Learning to Segment Blob-like Objects by Image-Level Counting
Abstract
:1. Introduction
- The development of a counting head as an extension that enables count training for segmentation networks;
- The introduction of a method to connect counting networks to classification-based CAM analysis without losing the counting information;
- The development of two losses, contrast loss and morphological loss, which enforce the outputs of a segmentation learned by count training to be suitable for subsequent blob detection by penalizing noise and favoring blob-like structures.
2. State-of-the-Art Research
3. Methods
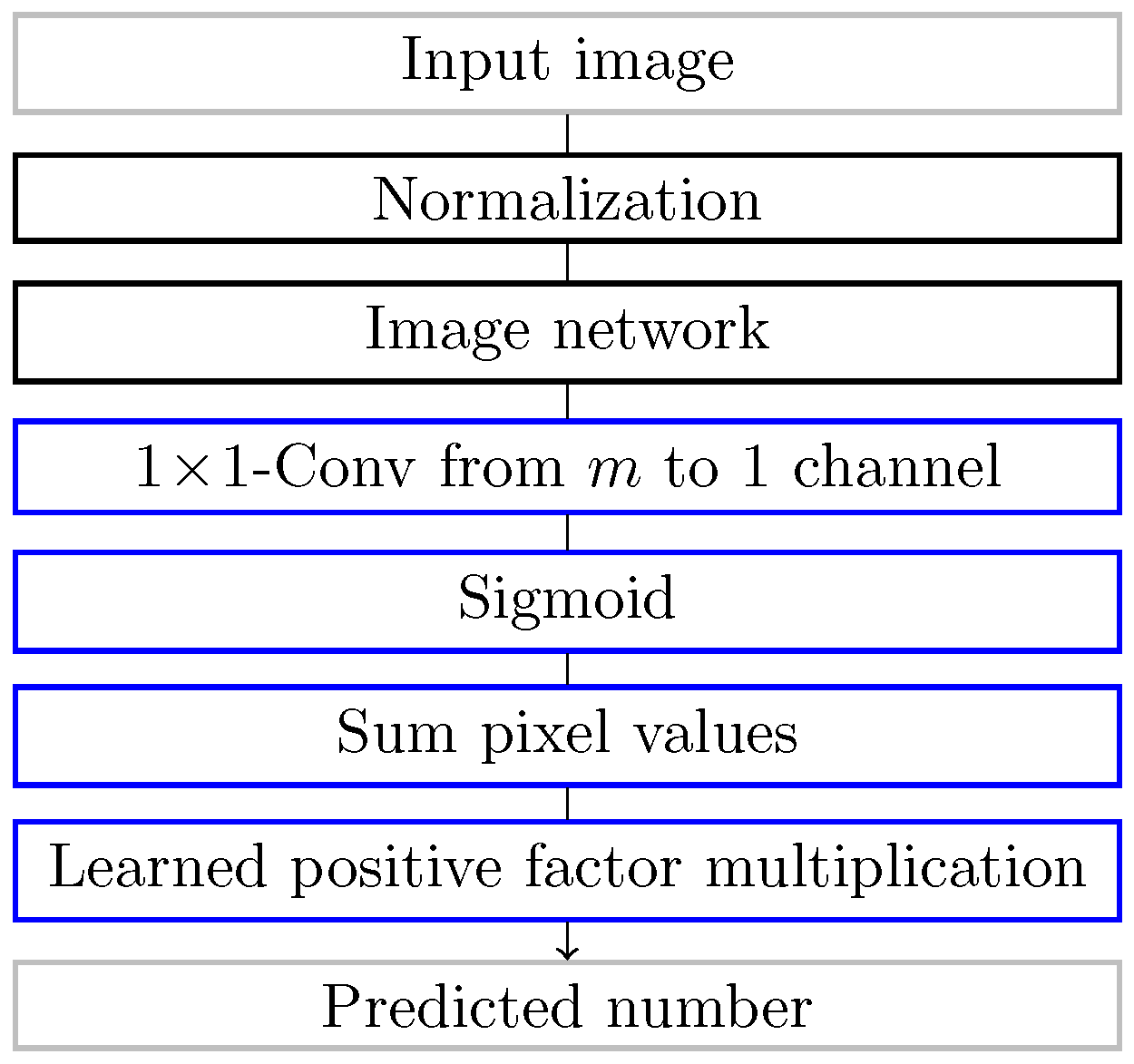
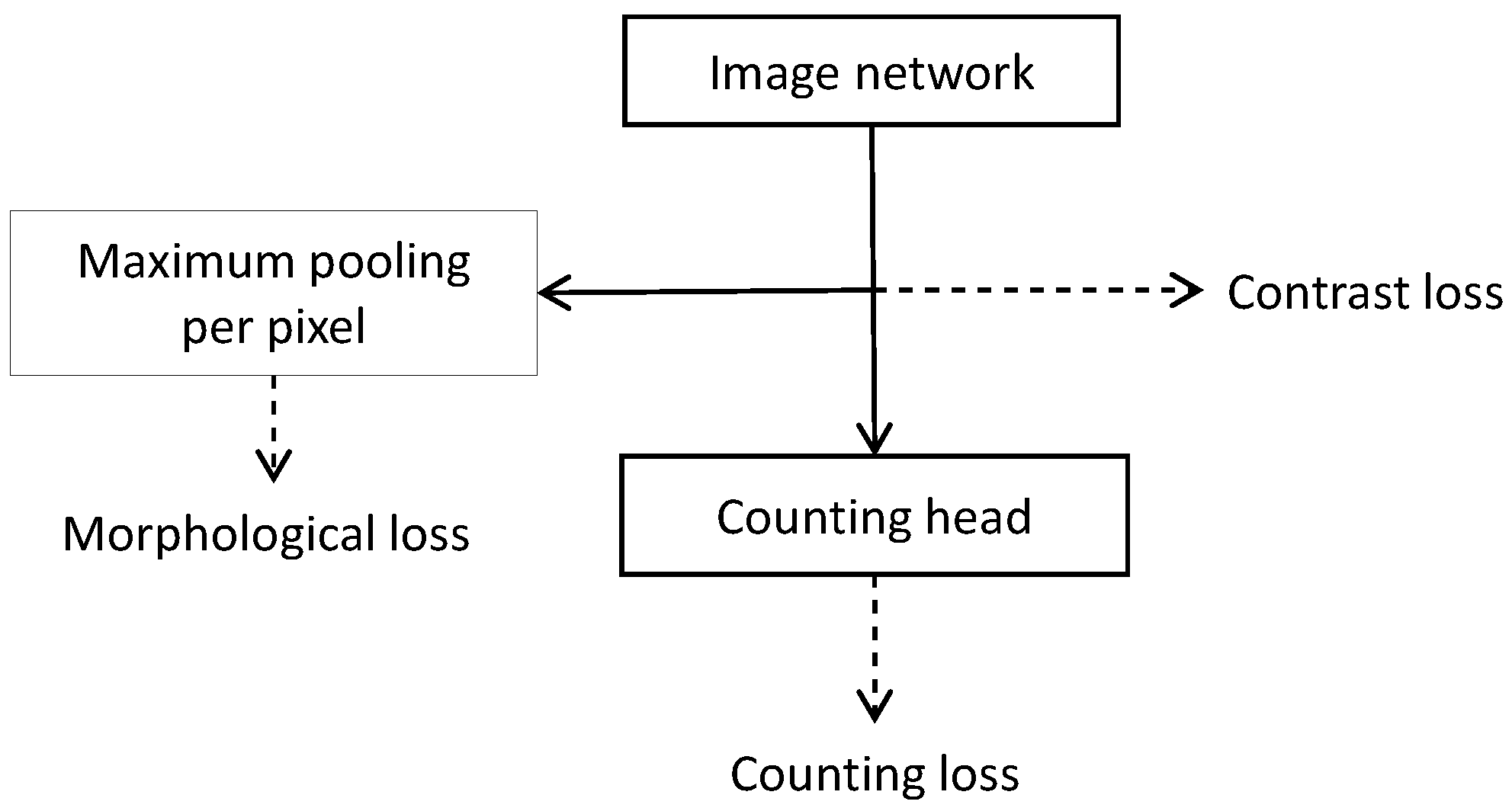
3.1. Counting Loss
3.2. Class-Activation-Mapping-Based Segmentation
3.3. Contrast Loss
3.4. Morphological Loss
| Algorithm 1 Computation of the morphological loss for object counting. The inputs are the image , the expected number of objects , the percentage p of extreme values to use for the smoothed minimum and maximum calculation, and the scaling factor m |
|
4. Experiments and Results
4.1. Hardware and Frameworks
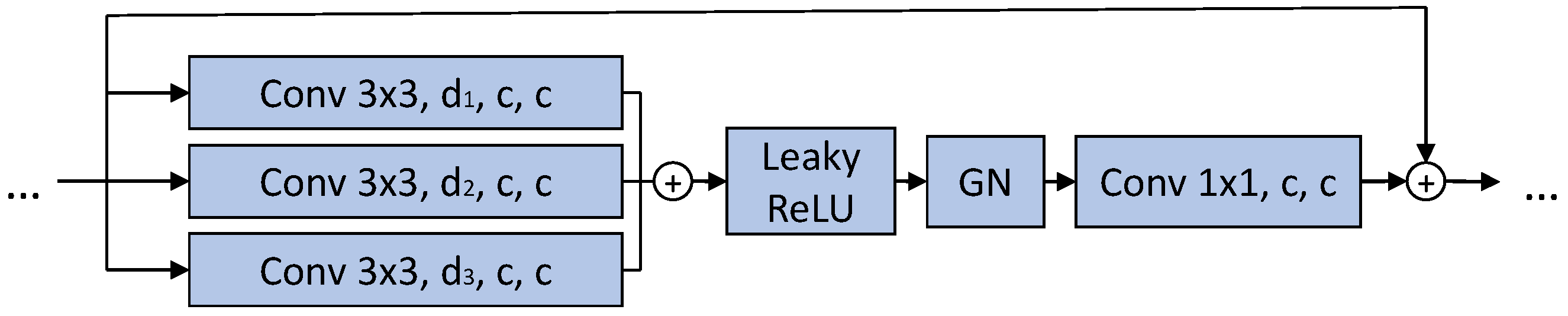
4.2. Architectures, Training, and Evaluation Settings
4.2.1. Natural Data
4.2.2. Synthetic Data Generation
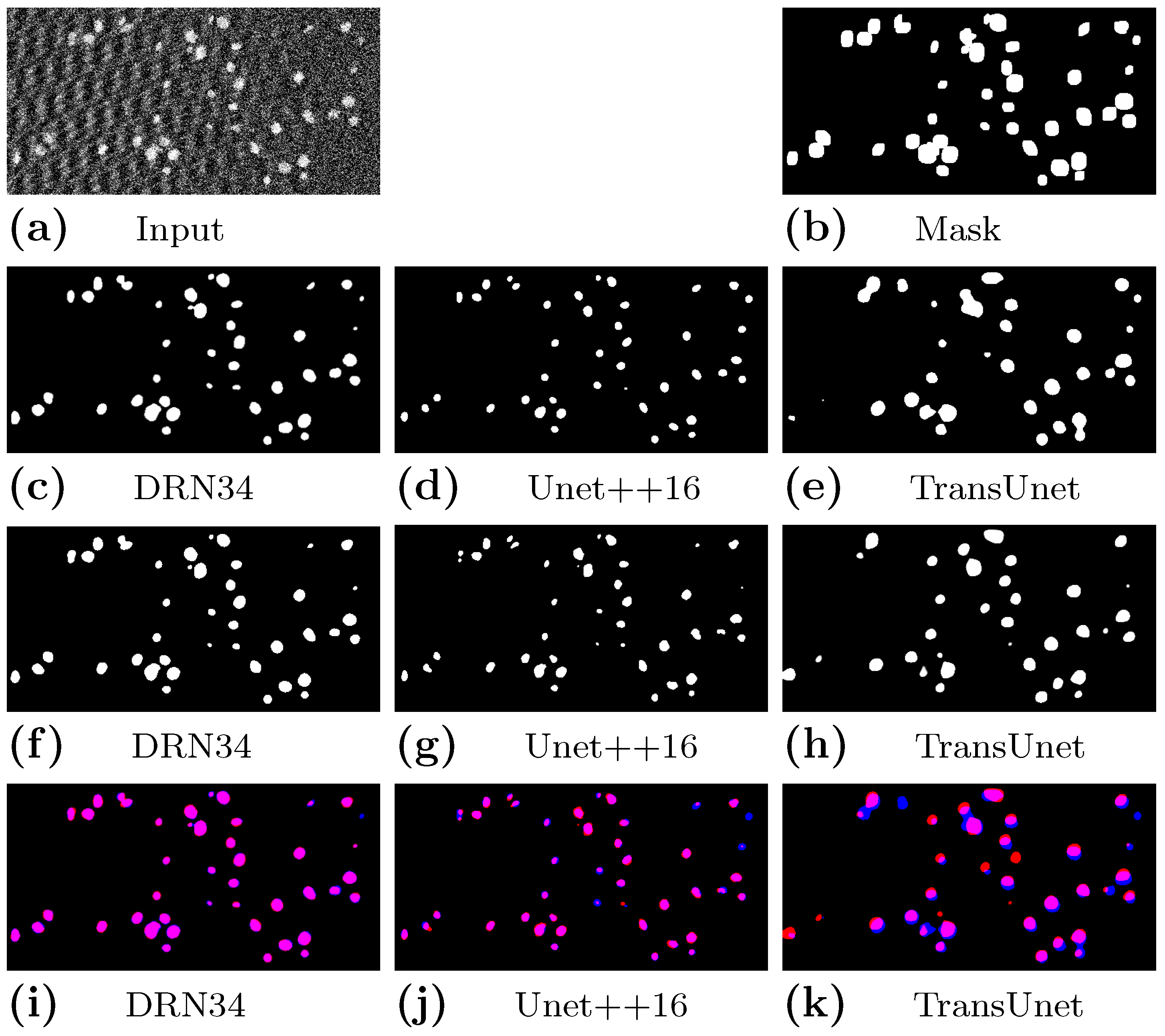
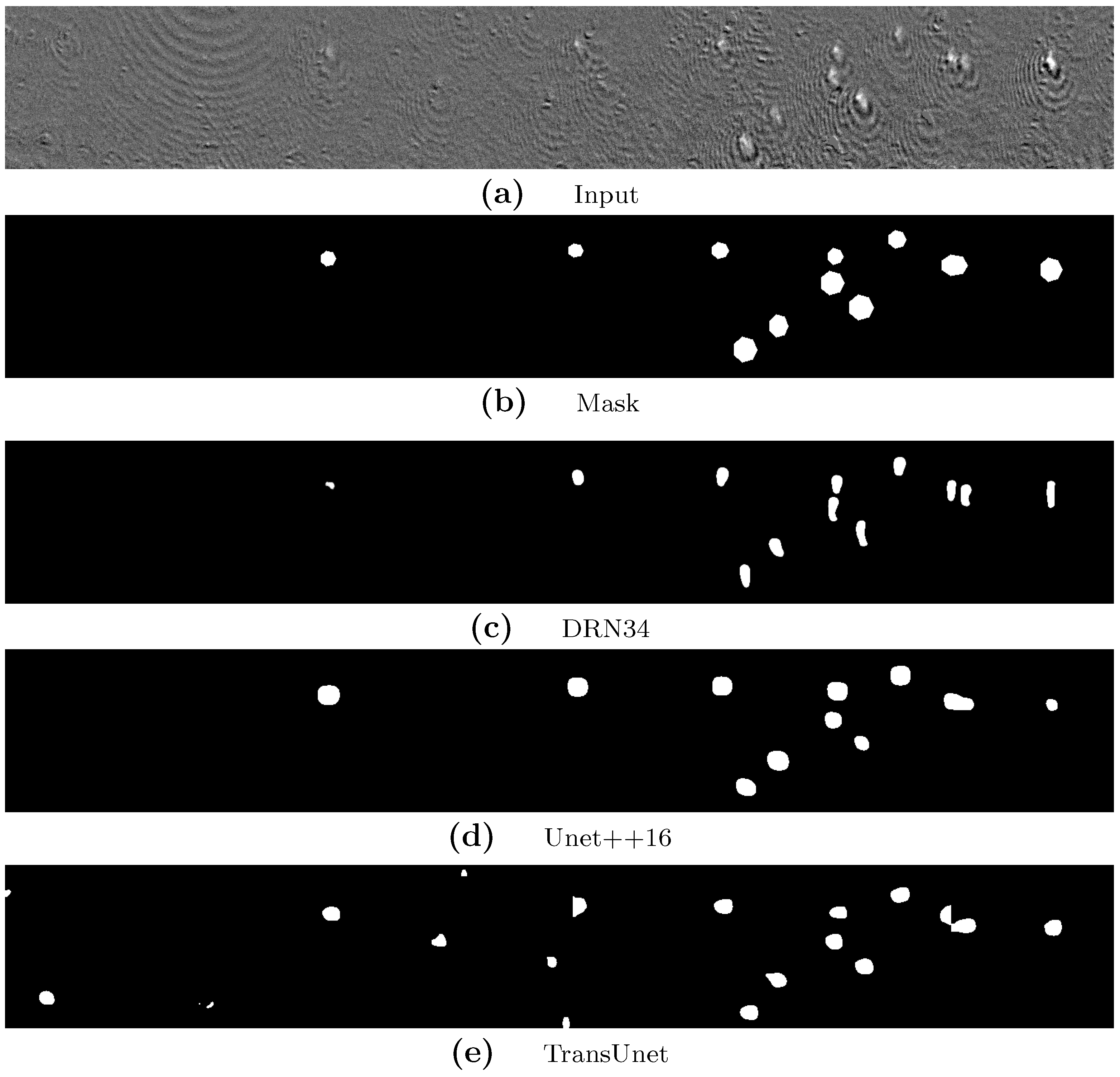
