Algorithms and Models for Automatic Detection and Classification of Diseases and Pests in Agricultural Crops: A Systematic Review
Abstract
1. Introduction
2. Related Work
3. Methodology
- Intended goals of the review (Section 1).
- How the search was conducted (Section 3.1).
- Screening for inclusion (Section 3.2).
- Screening for exclusion (Section 3.3).
- Data extraction (Section 4).
- Analysis and Discussion (Section 5).
- Writing the review.
3.1. Search Strategy
3.2. Screening for Inclusion
- (1)
- Studies that presented a solution for automatic detection and identification systems for diseases and pests in crops;
- (2)
- Studies with full text available.
3.3. Screening for Exclusion
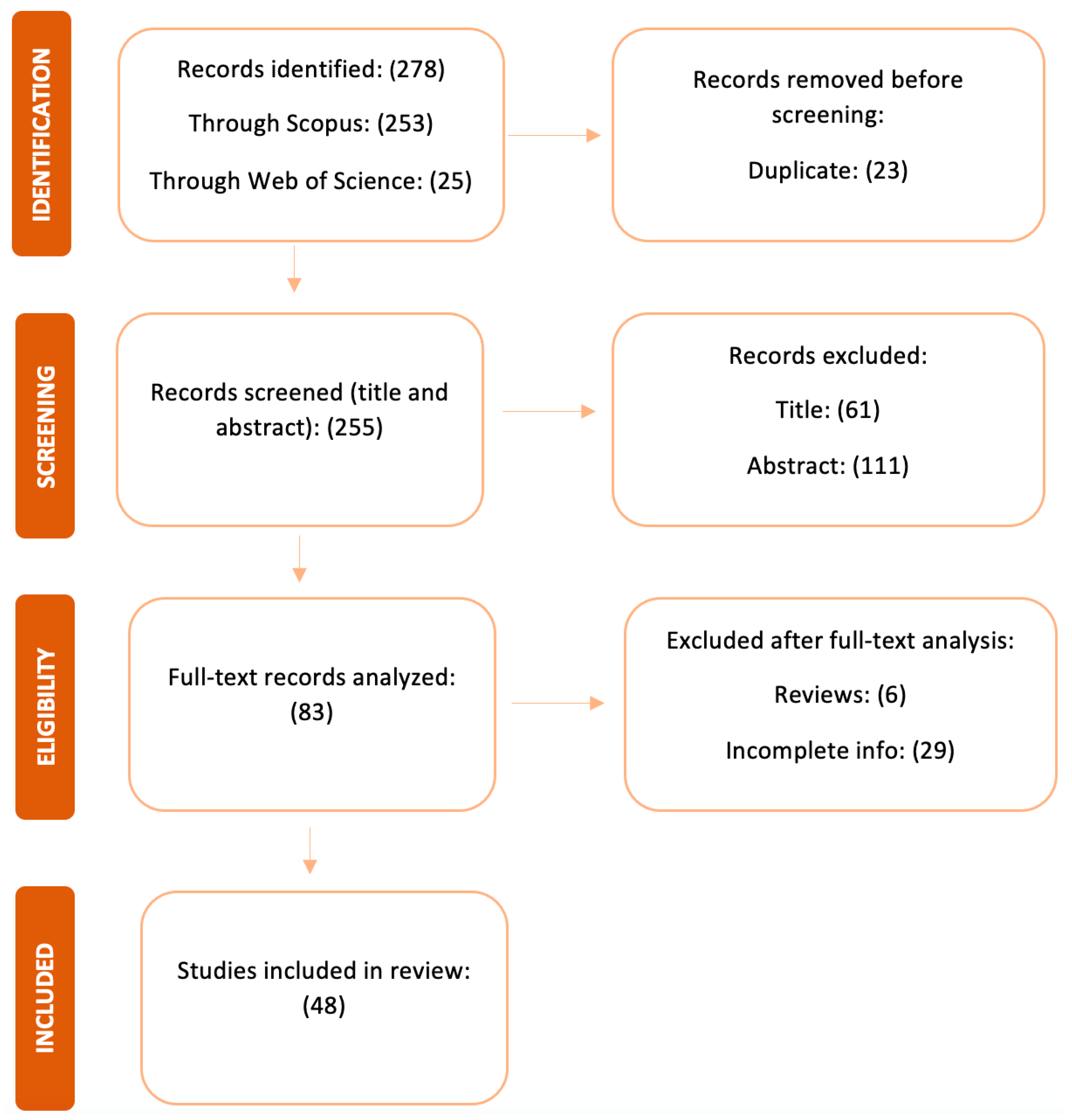
3.4. Results Summary
4. Data Extraction and Analysis
- Year of publication.
- What type of approach is described (algorithm or end user application)?
- What types of crops is it intended for?
- What are the inputs for the proposed algorithms?
- What algorithms are used?
- What information is used for the training and validation of the algorithms?
- What results were obtained in terms of accuracy and diseases or pests identified?
