Predicting the Composition and Mechanical Properties of Seaweed Bioplastics from the Scientific Literature: A Machine Learning Approach for Modeling Sparse Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Selection
2.2. Data Extraction
- Criterion 1: The concentrations of the components as a percentage of the total mass or volume of the manufacture.
- Criterion 2: The existence of no more than 5 components for manufacturing.
- Criterion 3: Method of manufacturing biopolymer films.
- Criterion 1: Relevant property report.
- Criterion 2: Values of the results in tabular form and not in graphs.
2.3. Data Preprocessing
2.4. Data Analysis
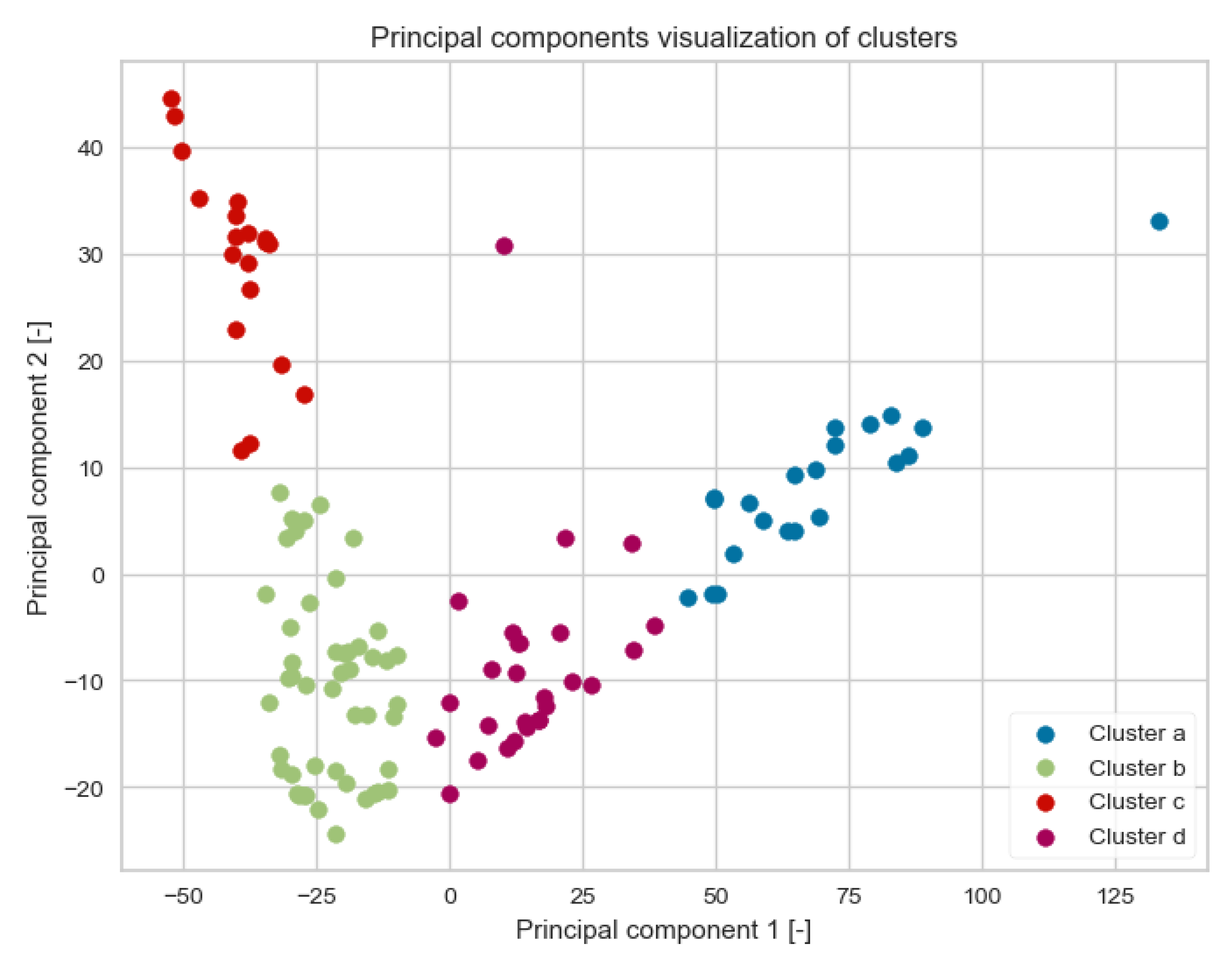
2.4.1. Unsupervised Algorithms
Clustering Algorithm
2.4.2. Supervised Algorithms
Decision Tree Regression
Random Forest Regression
Gradient Boosting Regression
3. Results
3.1. Clustering Analysis
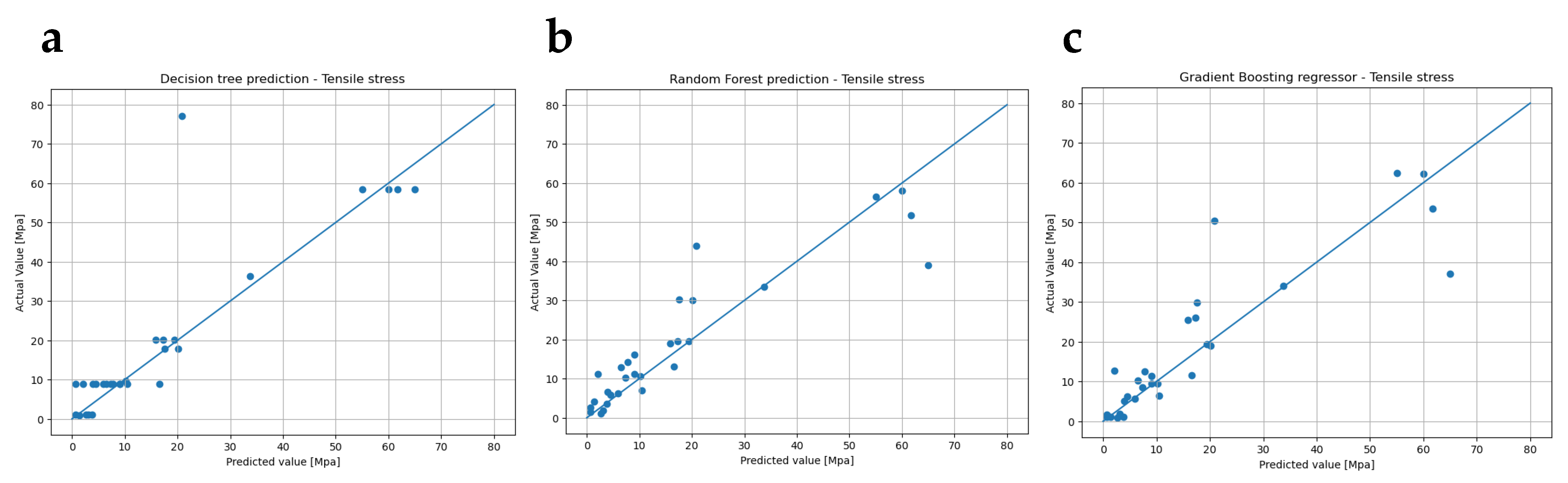
3.2. Regressions Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EUR-Lex—52018DC0028—EN—EUR-Lex. (n.d.). Europa.Eu. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=COM%3A2018%3A28%3AFIN (accessed on 25 August 2023).
- EUR-Lex—52019DC0190—EN—EUR-Lex. (n.d.). Europa.Eu. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A52019DC0190 (accessed on 25 August 2023).
- Matthews, C.; Moran, F.; Jaiswal, A.K. A review on European Union’s strategy for plastics in a circular economy and its impact on food safety. J. Clean. Prod. 2021, 283, 125263. [Google Scholar] [CrossRef]
- Ahamed, A.; Veksha, A.; Giannis, A.; Lisak, G. Flexible packaging plastic waste—Environmental implications, management solutions, and the way forward. Curr. Opin. Chem. Eng. 2021, 32, 100684. [Google Scholar] [CrossRef]
- Prata, J.C.; Silva AL, P.; da Costa, J.P.; Mouneyrac, C.; Walker, T.R.; Duarte, A.C.; Rocha-Santos, T. Solutions and integrated strategies for the control and mitigation of plastic and microplastic pollution. Int. J. Environ. Res. Public Health 2019, 16, 2411. [Google Scholar] [CrossRef]
- Foschi, E.; Bonoli, A. The commitment of packaging industry in the framework of the European Strategy for plastics in a circular economy. Adm. Sci. 2019, 9, 18. [Google Scholar] [CrossRef]
- Svanes, E.; Vold, M.; Møller, H.; Pettersen, M.K.; Larsen, H.; Hanssen, O.J. Sustainable packaging design: A holistic methodology for packaging design: Sustainable packaging design. Packag. Technol. Sci. 2010, 23, 161–175. [Google Scholar] [CrossRef]
- George, A.; Sanjay, M.R.; Srisuk, R.; Parameswaranpillai, J.; Siengchin, S. A comprehensive review on chemical properties and applications of biopolymers and their composites. Int. J. Biol. Macromol. 2020, 154, 329–338. [Google Scholar] [CrossRef]