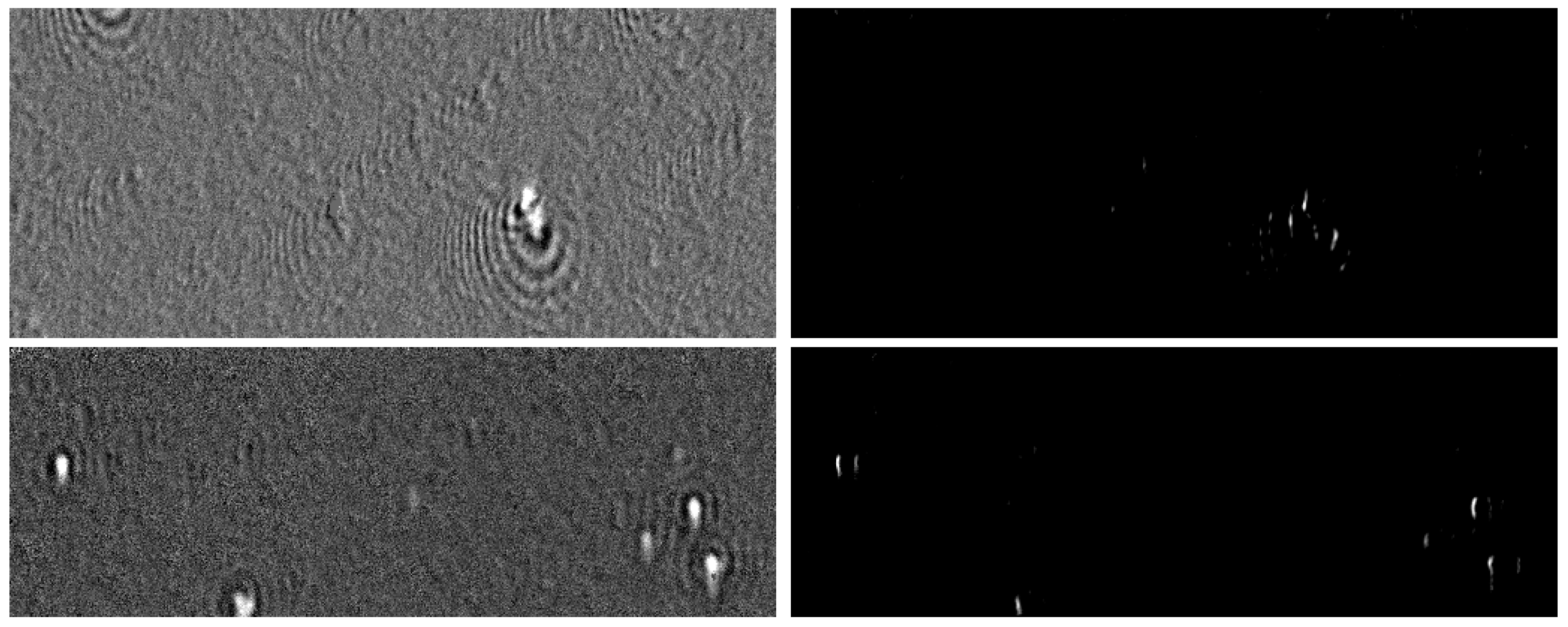
4.3. Results
4.3.1. Results on Synthetic Images
4.3.2. Results on Natural Images
4.3.3. Undesirable Effects
5. Discussion
6. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vădineanu, C.; Pelt, D.M.; Dzyubachyk, O.; Batenburg, K.J. An Analysis of the Impact of Annotation Errors on the Accuracy of Deep Learning for Cell Segmentation. In Proceedings of the 5th International Conference on Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022; Volume 172, pp. 1251–1267. [Google Scholar]
- Zhang, D.; Han, J.; Cheng, G.; Yang, M. Weakly Supervised Object Localization and Detection: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 5866–5885. [Google Scholar] [CrossRef]
- Khan, M.A.; Menouar, H.; Hamila, R. Revisiting Crowd Counting: State-of-the-Art, Trends, and Future Perspectives. Image Vis. Comput. 2023, 129, 104597. [Google Scholar] [CrossRef]
- Pandey, G.; Dukkipati, A. Learning to Segment with Image-Level Supervision. In Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa Village, HI, USA, 7–11 January 2019; pp. 1856–1865. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, À.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2016; pp. 2921–2929. [Google Scholar]
- Caicedo, J.C.; Roth, J.; Goodman, A.; Becker, T.; Karhohs, K.W.; Broisin, M.; Molnar, C.; McQuin, C.; Singh, S.; Theis, F.J.; et al. Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images. Cytom. Part A 2019, 95, 952–965. [Google Scholar] [CrossRef]