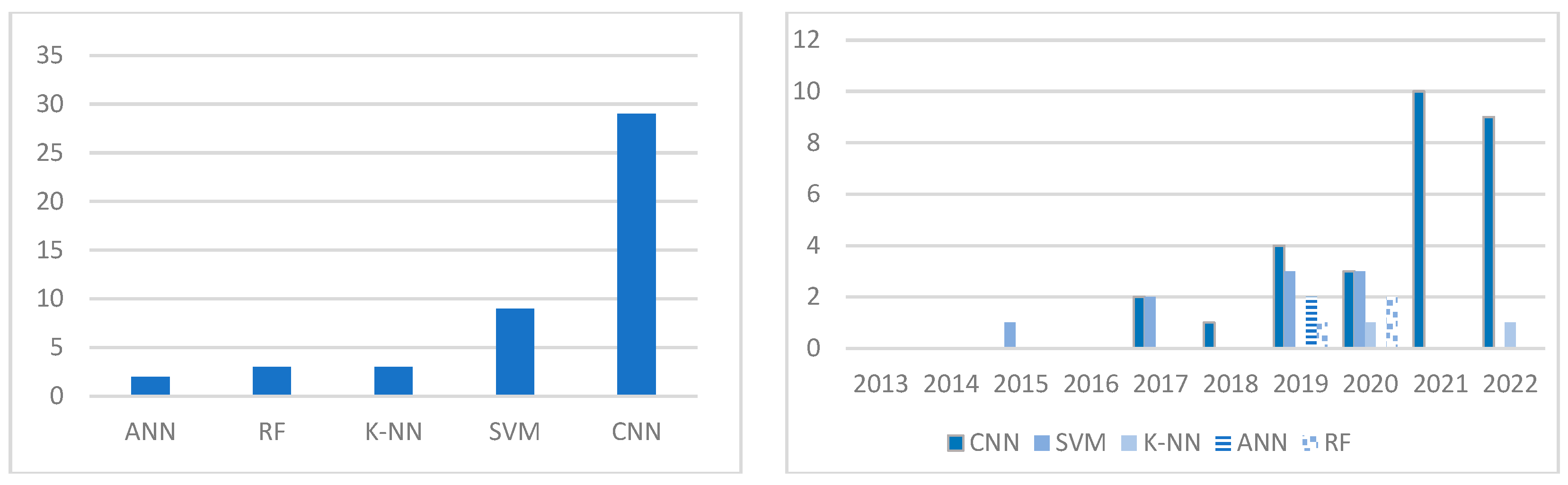
5. Discussion
6. Strengths and Limitations of this Review
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Altieri, M.A. Agroecology: The Science Of Sustainable Agriculture; CRC Press LLC: Boca Raton, FL, USA, 2018. [Google Scholar]
- Ahmed, A.A.; Al Omari, S.; Awal, R.; Fares, A.; Chouikha, M. A distributed system for supporting smart irrigation using Internet of Things technology. Eng. Rep. 2020, 3, e12352. [Google Scholar] [CrossRef]
- Sun, J.; Yang, Y.; He, X.; Wu, X. Northern Maize Leaf Blight Detection Under Complex Field Environment Based on Deep Learning. IEEE Access 2020, 8, 33679–33688. [Google Scholar] [CrossRef]
- Sinha, A.; Shekhawat, R.S. Review of image processing approaches for detecting plant diseases. IET Image Process. 2020, 14, 1427–1439. [Google Scholar] [CrossRef]
- Su, J.; Yi, D.; Su, B.; Mi, Z.; Liu, C.; Hu, X.; Xu, X.; Guo, L.; Chen, W.-H. Aerial Visual Perception in Smart Farming: Field Study of Wheat Yellow Rust Monitoring. IEEE Trans. Ind. Inform. 2021, 17, 2242–2249. [Google Scholar] [CrossRef]
- Karthick, M.; Vijayalakshmi, D.; Kumar Nath, M.; Mathumathi, M. Machine Learning Approaches for Automatic Disease Detection from Paddy Crops—A Review. Int. J. Eng. Trends Technol. 2022, 70, 392–405. [Google Scholar] [CrossRef]
- Sharma, T.; Kaur, P.; Chahal, J.; Sharma, H. Classification of rice leaf diseases based on the deep convolutional neural network architectures: Review. AIP Conf. Proc. 2022, 2451, 020086. [Google Scholar] [CrossRef]
- Domingues, T.; Brandão, T.; Ferreira, J.C. Machine Learning for Detection and Prediction of Crop Diseases and Pests: A Comprehensive Survey. Agriculture 2022, 12, 1350. [Google Scholar] [CrossRef]
- Manavalan, R. Towards an intelligent approaches for cotton diseases detection: A review. Comput. Electron. Agric. 2022, 200, 107255. [Google Scholar] [CrossRef]
- Tugrul, B.; Elfatimi, E.; Eryigit, R. Convolutional Neural Networks in Detection of Plant Leaf Diseases: A Review. Agriculture 2022, 12, 1192. [Google Scholar] [CrossRef]
- Thangaraj, R.; Anandamurugan, S.; Pandiyan, P.; Kaliappan, V.K. Artificial intelligence in tomato leaf disease detection: A comprehensive review and discussion. J. Plant Dis. Prot. 2022, 129, 469–488. [Google Scholar] [CrossRef]
- Bhise, D.; Kumar, S.; Mohapatra, H. Review on deep learning-based plant disease detection. In Proceedings of the 2022 6th International Conference on Electronics, Communication and Aerospace Technology, Coimbatore, India, 1–3 December 2022; pp. 1106–1111. [Google Scholar] [CrossRef]
- Setiawan, W.; Rochman, E.M.S.; Satoto, B.D.; Rachmad, A. Machine Learning and Deep Learning for Maize Leaf Disease Classification: A Review. J. Phys. Conf. Ser. 2022, 2406, 012019. [Google Scholar] [CrossRef]
- Sinshaw, N.T.; Assefa, B.G.; Mohapatra, S.K.; Beyene, A.M. Applications of Computer Vision on Automatic Potato Plant Disease Detection: A Systematic Literature Review. Comput. Intell. Neurosci. 2022, 2022, 7186687. [Google Scholar] [CrossRef]
- Sharma, V.; Tripathi, A.K.; Mittal, H. Technological Advancements in Automated Crop Pest and Disease Detection: A Review & Ongoing Research. In Proceedings of the 2022 International Conference on Computing, Communication, Security and Intelligent Systems (IC3SIS), Kochi, India, 23–25 June 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Saifi, S.; Mukund, B.; Alam, D.; Khanam, R. A Review on Plant Leaf Disease Detection using Deep Learning. In Proceedings of the 2022 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES), Greater Noida, India, 20–21 May 2022; pp. 101–1073. [Google Scholar] [CrossRef]
- Kumar, R.; Chug, A.; Singh, A.P.; Singh, D. A Systematic Analysis of Machine Learning and Deep Learning Based Approaches for Plant Leaf Disease Classification: A Review. J. Sens. 2022, 2022, 3287561. [Google Scholar] [CrossRef]
- Kumar, V.; Laxmi, V. Pests Detection Using Artificial Neural Network and Image Processing: A Review. In Proceedings of the 2022 International Conference on Sustainable Computing and Data Communication Systems (ICSCDS), Erode, India, 7–9 April 2022; pp. 462–467. [Google Scholar] [CrossRef]
- Li, L.; Zhang, S.; Wang, B. Plant Disease Detection and Classification by Deep Learning—A Review. IEEE Access 2021, 9, 56683–56698. [Google Scholar] [CrossRef]