- Ebrahimzadeh, S.; Biswas, D.; Roy, S.; McClements, D.J. Incorporation of essential oils in edible seaweed-based films: A comprehensive review. Trends Food Sci. Technol. 2023, 135, 43–56. [Google Scholar] [CrossRef]
- Lomartire, S.; Marques, J.C.; Gonçalves, A.M.M. An overview of the alternative use of seaweeds to produce safe and sustainable bio-packaging. Appl. Sci. 2022, 12, 3123. [Google Scholar] [CrossRef]
- Thiruchelvi, R.; Das, A.; Sikdar, E. Bioplastics as better alternative to petro plastic. Mater. Today Proc. 2021, 37, 1634–1639. [Google Scholar] [CrossRef]
- Lim, C.; Yusoff, S.; Ng, C.G.; Lim, P.E.; Ching, Y.C. Bioplastic made from seaweed polysaccharides with green production methods. J. Environ. Chem. Eng. 2021, 9, 105895. [Google Scholar] [CrossRef]
- Rioux, L.-E.; Turgeon, S.L. Seaweed carbohydrates. In Seaweed Sustainability; Elsevier: Amsterdam, The Netherlands, 2015; pp. 141–192. [Google Scholar]
- Schmitz, C.; Auza, L.G.; Koberidze, D.; Rasche, S.; Fischer, R.; Bortesi, L. Conversion of chitin to defined chitosan oligomers: Current status and future prospects. Mar. Drugs 2019, 17, 452. [Google Scholar] [CrossRef]
- Chen, H.; Xiao, Q.; Weng, H.; Zhang, Y.; Yang, Q.; Xiao, A. Extraction of sulfated agar from Gracilaria lemaneiformis using hydrogen peroxide-assisted enzymatic method. Carbohydr. Polym. 2020, 232, 115790. [Google Scholar] [CrossRef]
- Kadam, S.U.; Álvarez, C.; Tiwari, B.K.; O’Donnell, C.P. Extraction of biomolecules from seaweeds. In Seaweed Sustainability; Elsevier: Amsterdam, The Netherlands, 2015; pp. 243–269. [Google Scholar]
- Kadam, S.U.; Álvarez, C.; Tiwari, B.K.; O’Donnell, C.P. Processing of seaweeds. In Seaweed Sustainability; Elsevier: Amsterdam, The Netherlands, 2015; pp. 61–78. [Google Scholar]
- Abdul Khalil HP, S.; Tye, Y.Y.; Saurabh, C.K.; Leh, C.P.; Lai, T.K.; Chong, E.W.N.; Nurul Fazita, M.R.; Mohd Hafiidz, J.; Banerjee, A.; Syakir, M.I. Biodegradable polymer films from seaweed polysaccharides: A review on cellulose as a reinforcement material. Express Polym. Lett. 2017, 11, 244–265. [Google Scholar] [CrossRef]
- Jumaidin, R.; Sapuan, S.M.; Jawaid, M.; Ishak, M.R.; Sahari, J. Effect of seaweed on mechanical, thermal, and biodegradation properties of thermoplastic sugar palm starch/agar composites. Int. J. Biol. Macromol. 2017, 99, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Abdul Khalil HP, S.; Tye, Y.Y.; Ismail, Z.; Leong, J.Y.; Saurabh, C.K.; Lai, T.K.; Chong, E.W.N.; Aditiawati, P.; Tahir, P.M.; Dungani, R. Oil palm shell nanofiller in seaweed-based composite film: Mechanical, physical, and morphological properties. Bioresources 2017, 12, 5996–6010. [Google Scholar] [CrossRef]
