Machine Learning Algorithm Accuracy Using Single- versus Multi-Institutional Image Data in the Classification of Prostate MRI Lesions
Abstract
Featured Application
Abstract
1. Introduction
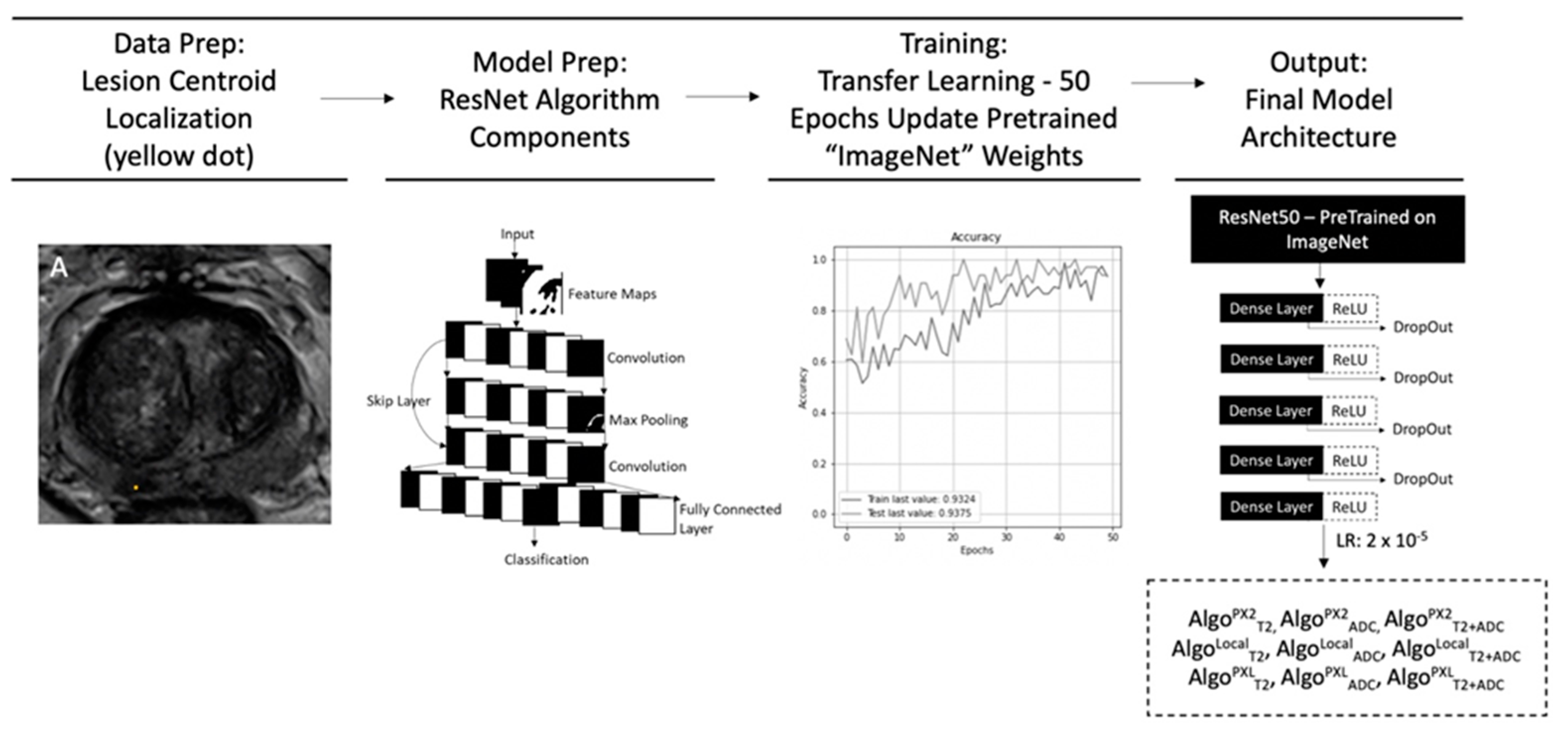
2. Materials and Methods
2.1. ProstateX-2 Patient Population
2.2. ProstateX-2 MR Imaging and Image Data
2.3. Local Institutional Patient Population
2.4. Local Institutional MR Imaging and Image Data
2.5. Image Preparation and Lesion Segmentation
2.6. Convolutional Neural Network Training and Testing
3. Results
3.1. Model Results for Classification of hrPCA on PX2, Local, and PXL Data
3.2. Sub-Analysis Results on PZ or TZ Lesions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bouchelouche, K.; Turkbey, B.; Choyke, P.L. Advances in imaging modalities in prostate cancer. Curr. Opin. Oncol. 2015, 27, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, J.C.; Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.; Schnall, M.D.; Shtern, F.; Tempany, C.M.; et al. PI-RADS Prostate Imaging-Reporting and Data System: 2015, Version 2. Eur. Urol. 2016, 69, 16–40. [Google Scholar] [CrossRef]
- Litjens, G.; Debats, O.; Barentsz, J.; Karssemeijer, N.; Huisman, H. Computer-aided detection of prostate cancer in MRI. IEEE Trans. Med. Imaging 2014, 33, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Mata, L.A.; Retamero, J.A.; Gupta, R.T.; Garcia Figueras, R.; Luna, A. Artificial Intelligence-assisted Prostate Cancer Diagnosis, Radiologic-Pathologic Correlation. Radiographics 2021, 41, 1676–1697. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lee, C.; Chia, D.; Lin, Z.; Huang, W.; Tan, C. Machine Learning in Prostate MRI for Prostate Cancer. Curr. Status Future Oppor. 2022, 12, 289. [Google Scholar]
- Ahmed, H.U.; El-Shater Bosaily, A.; Brown, L.C.; Gabe, R.; Kaplan, R.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS), a paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef]
- Siddiqui, M.M.; Rais-Bahrami, S.; Turkbey, B.; George, A.K.; Rothwax, J.; Shakir, N.; Okoro, C.; Raskolnikov, D.; Parnes, H.L.; Linehan, W.M.; et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015, 313, 390–397. [Google Scholar] [CrossRef]
- Wilt, T.J.; Jones, K.M.; Barry, M.J.; Andriole, G.L.; Culkin, D.; Wheeler, T.; Aronson, W.J.; Brawer, M.K. Follow-up of Prostatectomy versus Observation for Early Prostate Cancer. N. Engl. J. Med. 2017, 377, 132–142. [Google Scholar] [CrossRef]