- Jogekar, R.N.; Tiwari, N. A Review of Deep Learning Techniques for Identification and Diagnosis of Plant Leaf Disease. Smart Innov. Syst. Technol. 2020, 183, 435–441. [Google Scholar] [CrossRef]
- Ngugi, L.C.; Abelwahab, M.; Abo-Zahhad, M. Recent advances in image processing techniques for automated leaf pest and disease recognition—A review. Inf. Process. Agric. 2020, 8, 27–51. [Google Scholar] [CrossRef]
- Pardede, H.F.; Suryawati, E.; Krisnandi, D.; Yuwana, R.S.; Zilvan, V. Machine Learning Based Plant Diseases Detection: A Review. In Proceedings of the 2020 International Conference on Radar, Antenna, Microwave, Electronics, and Telecommunications (ICRAMET), Tangerang, Indonesia, 8–20 November 2020; pp. 212–217. [Google Scholar] [CrossRef]
- Manavalan, R. Automatic identification of diseases in grains crops through computational approaches: A review. Comput. Electron. Agric. 2020, 178, 105802. [Google Scholar] [CrossRef]
- Nagaraju, M.; Chawla, P. Systematic review of deep learning techniques in plant disease detection. Int. J. Syst. Assur. Eng. Manag. 2020, 11, 547–560. [Google Scholar] [CrossRef]
- Lima, M.C.F.; Leandro, M.E.D.D.A.; Valero, C.; Coronel, L.C.P.; Bazzo, C.O.G. Automatic Detection and Monitoring of Insect Pests—A Review. Agriculture 2020, 10, 161. [Google Scholar] [CrossRef]
- Scopus, E. Scopus. Available online: https://www.scopus.com/search/form.uri?display=basic#basic (accessed on 5 December 2022).
- Clarivate: Web of Science. Available online: https://www.webofscience.com/wos/woscc/basic-search (accessed on 5 December 2022).
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 88, 105906. [Google Scholar] [CrossRef]
- Zhao, N.; Zhou, L.; Huang, T.; Taha, M.F.; He, Y.; Qiu, Z. Development of an automatic pest monitoring system using a deep learning model of DPeNet. Measurement 2022, 203, 111970. [Google Scholar] [CrossRef]
- Lee, S.; Choi, G.; Park, H.-C.; Choi, C. Automatic Classification Service System for Citrus Pest Recognition Based on Deep Learning. Sensors 2022, 22, 8911. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liang, Y.; Zhou, L.; Tang, X.; Dai, M. An automatic inspection system for pest detection in granaries using YOLOv4. Comput. Electron. Agric. 2022, 201, 107302. [Google Scholar] [CrossRef]
- Yakkundimath, R.; Saunshi, G.; Anami, B.; Palaiah, S. Classification of Rice Diseases using Convolutional Neural Network Models. J. Inst. Eng. Ser. B 2022, 103, 1047–1059. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kodamana, H.; Chakraborty, S. Deep learning aided automatic and reliable detection of tomato begomovirus infections in plants. J. Plant Biochem. Biotechnol. 2021, 31, 573–580. [Google Scholar] [CrossRef]
- Acar, E.; Ertugrul, O.F.; Aldemir, E.; Oztekin, A. Automatic identification of cassava leaf diseases utilizing morphological hidden patterns and multi-feature textures with a distributed structure-based classification approach. J. Plant Dis. Prot. 2022, 129, 605–621. [Google Scholar] [CrossRef]
- Zhang, Y.; Wa, S.; Zhang, L.; Lv, C. Automatic Plant Disease Detection Based on Tranvolution Detection Network With GAN Modules Using Leaf Images. Front. Plant Sci. 2022, 13, 875693. [Google Scholar] [CrossRef] [PubMed]
- Sathiya, V.; Josephine, M.; Jeyabalaraja, V. An automatic classification and early disease detection technique for herbs plant. Comput. Electr. Eng. 2022, 100, 108026. [Google Scholar] [CrossRef]
- Dawod, R.G.; Dobre, C. Automatic Segmentation and Classification System for Foliar Diseases in Sunflower. Sustainability 2022, 14, 11312. [Google Scholar] [CrossRef]
- Brindha, G.M.; Karishma, K.K.; Nivetha, J.; Vidhya, B. Automatic Detection of Citrus Fruit Diseases Using MIB Classifier. In Proceedings of the 2022 3rd International Conference on Electronics and Sustainable Communication Systems (ICESC), Coimbatore, India, 17–19 August 2022; pp. 1111–1116. [Google Scholar] [CrossRef]
- Mustafa, H.; Umer, M.; Hafeez, U.; Hameed, A.; Sohaib, A.; Ullah, S.; Madni, H.A. Pepper bell leaf disease detection and classification using optimized convolutional neural network. Multimedia Tools Appl. 2022, 82, 12065–12080. [Google Scholar] [CrossRef]
- Verma, A.; Shekhar, S.; Garg, H. Plant Disease Classification Using Deep Learning Framework. In Proceedings of the 2022 International Conference on Computational Intelligence and Sustainable Engineering Solutions (CISES), Greater Noida, India, 20–21 May 2022; pp. 512–518. [Google Scholar] [CrossRef]