- Aloui, H.; Deshmukh, A.R.; Khomlaem, C.; Kim, B.S. Novel composite films based on sodium alginate and gallnut extract with enhanced antioxidant, antimicrobial, barrier and mechanical properties. Food Hydrocoll. 2021, 113, 106508. [Google Scholar] [CrossRef]
- Nanda, S.; Patra, B.R.; Patel, R.; Bakos, J.; Dalai, A.K. Innovations in applications and prospects of bioplastics and biopolymers: A review. Environ. Chem. Lett. 2022, 20, 379–395. [Google Scholar] [CrossRef]
- Nanda, N.; Bharadvaja, N. Algal bioplastics: Current market trends and technical aspects. Clean Technol. Environ. Policy 2022, 24, 2659–2679. [Google Scholar] [CrossRef]
- Chia, W.Y.; Ying Tang, D.Y.; Khoo, K.S.; Kay Lup, A.N.; Chew, K.W. Nature’s fight against plastic pollution: Algae for plastic biodegradation and bioplastics production. Environ. Sci. Ecotechnol. 2020, 4, 100065. [Google Scholar] [CrossRef]
- Pyzer-Knapp, E.O.; Pitera, J.W.; Staar PW, J.; Takeda, S.; Laino, T.; Sanders, D.P.; Sexton, J.; Smith, J.R.; Curioni, A. Accelerating materials discovery using artificial intelligence, high performance computing and robotics. npj Comput. Mater. 2022, 8, 84. [Google Scholar] [CrossRef]
- Himanen, L.; Geurts, A.; Foster, A.S.; Rinke, P. Data-driven materials science: Status, challenges, and perspectives. Adv. Sci. 2019, 6, 1900808. [Google Scholar] [CrossRef] [PubMed]
- Suh, C.; Fare, C.; Warren, J.A.; Pyzer-Knapp, E.O. Evolving the materials genome: How machine learning is fueling the next generation of materials discovery. Annu. Rev. Mater. Res. 2020, 50, 1–25. [Google Scholar] [CrossRef]
- Sha, W.; Li, Y.; Tang, S.; Tian, J.; Zhao, Y.; Guo, Y.; Zhang, W.; Zhang, X.; Lu, S.; Cao, Y.-C.; et al. Machine learning in polymer informatics. InfoMat 2021, 3, 353–361. [Google Scholar] [CrossRef]
- Gormley, A.J.; Webb, M.A. Machine learning in combinatorial polymer chemistry. Nat. Rev. Mater. 2021, 6, 642–644. [Google Scholar] [CrossRef]
- Martin, T.B.; Audus, D.J. Emerging trends in machine learning: A polymer perspective. ACS Polym. Au 2023, 3, 239–258. [Google Scholar] [CrossRef]
- Kusne, A.G.; Yu, H.; Wu, C.; Zhang, H.; Hattrick-Simpers, J.; DeCost, B.; Sarker, S.; Oses, C.; Toher, C.; Curtarolo, S.; et al. On-the-fly closed-loop materials discovery via Bayesian active learning. Nat. Commun. 2020, 11, 5966. [Google Scholar] [CrossRef]
- Unsupervised Learning. In Encyclopedia of Machine Learning and Data Mining; Springer: New York, NY, USA, 2017; p. 1304.
