] [CrossRef]
- Cullen, N.P.; Fetters, A.M.; Ashman, T.L. Integrating microbes into pollination. Curr. Opin. Insect Sci. 2021, 44, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Sahu, P.K.; Kumar, K.; Pal, G.; Gond, S.K.; Kharwar, R.N.; White, J.F. Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J. Appl. Microbiol. 2021, 131, 2161–2177. [Google Scholar] [CrossRef]
- Mayhood, P.; Mirza, B.S. Soybean Root Nodule and Rhizosphere Microbiome: Distribution of Rhizobial and Nonrhizobial Endophytes. Appl. Environ. Microbiol. 2021, 87, e02884-20. [Google Scholar] [CrossRef]
- Ovchinnikova, E.; Journet, E.P.; Chabaud, M.; Cosson, V.; Ratet, P.; Duc, G.; Fedorova, E.; Liu, W.; den Camp, R.O.; Zhukov, V.; et al. IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol. Plant-Microbe Interact. 2011, 24, 1333–1344. [Google Scholar] [CrossRef]
- Terpolilli, J.J.; Masakapalli, S.K.; Karunakaran, R.; Webb, I.U.; Green, R.; Watmough, N.J.; Kruger, N.J.; Ratcliffe, R.G.; Poole, P.S. Lipogenesis and Redox Balance in Nitrogen-Fixing Pea Bacteroids. J. Bacteriol. 2016, 198, 2864–2875. [Google Scholar] [CrossRef]
- Susniak, K.; Krysa, M.; Kidaj, D.; Szymanska-Chargot, M.; Komaniecka, I.; Zamlynska, K.; Choma, A.; Wielbo, J.; Ilag, L.L.; Sroka-Bartnicka, A. Multimodal Spectroscopic Imaging of Pea Root Nodules to Assess the Nitrogen Fixation in the Presence of Biofertilizer Based on Nod-Factors. Int. J. Mol. Sci. 2021, 22, 12991. [Google Scholar] [CrossRef]
- Westhoek, A.; Clark, L.J.; Culbert, M.; Dalchau, N.; Griffiths, M.; Jorrin, B.; Karunakaran, R.; Ledermann, R.; Tkacz, A.; Webb, I.; et al. Conditional sanctioning in a legume-Rhizobium mutualism. Proc. Natl. Acad. Sci. USA 2021, 118, e2025760118. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Sahu, P.K. Vertical Transmission of Diverse Cultivation-Recalcitrant Endophytic Bacteria Elucidated Using Watermelon Seed Embryos. Front. Microbiol. 2021, 12, 635810. [Google Scholar] [CrossRef] [PubMed]
- Passera, A.; Follador, A.; Morandi, S.; Miotti, N.; Ghidoli, M.; Venturini, G.; Quaglino, F.; Brasca, M.; Casati, P.; Pilu, R.; et al. Bacterial Communities in the Embryo of Maize Landraces: Relation with Susceptibility to Fusarium Ear Rot. Microorganisms 2021, 9, 2388. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Wang, Q.; Zhang, X.; Hao, J.; Li, L.; Chen, W.; Li, H.; Wang, Y.; Ma, C.; Wang, J.; et al. Community structure and associated networks of endophytic bacteria in pea roots throughout plant life cycle. Plant Soil 2021, 468, 225–238. [Google Scholar] [CrossRef]
- Chen, S.K.; Lin, H.F.; Wang, X.; Yuan, Y.; Yin, J.Y.; Song, X.X. Comprehensive analysis in the nutritional composition, phenolic species and in vitro antioxidant activities of different pea cultivars. Food Chem. X 2023, 17, 100599. [Google Scholar] [CrossRef]
- Wu, D.T.; Li, W.X.; Wan, J.J.; Hu, Y.C.; Gan, R.Y.; Zou, L. A Comprehensive Review of Pea (Pisum sativum L.): Chemical Composition, Processing, Health Benefits, and Food Applications. Foods 2023, 12, 2527. [Google Scholar] [CrossRef]
- Farshi, P.; Mirmohammadali, S.N.; Rajpurohit, B.; Smith, J.S.; Li, Y. Pea protein and starch: Functional properties and applications in edible films. J. Agric. Food Res. 2024, 15, 100927. [Google Scholar] [CrossRef]
- Stal, L.J. The effect of oxygen concentration and temperature on nitrogenase activity in the heterocystous cyanobacterium Fischerella sp. Sci. Rep. 2017, 7, 5402. [Google Scholar] [CrossRef]
- Bytnerowicz, T.A.; Min, E.; Griffin, K.L.; Menge, D.N.L. Repeatable, continuous and real-time estimates of coupled nitrogenase activity and carbon exchange at the whole-plant scale. Methods Ecol. Evol. 2019, 10, 960–970. [Google Scholar] [CrossRef]