- Qian, S.; Du, J.; Zhou, J.; Xie, C.; Jiao, L.; Li, R. An effective pest detection method with automatic data augmentation strategy in the agricultural field. Signal Image Video Process. 2022, 17, 563–571. [Google Scholar] [CrossRef]
- Sharma, P. Automatic Classification and Diseases Identification of Indian Medicinal Plants Using Deep Learning Models. ECS Trans. 2022, 107, 4631–4639. [Google Scholar] [CrossRef]
- Jwo, D.-J.; Chiu, S.-F. Deep Learning Based Automated Detection of Diseases from Apple Leaf Images. Comput. Mater. Contin. 2022, 71, 1849–1866. [Google Scholar] [CrossRef]
- Abisha, S.; Jayasree, T. Application of Image Processing Techniques and Artificial Neural Network for Detection of Diseases on Brinjal Leaf. IETE J. Res. 2019, 68, 2246–2258. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Suzauddola; Zeb, A. Identifying crop diseases using attention embedded MobileNet-V2 model. Appl. Soft Comput. 2021, 113, 107901. [Google Scholar] [CrossRef]
- Hemalatha, A.; Vijayakumar, J. Automatic Tomato Leaf Diseases Classification and Recognition using Transfer Learning Model with Image Processing Techniques. In Proceedings of the 2021 Smart Technologies, Communication and Robotics (STCR), Sathyamangalam, India, 9–10 October 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Chowdhury, M.; Rahman, T.; Khandakar, A.; Ayari, M.; Khan, A.; Khan, M.; Al-Emadi, N.; Reaz, M.; Islam, M.; Ali, S. Automatic and Reliable Leaf Disease Detection Using Deep Learning Techniques. Agriengineering 2021, 3, 294–312. [Google Scholar] [CrossRef]
- Li, W.; Wang, D.; Li, M.; Gao, Y.; Wu, J.; Yang, X. Field detection of tiny pests from sticky trap images using deep learning in agricultural greenhouse. Comput. Electron. Agric. 2021, 183, 106048. [Google Scholar] [CrossRef]
- Latha, R.S.; Sreekanth, G.R.; Suganthe, R.C.; Rajadevi, R.; Karthikeyan, S.; Kanivel, S.; Inbaraj, B. Automatic Detection of Tea Leaf Diseases using Deep Convolution Neural Network. In Proceedings of the 2021 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 27–29 January 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Nagar, H.; Sharma, R.S. Pest Detection on Leaf using Image Processing. In Proceedings of the 2021 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 21 April 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Yatoo, A.A.; Sharma, A. A Novel Model for Automatic Crop Disease Detection. In Proceedings of the 2021 Sixth International Conference on Image Information Processing (ICIIP), Shimla, India, 26–28 November 2021; pp. 310–313. [Google Scholar] [CrossRef]
- Saini, A.K.; Bhatnagar, R.; Srivastava, D.K. AI based automatic detection of citrus fruit and leaves diseases using deep neural network model. J. Discret. Math. Sci. Cryptogr. 2021, 24, 2181–2193. [Google Scholar] [CrossRef]
- Rehman, M.Z.U.; Ahmed, F.; Khan, M.A.; Tariq, U.; Jamal, S.S.; Ahmad, J.; Hussain, I. Classification of Citrus Plant Diseases Using Deep Transfer Learning. Comput. Mater. Contin. 2022, 70, 1401–1417. [Google Scholar] [CrossRef]
- Khattak, A.; Asghar, M.U.; Batool, U.; Ullah, H.; Al-Rakhami, M.; Gumaei, A. Automatic Detection of Citrus Fruit and Leaves Diseases Using Deep Neural Network Model. IEEE Access 2021, 9, 112942–112954. [Google Scholar] [CrossRef]
- Afifi, A.; Alhumam, A.; Abdelwahab, A. Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data. Plants 2020, 10, 28. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Talukder, K.H. Detection of Potato Disease Using Image Segmentation and Machine Learning. In Proceedings of the 2020 International Conference on Wireless Communications Signal Processing and Networking (WiSPNET), Chennai, India, 4–6 August 2020; pp. 43–47. [Google Scholar] [CrossRef]
- Argüeso, D.; Picon, A.; Irusta, U.; Medela, A.; San-Emeterio, M.G.; Bereciartua, A.; Alvarez-Gila, A. Few-Shot Learning approach for plant disease classification using images taken in the field. Comput. Electron. Agric. 2020, 175, 105542. [Google Scholar] [CrossRef]
- Das, D.; Singh, M.; Mohanty, S.S.; Chakravarty, S. Leaf Disease Detection using Support Vector Machine. In Proceedings of the 2020 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 28–30 July 2020; pp. 1036–1040. [Google Scholar] [CrossRef]