- Goldstein, M.; Uchida, S. A comparative evaluation of unsupervised anomaly detection algorithms for multivariate data. PLoS ONE 2016, 11, e0152173. [Google Scholar] [CrossRef]
- Li, Z.; Liu, J.; Yang, Y.; Zhou, X.; Lu, H. Clustering-guided sparse structural learning for unsupervised feature selection. IEEE Trans. Knowl. Data Eng. 2014, 26, 2138–2150. [Google Scholar] [CrossRef]
- Pandit, A.A.; Pimpale, B.; Dubey, S. A comprehensive review on unsupervised feature selection algorithms. In International Conference on Intelligent Computing and Smart Communication 2019; Springer: Singapore, 2020; pp. 255–266. [Google Scholar]
- Syakur, M.A.; Khotimah, B.K.; Rochman EM, S.; Satoto, B.D. Integration K-means clustering method and elbow method for identification of the best customer profile cluster. IOP Conf. Ser. Mater. Sci. Eng. 2018, 336, 012017. [Google Scholar] [CrossRef]
- Umargono, E.; Suseno, J.E.; Vincensius Gunawan, S.K. K-means clustering optimization using the elbow method and early centroid determination based on mean and median formula. In Proceedings of the 2nd International Seminar on Science and Technology (ISSTEC 2019), Yogyakarta, Indonesia, 25–26 November 2019. [Google Scholar]
- Nainggolan, R.; Perangin-angin, R.; Simarmata, E.; Tarigan, A.F. Improved the performance of the K-means cluster using the Sum of Squared Error (SSE) optimized by using the elbow method. J. Phys. Conf. Ser. 2019, 1361, 012015. [Google Scholar] [CrossRef]
- Strehl, A.; Ghosh, J. Relationship-based clustering and visualization for high-dimensional data mining. INFORMS J. Comput. 2003, 15, 208–230. [Google Scholar] [CrossRef]
- Navada, A.; Ansari, A.N.; Patil, S.; Sonkamble, B.A. Overview of use of decision tree algorithms in machine learning. In Proceedings of the 2011 IEEE Control and System Graduate Research Colloquium, Shah Alam, Malaysia, 27–28 June 2011. [Google Scholar]
- Talekar, B. A detailed review on decision tree and random forest. Biosci. Biotechnol. Res. Commun. 2020, 13, 245–248. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, X.; Liu, D. Model of gradient boosting random forest prediction. In Proceedings of the 2022 IEEE International Conference on Networking, Sensing and Control (ICNSC), Shanghai, China, 15–18 December 2022. [Google Scholar]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Roy, S.; Kim, H.-J.; Rhim, J.-W. Effect of blended colorants of anthocyanin and shikonin on carboxymethyl cellulose/agar-based smart packaging film. Int. J. Biol. Macromol. 2021, 183, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, K.M.; Cavalheiro, J.B.; Netto, A.M.; Scapim, M.R.d.S.; Bergamasco, R.d.C. Properties of alginate films incorporated with free and microencapsulated Stryphnodendron adstringens extract (barbatimão). Food Packag. Shelf Life 2021, 28, 100637. [Google Scholar] [CrossRef]
- Syafiq, R.; Sapuan, S.M.; Zuhri, M.R.M. Antimicrobial activity, physical, mechanical and barrier properties of sugar palm based nanocellulose/starch biocomposite films incorporated with cinnamon essential oil. J. Mater. Res. Technol. 2021, 11, 144–157. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, B.; Zhao, S.; Qiao, D.; Xie, F. Plasticized starch/agar composite films: Processing, morphology, structure, mechanical properties and surface hydrophilicity. Coatings 2021, 11, 311. [Google Scholar] [CrossRef]
- Park, J.; Nam, J.; Yun, H.; Jin, H.-J.; Kwak, H.W. Aquatic polymer-based edible films of fish gelatin crosslinked with alginate dialdehyde having enhanced physicochemical properties. Carbohydr. Polym. 2021, 254, 117317. [Google Scholar] [CrossRef]
- Roy, S.; Kim, H.-J.; Rhim, J.-W. Synthesis of carboxymethyl cellulose and agar-based multifunctional films reinforced with cellulose nanocrystals and shikonin. ACS Appl. Polym. Mater. 2021, 3, 1060–1069. [Google Scholar] [CrossRef]
- Cheng, T.; Xu, J.; Li, Y.; Zhao, Y.; Bai, Y.; Fu, X.; Gao, X.; Mao, X. Effect of gum ghatti on physicochemical and microstructural properties of biodegradable sodium alginate edible films. J. Food Meas. Charact. 2021, 15, 107–118. [Google Scholar] [CrossRef]