- Melanthota, S.K.; Gopal, D.; Chakrabarti, S.; Kashyap, A.A.; Radhakrishnan, R.; Mazumder, N. Deep Learning-Based Image Processing in Optical Microscopy. Biophys. Rev. 2022, 14, 463–481. [Google Scholar] [CrossRef]
- Huo, Z.; Li, Y.; Chen, B.; Zhang, W.; Yang, X.; Yang, X. Recent Advances in Surface Plasmon Resonance Imaging and Biological Applications. Talanta 2023, 255, 124213. [Google Scholar] [CrossRef]
- Hergenröder, R.; Weichert, F.; Wüstefeld, K.; Shpacovitch, V. 2.2 Virus Detection. In Volume 3 Applications; De Gruyter: Berlin, Germany; Boston, MA, USA, 2023; pp. 21–42. [Google Scholar] [CrossRef]
- Libuschewski, P. Exploration of Cyber-Physical Systems for GPGPU Computer Vision-Based Detection of Biological Viruses. Ph.D. Thesis, TU Dortmund, Dortmund, Germany, 2017. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Is Object Localization for Free?—Weakly-Supervised Learning with Convolutional Neural Networks. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 685–694. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Gao, G.; Gao, J.; Liu, Q.; Wang, Q.; Wang, Y. CNN-based Density Estimation and Crowd Counting: A Survey. arXiv 2020, arXiv:2003.12783. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Li, Y.; Zhang, X.; Chen, D. CSRNet: Dilated Convolutional Neural Networks for Understanding the Highly Congested Scenes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1091–1100. [Google Scholar] [CrossRef]
- Liang, D.; Chen, X.; Xu, W.; Zhou, Y.; Bai, X. Transcrowd: Weakly-Supervised Crowd Counting With Transformers. Sci. China Inf. Sci. 2022, 65, 160104. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings Part III 18; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Che, H.; Yin, X.X.; Sun, L.; Fu, Y.; Lu, R.; Zhang, Y. U-Net-Based Medical Image Segmentation. J. Healthc. Eng. 2022, 2022, 4189781. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 31, 6000–6010. [Google Scholar]
- Gohel, P.; Singh, P.; Mohanty, M. Explainable AI: Current Status and Future Directions. arXiv 2021, arXiv:2107.07045. [Google Scholar]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations From Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Generalized Gradient-based Visual Explanations for Deep Convolutional Networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar] [CrossRef]
- Srinivas, S.; Fleuret, F. Full-Gradient Representation for Neural Network Visualization. Adv. Neural Inf. Process. Syst. 2019, 33, 4124–4133. [Google Scholar]
- Draelos, R.L.; Carin, L. Use HiResCAM Instead of Grad-CAM for Faithful Explanations of Convolutional Neural Networks. arXiv 2020, arXiv:2011.08891. [Google Scholar] [CrossRef]