- Mugithe, P.K.; Mudunuri, R.V.; Rajasekar, B.; Karthikeyan, S. Image Processing Technique for Automatic Detection of Plant Diseases and Alerting System in Agricultural Farms. In Proceedings of the 2020 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 28–30 July 2020; pp. 1603–1607. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.; Zhang, D.; Sun, Y.; Nanehkaran, Y. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, 105393. [Google Scholar] [CrossRef]
- Tetila, E.C.; Machado, B.B.; Menezes, G.K.; Oliveira, A.D.S.; Alvarez, M.; Amorim, W.P.; Belete, N.A.; Da Silva, G.G.; Pistori, H. Automatic Recognition of Soybean Leaf Diseases Using UAV Images and Deep Convolutional Neural Networks. IEEE Geosci. Remote Sens. Lett. 2019, 17, 903–907. [Google Scholar] [CrossRef]
- Gangadharan, K.; Rosline, G.; Dhanasekaran, D.; Malathi, K. Automatic Detection of Plant Disease and Insect Attack using EFFTA Algorithm. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 160–169. [Google Scholar] [CrossRef]
- Ozguven, M.M.; Adem, K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Phys. A Stat. Mech. Its Appl. 2019, 535, 122537. [Google Scholar] [CrossRef]
- Doh, B.; Zhang, D.; Shen, Y.; Hussain, F.; Doh, R.F.; Ayepah, K. Automatic Citrus Fruit Disease Detection by Phenotyping Using Machine Learning. In Proceedings of the 2019 25th International Conference on Automation and Computing (ICAC), Lancaster, UK, 5–7 September 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Thenmozhi, K.; Reddy, U.S. Crop pest classification based on deep convolutional neural network and transfer learning. Comput. Electron. Agric. 2019, 164, 104906. [Google Scholar] [CrossRef]
- Elhassouny, A.; Smarandache, F. Smart mobile application to recognize tomato leaf diseases using Convolutional Neural Networks. In Proceedings of the 2019 International Conference of Computer Science and Renewable Energies (ICCSRE), Agadir, Morocco, 22–24 July 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Howlader, M.R.; Habiba, U.; Faisal, R.H.; Rahman, M.M. Automatic Recognition of Guava Leaf Diseases using Deep Convolution Neural Network. In Proceedings of the 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’sBazar, Bangladesh, 7–9 February 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Bhimte, N.R.; Thool, V.R. Diseases Detection of Cotton Leaf Spot Using Image Processing and SVM Classifier. In Proceedings of the 2018 Second International Conference on Intelligent Computing and Control Systems (ICICCS), Madurai, India, 14–15 June 2018; pp. 340–344. [Google Scholar] [CrossRef]
- Jaisakthi, S.M.; Mirunalini, P.; Thenmozhi, D.; Vatsala. Grape Leaf Disease Identification using Machine Learning Techniques. In Proceedings of the 2019 International Conference on Computational Intelligence in Data Science (ICCIDS), Chennai, India, 21–23 February 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Tm, P.; Pranathi, A.; SaiAshritha, K.; Chittaragi, N.B.; Koolagudi, S.G. Tomato Leaf Disease Detection Using Convolutional Neural Networks. In Proceedings of the 2018 Eleventh International Conference on Contemporary Computing (IC3), Noida, India, 2–4 August 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Roldán-Serrato, K.L.; Escalante-Estrada, J.; Rodríguez-González, M. Automatic pest detection on bean and potato crops by applying neural classifiers. Eng. Agric. Environ. Food 2018, 11, 245–255. [Google Scholar] [CrossRef]
- Shijie, J.; Peiyi, J.; Siping, H.; Haibo, S. Automatic detection of tomato diseases and pests based on leaf images. In Proceedings of the 2017 Chinese Automation Congress (CAC), Jinan, China, 20–22 October 2017; pp. 2510–2537. [Google Scholar] [CrossRef]
- Lu, Y.; Yi, S.; Zeng, N.; Liu, Y.; Zhang, Y. Identification of rice diseases using deep convolutional neural networks. Neurocomputing 2017, 267, 378–384. [Google Scholar] [CrossRef]
- Singh, V.; Misra, A. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef]
- Raza, S.-E.-A.; Prince, G.; Clarkson, J.P.; Rajpoot, N.M. Automatic Detection of Diseased Tomato Plants Using Thermal and Stereo Visible Light Images. PLoS ONE 2015, 10, e0123262. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Liang, Y.; Ding, Q.; Zou, J. Automatic Identification of Peanut-Leaf Diseases Based on Stack Ensemble. Appl. Sci. 2021, 11, 1950. [Google Scholar] [CrossRef]

| Review | Year | Focus | Crop | Studies | Timespan |
|---|---|---|---|---|---|
| [6] | 2022 | Artificial Intelligence-based disease diagnosis and categorization techniques for paddy crops. | Rice | 20 | 2016–2021 |