- Phinainitisatra, T.; Harnkarnsujarit, N. Development of starch-based peelable coating for edible packaging. Int. J. Food Sci. Technol. 2021, 56, 321–329. [Google Scholar] [CrossRef]
- Yaradoddi, J.S.; Banapurmath, N.R.; Ganachari, S.V.; Soudagar, M.E.M.; Mubarak, N.M.; Hallad, S.; Hugar, S.; Fayaz, H. Biodegradable carboxymethyl cellulose based material for sustainable packaging application. Sci. Rep. 2020, 10, 21960. [Google Scholar] [CrossRef] [PubMed]
- Avila, L.B.; Barreto, E.R.C.; de Souza, P.K.; Silva, B.D.Z.; Martiny, T.R.; Moraes, C.C.; Morais, M.M.; Raghavan, V.; da Rosa, G.S. Carrageenan-based films incorporated with jaboticaba peel extract: An innovative material for active food packaging. Molecules 2020, 25, 5563. [Google Scholar] [CrossRef] [PubMed]
- Fransiska, D.; Giyatmi Basmal, J.; Susanti, E. The effect of organic powdered cottonii concentration and types of plasticizers on the characteristics of edible film. IOP Conf. Ser. Earth Environ. Sci. 2020, 483, 012008. [Google Scholar] [CrossRef]
- Chowdhury, S.; Teoh, Y.L.; Ong, K.M.; Rafflisman Zaidi, N.S.; Mah, S.-K. Poly(vinyl) alcohol crosslinked composite packaging film containing gold nanoparticles on shelf life extension of banana. Food Packag. Shelf Life 2020, 24, 100463. [Google Scholar] [CrossRef]
- Nagar, M.; Sharanagat, V.S.; Kumar, Y.; Singh, L. Development and characterization of elephant foot yam starch–hydrocolloids based edible packaging film: Physical, optical, thermal and barrier properties. J. Food Sci. Technol. 2020, 57, 1331–1341. [Google Scholar] [CrossRef]
- Mahcene, Z.; Khelil, A.; Hasni, S.; Akman, P.K.; Bozkurt, F.; Birech, K.; Goudjil, M.B.; Tornuk, F. Development and characterization of sodium alginate based active edible films incorporated with essential oils of some medicinal plants. Int. J. Biol. Macromol. 2020, 145, 124–132. [Google Scholar] [CrossRef]
- Ma, D.; Jiang, Y.; Ahmed, S.; Qin, W.; Liu, Y. Antilisterial and physical properties of polysaccharide-collagen films embedded with cell-free supernatant of Lactococcus lactis. Int. J. Biol. Macromol. 2020, 145, 1031–1038. [Google Scholar] [CrossRef]
- Marismandani, A.D.P.; Husni, A. Development and characterization of biobased alginate/glycerol/virgin coconut oil as biodegradable packaging. E3S Web Conf. 2020, 147, 03016. [Google Scholar] [CrossRef]
- Racmayani, N.; Husni, A. Effect of different formulations on characteristic of biobased alginate edible films as biodegradable packaging. E3S Web Conf. 2020, 147, 03003. [Google Scholar] [CrossRef]
- Dewi, M.Y.; Husni, A. Characterization of biobased alginate/glycerol/sunflower oil as biodegradable packaging. E3S Web Conf. 2020, 147, 03004. [Google Scholar] [CrossRef]
- Nazurah, N.F.; Nur Hanani, Z.A. Physicochemical characterization of kappa-carrageenan (Euchema cottoni) based films incorporated with various plant oils. Carbohydr. Polym. 2017, 157, 1479–1487. [Google Scholar] [CrossRef]
- Praseptiangga, D.; Fatmala, N.; Manuhara, G.J.; Utami, R.; Khasanah, L.U. Preparation and preliminary characterization of semi refined kappa carrageenan-based edible film incorporated with cinnamon essential oil. AIP Conf. Proc. 2016, 1746, 020036. [Google Scholar]
- Eghbalifam, N.; Frounchi, M.; Dadbin, S. Antibacterial silver nanoparticles in polyvinyl alcohol/sodium alginate blend produced by gamma irradiation. Int. J. Biol. Macromol. 2015, 80, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Amariei, S.; Ursachi, F.; Petraru, A. Development of new biodegradable agar-alginate membranes for food packaging. Membranes 2022, 12, 576. [Google Scholar] [CrossRef]
- Ho, B.K.X.; Azahari, B.; Yhaya, M.F.B.; Talebi, A.; Ng, C.W.C.; Tajarudin, H.A.; Ismail, N. Green technology approach for reinforcement of calcium chloride cured sodium alginate films by isolated bacteria from palm oil mill effluent (POME). Sustainability 2020, 12, 9468. [Google Scholar] [CrossRef]
- Kim, H.-J.; Roy, S.; Rhim, J.-W. Gelatin/agar-based color-indicator film integrated with Clitoria ternatea flower anthocyanin and zinc oxide nanoparticles for monitoring freshness of shrimp. Food Hydrocoll. 2022, 124, 107294. [Google Scholar] [CrossRef]