- Jiang, P.T.; Zhang, C.B.; Hou, Q.; Cheng, M.M.; Wei, Y. LayerCAM: Exploring Hierarchical Class Activation Maps for Localization. IEEE Trans. Image Process. 2021, 30, 5875–5888. [Google Scholar] [CrossRef]
- Muhammad, M.B.; Yeasin, M. Eigen-CAM: Class Activation Map Using Principal Components. In Proceedings of the 2020 International Joint Conference on Neural Networks, Glasgow, UK, 29–24 July 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Desai, S.; Ramaswamy, H.G. Ablation-CAM: Visual Explanations for Deep Convolutional Network via Gradient-free Localization. In Proceedings of the 2020 IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, CO, USA, 1–5 March 2020; pp. 972–980. [Google Scholar] [CrossRef]
- Wang, H.; Du, M.; Yang, F.; Zhang, Z. Score-CAM: Improved Visual Explanations via Score-Weighted Class Activation Mapping. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Seattle, WA, USA, 13–19 June 2020; pp. 24–25. [Google Scholar]
- Zheng, H.; Yang, Z.; Liu, W.; Liang, J.; Li, Y. Improving Deep Neural Networks Using Softplus Units. In Proceedings of the 2015 International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–17 July 2015; pp. 1–4. [Google Scholar] [CrossRef]