| [7] | 2022 | Rice leaf diseases based on the Deep Convolutional Neural Network (D-CNN) architectures | Rice | 8 | 2019–2020 |
| [8] | 2022 | Machine Learning (ML) based techniques for forecasting, detection, and classification of diseases and pests, with an emphasis on tomato crops | Tomato | 16 | 2006–2020 |
| [9] | 2022 | Computational methods operated in different stages of plant-pathogen systems such as image preprocessing, segmentation, feature extraction and selection, and classification to diagnose the diseases | Cotton | 6 | 2012–2018 |
| [10] | 2022 | Review of 100 Convolutional Neural Network (CNN) approaches to detect plant leaf diseases. | Multiple | 100 | 2015–2022 |
| [11] | 2022 | Tomato leaf disease identification using image processing, ML, and deep learning approaches | Tomato | 44 | 2015–2022 |
| [12] | 2022 | Plant leaf disease recognition and classification using deep learning and advanced imaging techniques | Multiple | 50 | NA |
| [13] | 2022 | ML and deep learning for maize leaf disease classification | Maize | 62 | 2015–2022 |
| [14] | 2022 | Diseases that harm potato crops and a survey on computer vision-based techniques and types of ML algorithms used. | Potato | 39 | 2016–2022 |
| [15] | 2022 | Crop disease and pest recognition using image processing to extract the features and algorithms used in prediction studies | Multiple | 10 | 2015–2020 |
| [16] | 2022 | Plant disease detection using ML | Multiple | 15 | 2014–2021 |
| [17] | 2022 | ML and Deep Learning based approaches for plant leaf disease classification | Multiple | 30 | 1970–2022 |
| [18] | 2022 | Image processing techniques with the help of artificial neural networks for automatic pest detection | Multiple | 16 | 2012–2021 |
| [19] | 2021 | Trends and challenges for the detection of plant leaf disease using deep learning and advanced imaging techniques | Multiple | 42 | 2019–2020 |
| [20] | 2021 | D-CNN approaches for identification and diagnosis of various diseases on plant leaves | Multiple | 7 | 2016–2019 |
| [21] | 2021 | Image processing techniques employed in pest and disease recognition using visible light (RGB) images | Multiple | 51 | 2015–2020 |
| [22] | 2020 | ML technologies for plant disease detection | Multiple | 25 | 2016–2022 |
| [23] | 2020 | Grains disease detection, quantification, and classification using computational techniques | Grains | 109 | 2001–2020 |
| [24] | 2020 | Deep learning techniques in plant disease detection | Multiple | 18 | 2015–2018 |
| [25] | 2020 | Sensors for automatic detection and monitoring of insect pests | Multiple | 63 | 2004–2020 |
| Paper | Year | Application | Disease or Pest | Crop | Disease or Pest Type Found | Input |
|---|---|---|---|---|---|---|
| [29] | 2022 | Web & Mobile | Pest | N/A | N/A | Insect images from the sticky trap |
| [30] | 2022 | Web | Pest | Citrus | Citrus bacterial canker (CBC), Toxoptera citricida & Panonychus citri | Fruit and leaf image |
| [31] | 2022 | Robotic System | Pest | N/A | Red Flour Beetle and Rice Weevil | Leaf images from a small car with a camera and a supplementary light |
| [32] | 2022 | Algorithm | Disease | Rice | Grassy stunt virus, tungro virus, and yellowing mottle | Plant images |
| [33] | 2022 | Algorithm | Disease | Tomato | ToLCNDV & ToLCGV (begomovirus infections) | Leaf images |
| [34] | 2022 | Algorithm | Disease | Cassava | Bacterial blight, Brown streak disease | Leaf imagens |
| [35] | 2022 | Robotic System | Disease | N/A | 17 types of diseases (Bell Pepper Bacterial, Apple Black Rot, and others) | Leaf images |
| [36] | 2022 | Algorithm | Disease | Herbs | Gray mold, rust, and scab disease | Leaf images |
| [37] | 2022 | Algorithm | Disease | Sunflower | Alternaria, Rust Powdery mildew, downy mildew | Leaf images |
| [38] | 2022 | Algorithm | Disease | Citrus | Black spot, Melanose, Scab, Greening, Canker | Leaf Image |
| [39] | 2022 | Algorithm | Disease | Pepper | Pepper bell leaf bacterial | Leaf images |
| [40] | 2022 | Algorithm | Disease | Apple | Fungal, bacterial, and viral | Leaf images |
| Corn | ||||||
| Grape | ||||||
| Peach | ||||||
| Pepper | ||||||
| Potato | ||||||
| Strawberry | ||||||
| Tomato | ||||||
| [41] | 2022 | Algorithm | Pest | Maize | Wheat mites, wheat aphids, wheat sawflies, rice plant hopper | Leaf images |
| [42] | 2021 | Algorithm | Disease | N/A | N/A | Plant images |