- Hoque, M.S.; Benjakul, S.; Prodpran, T. Properties of film from cuttlefish (Sepia pharaonis) skin gelatin incorporated with cinnamon, clove and star anise extracts. Food Hydrocoll. 2011, 25, 1085–1097. [Google Scholar] [CrossRef]
- Thakur, R.; Pristijono, P.; Golding, J.B.; Stathopoulos, C.E.; Scarlett, C.; Bowyer, M.; Singh, S.P.; Vuong, Q.V. Effect of starch physiology, gelatinization, and retrogradation on the attributes of rice starch-ι-carrageenan film. Die Starke 2018, 70, 1700099. [Google Scholar] [CrossRef]
- de Lima Barizão, C.; Crepaldi, M.I.; de Oliveira S. Junior, O.; de Oliveira, A.C.; Martins, A.F.; Garcia, P.S.; Bonafé, E.G. Biodegradable films based on commercial κ-carrageenan and cassava starch to achieve low production costs. Int. J. Biol. Macromol. 2020, 165, 582–590. [Google Scholar] [CrossRef]
- Lim, Z.Q.J.; Tong, S.Y.; Wang, K.; Lim, P.N.; Thian, E.S. Cinnamon oil incorporated polymeric films for active food packaging. Mater. Lett. 2022, 313, 131744. [Google Scholar] [CrossRef]
- Castaño, J.; Guadarrama-Lezama, A.Y.; Hernández, J.; Colín-Cruz, M.; Muñoz, M.; Castillo, S. Preparation, characterization and antifungal properties of polysaccharide–polysaccharide and polysaccharide–protein films. J. Mater. Sci. 2017, 52, 353–366. [Google Scholar] [CrossRef]
- Tyuftin, A.A.; Wang, L.; Auty, M.A.E.; Kerry, J.P. Development and assessment of duplex and triplex laminated edible films using whey protein isolate, gelatin and sodium alginate. Int. J. Mol. Sci. 2020, 21, 2486. [Google Scholar] [CrossRef] [PubMed]
- Sahraee, S.; Milani, J.M.; Ghanbarzadeh, B.; Hamishehkar, H. Effect of corn oil on physical, thermal, and antifungal properties of gelatin-based nanocomposite films containing nano chitin. LWT Food Sci. Technol. 2017, 76, 33–39. [Google Scholar] [CrossRef]
- Yang, M.; Shi, J.; Xia, Y. Effect of SiO2, PVA and glycerol concentrations on chemical and mechanical properties of alginate-based films. Int. J. Biol. Macromol. 2018, 107, 2686–2694. [Google Scholar] [CrossRef] [PubMed]
- Afshar, M.; Dini, G.; Vaezifar, S.; Mehdikhani, M.; Movahedi, B. Preparation and characterization of sodium alginate/polyvinyl alcohol hydrogel containing drug-loaded chitosan nanoparticles as a drug delivery system. J. Drug Deliv. Sci. Technol. 2020, 56, 101530. [Google Scholar] [CrossRef]
- Avila, L.B.; Barreto, E.R.C.; Moraes, C.C.; Morais, M.M.; da Rosa, G.S. Promising new material for food packaging: An active and intelligent carrageenan film with natural jaboticaba additive. Foods 2022, 11, 792. [Google Scholar] [CrossRef]
- Nehchiri, N.; Amiri, S.; Radi, M. Improving the water barrier properties of alginate packaging films by submicron coating with drying linseed oil. Packag. Technol. Sci. 2021, 34, 283–295. [Google Scholar] [CrossRef]
- Abdin, M.; El-Beltagy, A.E.; El-sayed, M.E.; Naeem, M.A. Production and characterization of sodium alginate/gum Arabic based films enriched with Syzygium cumini seeds extracts for food application. J. Polym. Environ. 2022, 30, 1615–1626. [Google Scholar] [CrossRef]
- Tongnuanchan, P.; Benjakul, S.; Prodpran, T.; Nilsuwan, K. Emulsion film based on fish skin gelatin and palm oil: Physical, structural and thermal properties. Food Hydrocoll. 2015, 48, 248–259. [Google Scholar] [CrossRef]
- de Campo, C.; Pagno, C.H.; Costa, T.M.H.; Rios, A.d.O.; Flôres, S.H. Gelatin capsule waste: New source of protein to develop a biodegradable film. Polímeros 2017, 27, 100–107. [Google Scholar] [CrossRef]
- Hilbig, J.; Hartlieb, K.; Gibis, M.; Herrmann, K.; Weiss, J. Rheological and mechanical properties of alginate gels and films containing different chelators. Food Hydrocoll. 2020, 101, 105487. [Google Scholar] [CrossRef]
- Gulati, K.; Lal, S.; Kumar, S.; Arora, S. Effect of agar and walnut (Juglans regia.L) shell fibre addition on thermal stability, water barrier, biodegradability and mechanical properties of corn starch composites. Indian Chem. Eng. 2022, 64, 314–325. [Google Scholar] [CrossRef]
- Gutiérrez-Jara, C.; Bilbao-Sainz, C.; McHugh, T.; Chiou, B.-S.; Williams, T.; Villalobos-Carvajal, R. Physical, mechanical and transport properties of emulsified films based on alginate with soybean oil: Effects of soybean oil concentration, number of passes and degree of surface crosslinking. Food Hydrocoll. 2020, 109, 106133. [Google Scholar] [CrossRef]