- Wüstefeld, K.; Weichert, F. An Automated Rapid Test for Viral Nanoparticles Based on Spatiotemporal Deep Learning. In Proceedings of the 2020 IEEE Sensors Conference, Virtual, 25–28 October 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Pare, S.; Kumar, A.; Singh, G.K.; Bajaj, V. Image Segmentation Using Multilevel Thresholding: A Research Review. Iran. J. Sci. Technol. Trans. Electr. Eng. 2020, 44, 1–29. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 33, 8026–8037. [Google Scholar]
- Riba, E.; Mishkin, D.; Ponsa, D.; Rublee, E.; Bradski, G.R. Kornia: An Open Source Differentiable Computer Vision Library for PyTorch. In Proceedings of the 2020 IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass, CO, USA, 1–5 March 2020; pp. 3663–3672. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical Image Segmentation Review: The Success of U-Net. arXiv 2022, arXiv:2211.14830. [Google Scholar]
- Chang, M.; Li, Q.; Feng, H.; Xu, Z. Spatial-Adaptive Network for Single Image Denoising. arXiv 2020, arXiv:2001.10291. [Google Scholar]
- Wu, Y.; He, K. Group Normalization. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Liang, A.; Liu, Q.; Wen, G.; Jiang, Z. The Surface-Plasmon-Resonance Effect of Nanogold/Silver and Its Analytical Applications. TrAC Trends Anal. Chem. 2012, 37, 32–47. [Google Scholar] [CrossRef]
- Roth, A.; Wüstefeld, K.; Weichert, F. A Data-Centric Augmentation Approach for Disturbed Sensor Image Segmentation. J. Imaging 2021, 7, 206. [Google Scholar] [CrossRef] [PubMed]