| [43] | 2022 | Algorithm | Disease | Apple | Marsonina Coronaria, Apple Scab | Leaf images |
| [44] | 2019 | Algorithm | Disease | Brinjal | Pseudomonas solanacearum, Cercospora solani na others | Leaf images |
| [45] | 2021 | Algorithm | Disease | Paddy, Corn, and Cucumber | Paddy blast and others, Corn Eyespot, and others, Moderate (cucumber) and others | Leaf images |
| [46] | 2021 | Algorithm | Disease | Tomato | Early blight and late blight | Leaf images |
| [47] | 2021 | Algorithm | Disease | Tomato | Yellow Leaf Curl Virus, Mosaic Virus, Bacterial spot, and others | Leaf images |
| [48] | 2021 | Algorithm | Pest | N/A | Whitefly and Thrips | Insect images from a sticky trap |
| [49] | 2021 | Algorithm | Disease | Tea | Algae leaf spot, gray rust, white spot, and others | Leaf images |
| [50] | 2021 | Algorithm | Pest | Mustard and fava bean | N/A | Insects’ images |
| [51] | 2021 | Algorithm | Disease | N/A | Multiple diseases | Leaf images |
| [52] | 2021 | Algorithm | Disease | Citrus | N/A | Leaf images |
| [53] | 2021 | Algorithm | Disease | Citrus | Anthracnose, Canker, and Citrus scab | Leaf images |
| [54] | 2021 | Algorithm | Disease | Citrus | black spot, Canker, scab, greening, and Melanose | Leaf images |
| [55] | 2020 | Algorithm | Disease | Coffee | N/A | Leaf images |
| [56] | 2020 | Algorithm | Disease | Potato | N/A | Leaf images |
| [57] | 2020 | Algorithm | Disease | Cherry, Corn, Grape, Orange, Peach, Pepper, Potato, Raspberry, and Soybean | Several diseases | Leaf images |
| [58] | 2020 | Algorithm | Disease | Tomato | N/A | Leaf image |
| [59] | 2020 | Algorithm | Disease | N/A | Alternaria alternata | Leaf image |
| [60] | 2020 | Algorithm | Disease | Rice | Stackburn, Leaf Scald, Leaf Smut, White Tip, and Bacterial Leaf Streak. | Leaf images |
| [61] | 2020 | Algorithm | Disease | Soybean | N/A | Leaf images from UAV |
| [62] | 2020 | Algorithm | Disease | Peanut, Paddy, Cotton | Cercospora leaf spot, Bacterial Bligh, and others | Leaf image |
| [63] | 2019 | Algorithm | Disease | Sugar beet | Cercospora beticola Sacc. | Leaf images |
| [64] | 2019 | Algorithm | Disease | Citrus | Citrus scab, anthracnose, greening, and others. | Leaf images |
| [65] | 2019 | Algorithm | Pest | N/A | Several insects | Insects’ images |
| [66] | 2019 | Mobile Application | Disease | Tomato | Bacterial spot Xanthomonas campestris, mosaic virus, and others | Leaf imagens |
| [67] | 2019 | Algorithm | Disease | Guava | Whitefly, Algal leaf spot, and Rust | Leaf images |
| [68] | 2019 | Algorithm | Disease | Cotton | Bacterial Blight, Alternaria Leaf Spot, Gray Mildew, Magnesium Deficiency, and others | Leaf images |
| [69] | 2019 | Algorithm | Disease | Grape | Black rot, Esca, and leaf blight | Leaf images |
| [70] | 2018 | Algorithm | Disease | Tomato | Septoria leaf spot, Yellow leaf curl | Leaf images |
| [71] | 2018 | Algorithm | Pest | Bean, Potato | Mexican Bean Beetle and Colorado Potato Beetle | Fruit images |
| [72] | 2017 | Algorithm | Disease and pest | Tomato | Several tomato diseases | Leaf images |
| [73] | 2017 | Algorithm | Disease | Rice | Several diseases | Leaf images |
| [74] | 2017 | Algorithm | Disease | Bens, Lemon Banana | Several diseases | Leaf images |
| [75] | 2015 | Algorithm | Disease | Tomato | Powdery mildew fungus, Oidium neolycopersici | Leaf images |
| [76] | 2021 | Algorithm | Disease | Peanut | Rust, scorch, and leaf-spot | Leaf images |
| Paper | Dataset | Algorithm | Accuracy |
|---|---|---|---|
| [29] | Self-collected dataset | Faster R-CNN | 93.2% |
| [30] | Self-collected dataset (20k images) | EfficientNet-b0 model from CNN | 97% or more |
| [31] | N/A | YOLOv4 | 97.55% |
| [32] | Dataset from paddy farmlands situated at UAS, India | VGG-16 and GoogleNet CNN models | 92.24% and 91.28% |
| [33] | Self-collected dataset (12k images) | CNN (VGG 16 + ToLCNDV and ToLCGV) | 97.211% |
| [34] | Kaggle’s website—with 10.239 leaf images | k-NN approach (LBP, HOG, TEM, GWT, and GLCM) | 81.55%–89.7% |
| [35] | PlantDoc (2567 images) | Generative Adversarial Networks (GAN modules) | 51.7% (Precision), 48.1% (Recall), and 50.3% (mAP) |
| [36] | Self-collected Dataset | Fitness-distance balance deep neural network (FDB-DNN) | 2.8140%, 1.507%, 5.125%, and 0.402% higher than the existing state-of-art |
| [37] | Self-collected dataset | Faster R-CNN, Mask R-CNN | 31% to 96% |
| [38] | Self-collected dataset (3.900 images) | MIB Classifier | 98% |