- Sudhakar, M.P.; Magesh Peter, D.; Dharani, G. Studies on the development and characterization of bioplastic film from the red seaweed (Kappaphycus alvarezii). Environ. Sci. Pollut. Res. Int. 2021, 28, 33899–33913. [Google Scholar] [CrossRef]
- Davoodi, M.N.; Milani, J.M.; Farahmandfar, R. Preparation and characterization of a novel biodegradable film based on sulfated polysaccharide extracted from seaweed Ulva intestinalis. Food Sci. Nutr. 2021, 9, 4108–4116. [Google Scholar] [CrossRef]
- Roy, S.; Rhim, J.-W. Preparation of gelatin/carrageenan-based color-indicator film integrated with shikonin and Propolis for smart food packaging applications. ACS Appl. Bio Mater. 2021, 4, 770–779. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, X.; Shi, J.; Liu, L.; Zhang, X.; Zou, X.; Xiao, J.; Zhai, X.; Zhang, D.; Li, Y.; et al. A visual bi-layer indicator based on roselle anthocyanins with high hydrophobic property for monitoring griskin freshness. Food Chem. 2021, 355, 129573. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, H.; Yuan, M.; Qin, Y.; Chen, H. Effects of anthocyanin-rich Kadsura coccinea extract on the physical, antioxidant, and pH-sensitive properties of biodegradable film. Food Biophys. 2022, 17, 375–385. [Google Scholar] [CrossRef]
- Alves, M.C.M.A.; Nascimento, M.F.; de Almeida, B.M.; Alves, M.M.A.; Lima-Verde, I.B.; Costa, D.S.; Araújo, D.C.M.; de Paula, M.N.; de Mello, J.C.P.; Cano, A.; et al. Hydrophilic Scaffolds Containing Extracts of Stryphnodendron adstringens and Abarema cochliacarpa for Wound Healing: In Vivo Proofs of Concept. Pharmaceutics 2022, 14, 2150. [Google Scholar] [CrossRef]
- Wang, S.; Gao, Z.; Liu, L.; Li, M.; Zuo, A.; Guo, J. Preparation, in vitro and in vivo evaluation of chitosan-sodium alginate-ethyl cellulose polyelectrolyte film as a novel buccal mucosal delivery vehicle. Eur. J. Pharm. Sci. 2022, 168, 106085. [Google Scholar] [CrossRef] [PubMed]
- Genuer, R. Variance reduction in purely random forests. J. Nonparametr. Stat. 2012, 24, 543–562. [Google Scholar] [CrossRef]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobotics 2013, 7, 21. [Google Scholar] [CrossRef] [PubMed]
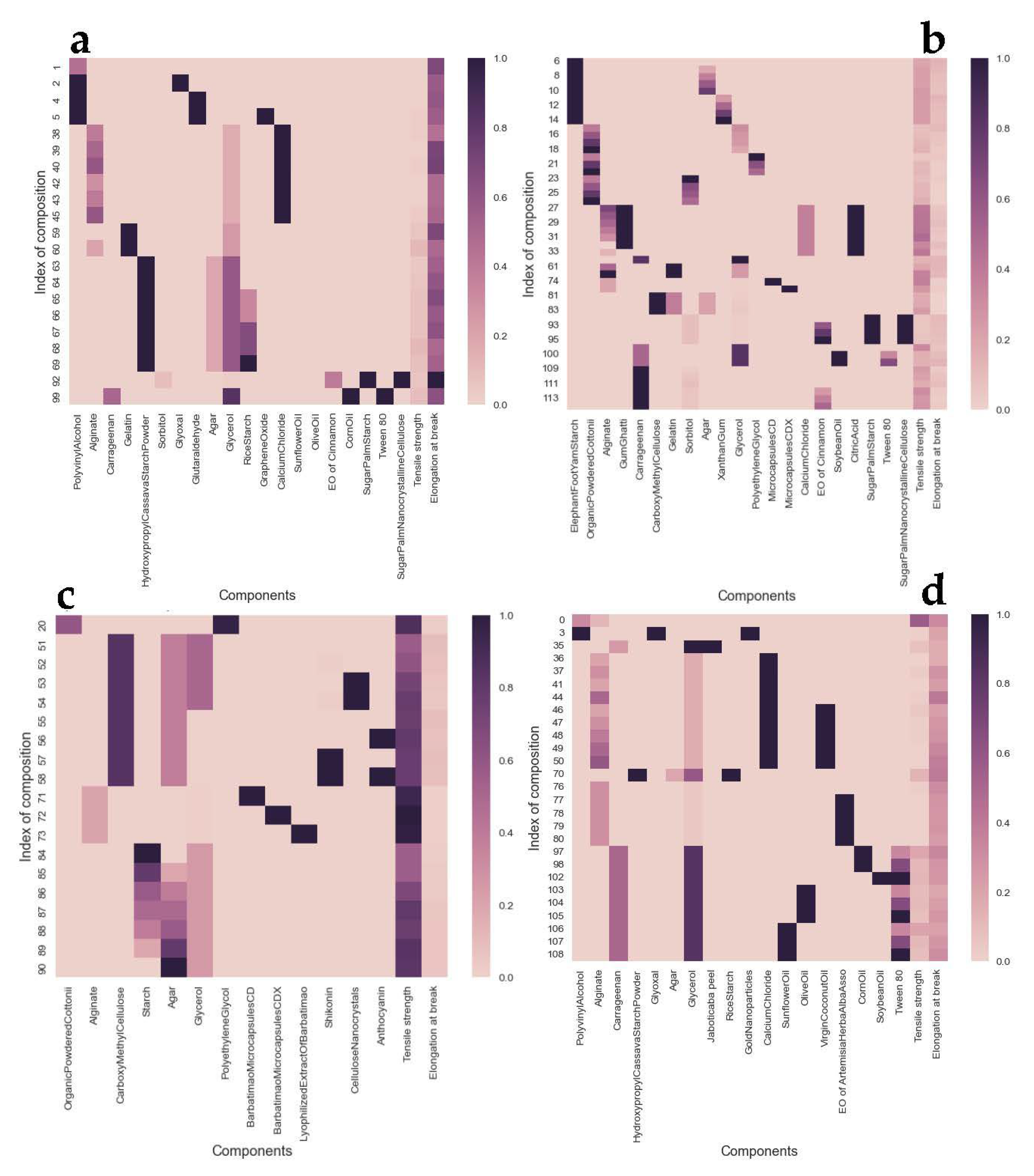
| Property | Ingredient | Description | References |
|---|---|---|---|
| Elongation at Break | Calcium chloride | The addition of calcium chloride of 0.08 g (1.6 wt.%) improves mechanical properties of membranes due to the network that is formed between calcium ions and carboxyl groups of alginate, but this is not necessarily the case in the elongation at break. | [65,66] |
| Gelatin | Intermolecular interaction of gelatin–agar strengthened the film, showing an increase in elongation at break due to the intermolecular forces between two polymer chains. | [67,68] | |
| Rice and cassava starch | Carrageenan films blended with rice or cassava starch showed significantly higher elongation at break due to strong binding forces in the compact crystalline region formed as a result of starch retrogradation. | [69,70] | |
| Essential oil of cinnamon | The incorporation of essential oil of cinnamon into poly-ε-caprolactone led to a reduction in the stretching ability of the film. Cinnamon agents tend to slightly lower values of elongation at break in polysaccharide films. | [71,72] | |
| Corn oil | The addition of corn oil improved mechanical properties of films based on protein isolate, gelatin, and sodium alginate, but this is not necessarily the case in the elongation at break. | [73,74] | |
| Polyvinyl alcohol (PVA) | Alginate-based films shown an increase in elongation at break due to the addition of PVA. | [75,76] | |
| Jaboticaba peel | The addition of jaboticaba peel in the polymeric matrix film based on carrageenan promoted a reduction in elongation at break. | [77] | |
| Sunflower oil | The addition of sunflower oil did not change the mechanical properties of alginate films. The highest concentration of Syzygium cumini seeds extract caused lower values of elongation at break in alginate/gum arabic films. | [78,79] | |
| Olive oil | The addition of plant oils to the formulation substantially increased elongation at break. | [62,80] | |