- Payá-Tormo, L.; Coroian, D.; Martín-Muñoz, S.; Badalyan, A.; Green, R.T.; Veldhuizen, M.; Jiang, X.; López-Torrejón, G.; Balk, J.; Seefeldt, L.C.; et al. A colorimetric method to measure in vitro nitrogenase functionality for engineering nitrogen fixation. Sci. Rep. 2022, 12, 10367. [Google Scholar] [CrossRef]
- Kamnev, A.; Shchelochkov, A.; Perfiliev, Y.D.; Tarantilis, P.A.; Polissiou, M.G. Spectroscopic investigation of indole-3-acetic acid interaction with iron(III). J. Mol. Struct. 2001, 563, 565–572. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Gupta, D.K.; Chatterjee, S.; Datta, S.; Veer, V.; Walther, C. Role of phosphate fertilizers in heavy metal uptake and detoxification of toxic metals. Chemosphere 2014, 108, 134–144. [Google Scholar] [CrossRef]
- Sun, D.; Hale, L.; Kar, G.; Soolanayakanahally, R.; Adl, S. Phosphorus recovery and reuse by pyrolysis: Applications for agriculture and environment. Chemosphere 2018, 194, 682–691. [Google Scholar] [CrossRef]
- Boubekri, K.; Soumare, A.; Mardad, I.; Lyamlouli, K.; Hafidi, M.; Ouhdouch, Y.; Kouisni, L. The Screening of Potassium- and Phosphate-Solubilizing Actinobacteria and the Assessment of Their Ability to Promote Wheat Growth Parameters. Microorganisms 2021, 9, 470. [Google Scholar] [CrossRef]
- Meena, V.S.; Maurya, B.R.; Verma, J.P. Does a rhizospheric microorganism enhance K⁺ availability in agricultural soils? Microbiol. Res. 2014, 169, 337–347. [Google Scholar] [CrossRef]
- Çakmakçı, R.; Mosber, G.; Milton, A.H.; Alatürk, F.; Ali, B. The Effect of Auxin and Auxin-Producing Bacteria on the Growth, Essential Oil Yield, and Composition in Medicinal and Aromatic Plants. Curr. Microbiol. 2020, 77, 564–577. [Google Scholar] [CrossRef]
- Konopiński, M.K. Shannon diversity index: A call to replace the original Shannon’s formula with unbiased estimator in the population genetics studies. PeerJ 2020, 8, e9391. [Google Scholar] [CrossRef]
- Hughes, J.B.; Hellmann, J.J.; Ricketts, T.H.; Bohannan, B.J. Counting the Uncountable: Statistical Approaches to Estimating Microbial Diversity. Appl. Environ. Microbiol. 2001, 67, 4399–4406. [Google Scholar] [CrossRef]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef]
- Sankaran, K.; Holmes, S.P. Multitable Methods for Microbiome Data Integration. Front. Genet. 2019, 10, 627. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Chang, F.; He, S.; Dang, C. Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data. J. Vis. Exp. 2022, 183, e61715. [Google Scholar]
- Benhamou, N.; Kloepper, J.W.; Quadt-Hallman, A.; Tuzun, S. Induction of Defense-Related Ultrastructural Modifications in Pea Root Tissues Inoculated with Endophytic Bacteria. Plant Physiol. 1996, 112, 919–929. [Google Scholar] [CrossRef]
- Naveed, M.; Mustafa, A.; Majeed, S.; Naseem, Z.; Saeed, Q.; Khan, A.; Nawaz, A.; Baig, K.S.; Chen, J.T. Enhancing Cadmium Tolerance and Pea Plant Health through Enterobacter sp. MN17 Inoculation Together with Biochar and Gravel Sand. Plants 2020, 9, 530. [Google Scholar] [CrossRef]
- Yang, C.; Bueckert, R.; Schoenau, J.; Diederichsen, A.; Zakeri, H.; Warkentin, T.D. Evaluation of growth and nitrogen fixation of pea nodulation mutants in western Canada. Can. J. Plant Sci. 2017, 97, 1121–1129. [Google Scholar] [CrossRef]
- Dhillon, L.K.; Lindsay, D.; Yang, T.; Zakeri, H.; Tar’an, B.; Knight, J.D.; Warkentin, T.D. Biological nitrogen fixation potential of pea lines derived from crosses with nodulation mutants. Field Crops Res. 2022, 289, 108731. [Google Scholar] [CrossRef]