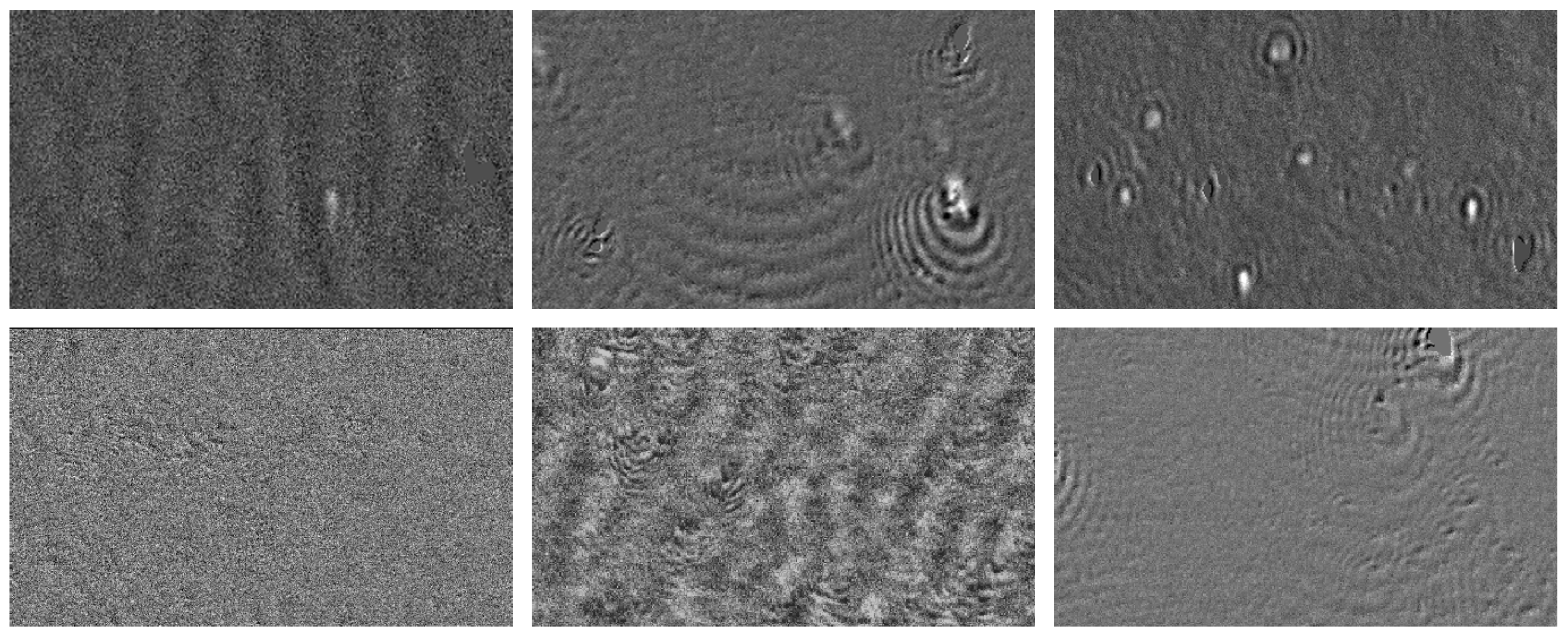


| Parameter | Interval | Description |
|---|---|---|
| number of ellipses | ||
| noise intensity | ||
| ellipse intensity | ||
| bounding box side lengths | ||
| wave intensity | ||
| waves probability | ||
| number of waves | ||
| wave fading rate |
| Network | F1-Score | Recall | Precision |
|---|---|---|---|
| Dilated ResNet-18 | 0.989 | 0.999 | 0.981 |
| Dilated ResNet-34 | 0.989 | 0.999 | 0.982 |
| UNet++-8 | 0.986 | 0.995 | 0.980 |
| UNet++-16 | 0.986 | 0.995 | 0.980 |
| TransUNet | 0.989 | 0.999 | 0.980 |
| Network | F1-Score | Recall | Precision |
|---|---|---|---|
| Dilated ResNet-18 | 0.978 | 0.967 | 0.992 |
| Dilated ResNet-34 | 0.985 | 0.997 | 0.979 |
| UNet++8 | 0.979 | 0.999 | 0.966 |
| UNet++16 | 0.993 | 0.996 | 0.992 |
| TransUNet | 0.983 | 0.992 | 0.982 |
| Network | F1-Score | Recall | Precision |
|---|---|---|---|
| Dilated ResNet-18 | 0.945 | 0.903 | 0.996 |
| Dilated ResNet-34 | 0.969 | 0.994 | 0.961 |
| UNet++8 | 0.969 | 0.980 | 0.973 |
| UNet++16 | 0.987 | 0.985 | 0.992 |
| TransUNet | 0.987 | 0.984 | 0.993 |
| Network | Only Spatial | With Tracking | ||
|---|---|---|---|---|
| F1-Score | Recall | Precision | F1 Score | |
| Dilated ResNet-18 | 0.871 | 0.936 | 0.839 | 0.941 |