| Virgin coconut oil | Coconut oil provided films with higher flexibility and higher elongation at break values of gelatin-based films. | [81] | |
| Tensile Strength | Gum ghatti | The addition of gum ghatti in biodegradable sodium alginate edible films increased the tensile strength. | [50] |
| Citric acid | The addition of citric acid significantly decreased the TS of the casing of alginate films. | [82,83] | |
| Soybean oil | The tensile strength decreased with increasing oil concentrations due to the plasticizing effect from oil of alginate films. | [84] | |
| PolyethyleneGlycol (PEG) | PEG is used as a plasticizer, improving the mechanical properties of bioplastic film from seaweeds. | [85,86] | |
| Shikonin | Shikonin is used as a reinforcement. The gelatin/carrageenan film’s mechanical properties did not change significantly by shikonin. But the incorporation into carboxymetyl cellulose/agar films slightly improved tensile strength, showing a reinforcing effect. | [49,87] | |
| Anthocyanin | Addition of roselle anthocyanin showed a plasticizing effect in polyvinylidene fluoride films. However, Kadsura coccinea extract added to a chitosan, gelatin, and sodium alginate film increased tensile strength. | [88,89] | |
| Starch extract | A decrease in tensile strength was observed in starch/agar composite films. | [19,47] | |
| Barbatimao extract (Stryphnodendron adstringens) | The incorporation of Stryphnodendron adstringens extract improved mechanical properties of gelatin membranes. | [90] | |
| Cellulose extract | The chitosan-sodium alginate-ethyl cellulose polyelectrolyte films showed high tensile strength. | [91] | |
| Cottonni extract | Eucheuma cottonii extract was incorporated as a biofiller to improve tensile strength values of starch/agar composite films. | [19] |
| Predicted Variable(s) | Model Predictive | Train | Test | |||||
|---|---|---|---|---|---|---|---|---|
| Tensile strength | Decision tree | 0.961 | 0.661 | 121.998 | 11.045 | 4.626 | 2.491 | 0.559 |
| Random forest | 0.939 | 0.821 | 64.310 | 8.019 | 5.003 | 2.632 | 0.468 | |
| Gradient boosting | 0.999 | 0.778 | 79.891 | 8.938 | 5.201 | 2.162 | 0.465 | |
| Elongation at break | Decision tree | 0.506 | 0.276 | 588.675 | 24.263 | 17.973 | 13.945 | 0.764 |
| Random forest | 0.931 | 0.421 | 470.787 | 21.698 | 11.035 | 4.590 | 0.474 | |
| Gradient boosting | 0.997 | 0.156 | 686.075 | 26.193 | 11.886 | 3.881 | 0.490 | |
| Tensile strength–elongation at break | Decision tree | 0.536 | 0.555 | 281.606 | 16.086 | 11.333 | 6.950 | 0.609 |
| Random forest | 0.930 | 0.650 | 232.919 | 14.324 | 8.540 | 3.801 | 0.555 | |
| Gradient boosting | 0.996 | 0.467 | 408.516 | 17.086 | 8.341 | 2.428 | 0.433 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibarra-Pérez, D.; Faba, S.; Hernández-Muñoz, V.; Smith, C.; Galotto, M.J.; Garmulewicz, A. Predicting the Composition and Mechanical Properties of Seaweed Bioplastics from the Scientific Literature: A Machine Learning Approach for Modeling Sparse Data. Appl. Sci. 2023, 13, 11841. https://doi.org/10.3390/app132111841
Ibarra-Pérez D, Faba S, Hernández-Muñoz V, Smith C, Galotto MJ, Garmulewicz A. Predicting the Composition and Mechanical Properties of Seaweed Bioplastics from the Scientific Literature: A Machine Learning Approach for Modeling Sparse Data. Applied Sciences. 2023; 13(21):11841. https://doi.org/10.3390/app132111841
Chicago/Turabian StyleIbarra-Pérez, Davor, Simón Faba, Valentina Hernández-Muñoz, Charlene Smith, María José Galotto, and Alysia Garmulewicz. 2023. "Predicting the Composition and Mechanical Properties of Seaweed Bioplastics from the Scientific Literature: A Machine Learning Approach for Modeling Sparse Data" Applied Sciences 13, no. 21: 11841. https://doi.org/10.3390/app132111841
APA StyleIbarra-Pérez, D., Faba, S., Hernández-Muñoz, V., Smith, C., Galotto, M. J., & Garmulewicz, A. (2023). Predicting the Composition and Mechanical Properties of Seaweed Bioplastics from the Scientific Literature: A Machine Learning Approach for Modeling Sparse Data. Applied Sciences, 13(21), 11841. https://doi.org/10.3390/app132111841