- Laguerre, G.; Depret, G.; Bourion, V.; Duc, G. Rhizobium leguminosarum bv. viciae genotypes interact with pea plants in developmental responses of nodules, roots and shoots. New Phytol. 2007, 176, 680–690. [Google Scholar]
- Wadhwa, K.; Dudeja, S.S.; Yadav, R.K. Molecular diversity of native rhizobia trapped by five field pea genotypes in Indian soils. J. Basic Microbiol. 2011, 51, 89–97. [Google Scholar] [CrossRef]
- Li, R.; Knox, M.R.; Edwards, A.; Hogg, B.; Ellis, T.H.; Wei, G.; Downie, J.A. Natural variation in host-specific nodulation of pea is associated with a haplotype of the SYM37 LysM-type receptor-like kinase. Mol. Plant-Microbe Interact. 2011, 24, 1396–1403. [Google Scholar] [CrossRef]
- Yang, C.; Bueckert, R.; Schoenau, J.; Diederichsen, A.; Zakeri, H.; Warkentin, T. Symbiosis of selected Rhizobium leguminosarum bv. viciae strains with diverse pea genotypes: Effects on biological nitrogen fixation. Can. J. Microbiol. 2017, 63, 909–919. [Google Scholar]
- Sherpa, M.T.; Bag, N.; Das, S.; Haokip, P.; Sharma, L. Isolation and characterization of plant growth promoting rhizobacteria isolated from organically grown high yielding pole type native pea (Pisum sativum L.) variety Dentami of Sikkim, India. Curr. Res. Microb. Sci. 2021, 2, 100068. [Google Scholar] [CrossRef]
- Dudeja, S.S.; Giri, R.; Saini, R.; Suneja-Madan, P.; Kothe, E. Interaction of endophytic microbes with legumes. J. Basic Microbiol. 2012, 52, 248–260. [Google Scholar] [CrossRef]
- Devi, R.; Kaur, T.; Kour, D.; Yadav, A.; Yadav, A.N.; Suman, A.; Ahluwalia, A.S.; Saxena, A.K. Minerals solubilizing and mobilizing microbiomes: A sustainable approach for managing minerals’ deficiency in agricultural soil. J. Appl. Microbiol. 2022, 133, 1245–1272. [Google Scholar] [CrossRef]
- Negi, R.; Sharma, B.; Kumar, S.; Chaubey, K.K.; Kaur, T.; Devi, R.; Yadav, A.; Kour, D.; Yadav, A.N. Plant endophytes: Unveiling hidden applications toward agro-environment sustainability. Folia Microbiol. 2023. [Google Scholar] [CrossRef]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [PubMed]
- Varga, T.; Hixson, K.K.; Ahkami, A.H.; Sher, A.W.; Barnes, M.E.; Chu, R.K.; Battu, A.K.; Nicora, C.D.; Winkler, T.E.; Reno, L.R.; et al. Endophyte-Promoted Phosphorus Solubilization in Populus. Front. Plant Sci. 2020, 11, 567918. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Emami, S.; Alikhani, H.A. Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth, and future prospects—A review. J. Soil Sci. Plant Nutr. 2017, 17, 897–911. [Google Scholar] [CrossRef]
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial bacteria activate nutrients and promote wheat growth under conditions of reduced fertilizer application. BMC Microbiol. 2020, 20, 38. [Google Scholar] [CrossRef]
- Laird, T.S.; Flores, N.; Leveau, J.H.J. Bacterial catabolism of indole-3-acetic acid. Appl. Microbiol. Biotechnol. 2020, 104, 9535–9550. [Google Scholar] [CrossRef]
- Sah, S.; Krishnani, S.; Singh, R. Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. Curr. Res. Microb. Sci. 2021, 2, 100084. [Google Scholar] [CrossRef]
- Wippel, K. Plant and microbial features governing an endophytic lifestyle. Curr. Opin. Plant Biol. 2023, 76, 102483. [Google Scholar] [CrossRef]
- Kaur, G.; Patel, A.; Dwibedi, V.; Rath, S.K. Harnessing the action mechanisms of microbial endophytes for enhancing plant performance and stress tolerance: Current understanding and future perspectives. Arch. Microbiol. 2023, 205, 303. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