| [39] | From Kaggle’s website | Five-layered CNN model | 99.99% |
| [40] | PlantVillage | Own CNN model with 18 layers and 28 target classes | 99.12% |
| [41] | Self-collected dataset | Cascade R-CNN model with Res2Net | 81.0% (mAP) |
| [42] | Self-collected dataset | CNN models (VGG16 & VGG19) | VGG16 (98.52%) and VGG19 (98.08%) |
| [43] | Self-collected dataset (50k images) | CNN + k-NN Classifier + SVM Classifier + Random Forest Classifier | 99.2% |
| [44] | Self-collected dataset (brinjal leaf images) | ANN Classifiers | 91% (with FFNN) and 98.1% (with CFNN) |
| [45] | PlantVillage | CNN models (MobileNet-V1, MobileNet-V2, NASNetMobile, EfficientNet-B0, and DenseNet-121) | 98.44% and 99.46% |
| [46] | Self-collected dataset | Transfer Learning model | 99.86% |
| [47] | PlantVillage | EfficientNet families | 99.89& (more significant result with EfficientNet-b4) |
| [48] | Self-collected dataset | Faster R-CNN | 0.944 and 0.952 |
| [49] | CIFAR-10 dataset | CNN model with 1 input layer | 94.45% |
| [50] | Self-collected dataset | ORB algorithm | 91.89% |
| [51] | PlantPathology | VGG16 DL model | 85% |
| [52] | N/A | CNN model | More than 90% |
| [53] | Self-collected dataset | Models: MobileNetv2 and DenseNet201 | 95.7% |
| [54] | PlantVillage | Multi-Layer CNN | 94.55% |
| [55] | PlantVillage and Coffee Leaf dataset | ResNet18, ResNet34 e ResNet50 (models from CNN) | 99% and 81% |
| [56] | PlantVillage | ML classifiers (Random Forest, k-NN, Decision Trees, SVM, and others) | 97% (higher precision from RF) |
| [57] | PlantVillage | FSL method, SVM classfifier | 94.0% (higher precision) |
| [58] | Kaggle datasets | SVM, LR, RF | 87.6, 67.3 and 70.05 |
| [59] | N/A | K-Means Clustering (for alerts) | 95.1613% |
| [60] | ImageNet and PlantVillage | VGGNet | 91.82% and 92.0% |
| [61] | Images taken from UAV (drone) | DL models (ResNet-50, Xception, VGG-19 and Inception-v3) | 99.04% |
| [62] | PlantVillage | Enhanced Fusion Fractal Texture Analysis (EFFTA) | 97.69% |
| [63] | Self-collected dataset | Faster R-CNN models | 95.48% |
| [64] | Kaggle datasets | SVM + ANN | SVM (93.12%) and ANN (88.96) |
| [65] | NBAIR dataset, Xie1 and Xie2 dataset | Self-model proposed is based on CNN models with six convolutional layers | 96.75, 97.47, and 95.97% |
| [66] | Plant health (7176 images of tomatoes used) | CNN (MobileNet Model) | 90.3% |
| [67] | Self-collected dataset (BU_Guava_Leaf (BUGL2018)) | 11 layers-based D-CNN model | 98.74% |
| [68] | Self-collected dataset (images taken by digital camera) | SVM classifier | 98.46% |
| [69] | PlantVillage | SVM, adaboost, and Random Forest tree | 93% |
| [70] | PlantVillage | AlexNet, GoogleNet and LeNet | 94%–95% |
| [71] | Self-collected dataset | RSC (Random Subspace Classifier) and LIRA (Limited Receptive Area) | 88% (CPB with LIRA) and 89% (MBB with RSC) |
| [72] | Self-collected dataset | VGG16 + SVM | 89% |
| [73] | Heilongjiang Academy of Land Reclamation Sciences, China | CNN model | 95.48% |
| [74] | Self-collected dataset (Images from a digital camera or similar devices) | SVM Classifier | 95.71% |
| [75] | Middlebury dataset | SVM classifier | More than 90% |
| [76] | Self-collected dataset | CNN (ResNet50 and DenseNet121) | ResNet50 (97.59%) and DenseNet121 (90.50%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Francisco, M.; Ribeiro, F.; Metrôlho, J.; Dionísio, R. Algorithms and Models for Automatic Detection and Classification of Diseases and Pests in Agricultural Crops: A Systematic Review. Appl. Sci. 2023, 13, 4720. https://doi.org/10.3390/app13084720
Francisco M, Ribeiro F, Metrôlho J, Dionísio R. Algorithms and Models for Automatic Detection and Classification of Diseases and Pests in Agricultural Crops: A Systematic Review. Applied Sciences. 2023; 13(8):4720. https://doi.org/10.3390/app13084720
Chicago/Turabian StyleFrancisco, Mauro, Fernando Ribeiro, José Metrôlho, and Rogério Dionísio. 2023. "Algorithms and Models for Automatic Detection and Classification of Diseases and Pests in Agricultural Crops: A Systematic Review" Applied Sciences 13, no. 8: 4720. https://doi.org/10.3390/app13084720
APA StyleFrancisco, M., Ribeiro, F., Metrôlho, J., & Dionísio, R. (2023). Algorithms and Models for Automatic Detection and Classification of Diseases and Pests in Agricultural Crops: A Systematic Review. Applied Sciences, 13(8), 4720. https://doi.org/10.3390/app13084720