| Dilated ResNet-34 | 0.870 | 0.893 | 0.873 | 0.914 |
| UNet++-8 | 0.896 | 0.928 | 0.884 | 0.945 |
| UNet++-16 | 0.890 | 0.894 | 0.909 | 0.941 |
| TransUNet | 0.873 | 0.947 | 0.828 | 0.929 |
| Network | Only Spatial | With Tracking | ||
|---|---|---|---|---|
| F1-Score | Recall | Precision | F1 Score | |
| Dilated ResNet-18 | 0.879 | 0.925 | 0.854 | 0.937 |
| Dilated ResNet-34 | 0.844 | 0.830 | 0.894 | 0.922 |
| UNet++-8 | 0.899 | 0.929 | 0.890 | 0.955 |
| UNet++-16 | 0.897 | 0.942 | 0.871 | 0.952 |
| TransUNet | 0.817 | 0.946 | 0.744 | 0.868 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wüstefeld, K.; Ebbinghaus, R.; Weichert, F. Learning to Segment Blob-like Objects by Image-Level Counting. Appl. Sci. 2023, 13, 12219. https://doi.org/10.3390/app132212219
Wüstefeld K, Ebbinghaus R, Weichert F. Learning to Segment Blob-like Objects by Image-Level Counting. Applied Sciences. 2023; 13(22):12219. https://doi.org/10.3390/app132212219
Chicago/Turabian StyleWüstefeld, Konstantin, Robin Ebbinghaus, and Frank Weichert. 2023. "Learning to Segment Blob-like Objects by Image-Level Counting" Applied Sciences 13, no. 22: 12219. https://doi.org/10.3390/app132212219
APA StyleWüstefeld, K., Ebbinghaus, R., & Weichert, F. (2023). Learning to Segment Blob-like Objects by Image-Level Counting. Applied Sciences, 13(22), 12219. https://doi.org/10.3390/app132212219