- Hamdy, F.C.; Donovan, J.L.; Lane, J.; Mason, M.; Metcalfe, C.; Holding, P.; Davis, M.; Peters, T.J.; Turner, E.L.; Martin, R.M.; et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N. Engl. J. Med. 2016, 375, 1415–1424. [Google Scholar] [CrossRef]
- Sushentsev, N.; Moreira Da Silva, N.; Yeung, M.; Barrett, T.; Sala, E.; Roberts, M.; Rundo, L. Comparative performance of fully-automated and semi-automated artificial intelligence methods for the detection of clinically significant prostate cancer on MRI, a systematic review. Insights Imaging 2022, 13, 59. [Google Scholar] [CrossRef]
- Armato, S.G.; Huisman, H.; Drukker, K.; Hadjiiski, L.; Kirby, J.; Petrick, N.; Redmond, G.; Giger, M.L.; Cha, K.; Mamonov, A.; et al. PROSTATEx Challenges for computerized classification of prostate lesions from multiparametric magnetic resonance images. J. Med. Imaging 2018, 5, 044501. [Google Scholar] [CrossRef]
- Nolan, T.; Govindarajan, K. “SPIE-AAPM-NCI PROSTATEx Challenges (PROSTATEx).” The Cancer Imaging Archive (TCIA). Available online: https://doi.org/10.7937/K9TCIA.2017.MURS5CL (accessed on 23 August 2021).
- Abraham, B.; Nair, M.S. Automated grading of prostate cancer using convolutional neural network and ordinal class classifier. Inform. Med. Unlocked 2019, 17, 100256. [Google Scholar] [CrossRef]
- Chen, Q.; Hu, S.; Long, P.; Lu, F.; Shi, Y.; Li, Y. A Transfer Learning Approach for Malignant Prostate Lesion Detection on Multiparametric MRI. Technol. Cancer Res. Treat 2019, 18, 1533033819858363. [Google Scholar] [CrossRef] [PubMed]
- Hectors, S.; Cherny, M.; Yadav, K.K.; Beksaç, A.T.; Thulasidass, H.; Lewis, S.; Davicioni, E.; Wang, P.; Tewari, A.K.; Taouli, B. Radiomics Features Measured with Multiparametric Magnetic Resonance Imaging Predict Prostate Cancer Aggressiveness. J. Urol. 2019, 202, 498–505. [Google Scholar] [CrossRef]
- Schelb, P.; Kohl, S.; Radtke, J.P.; Wiesenfarth, M.; Kickingereder, P.; Bickelhaupt, S.; Kuder, T.A.; Stenzinger, A.; Hohenfellner, M.; Schlemmer, H.; et al. Classification of Cancer at Prostate MRI, Deep Learning versus Clinical PI-RADS Assessment. Radiology 2019, 293, 607–617. [Google Scholar] [CrossRef]
- Bernatz, S.; Ackermann, J.; Mandel, P.; Kaltenbach, B.; Zhdanovich, Y.; Harter, P.N.; Döring, C.; Hammerstingl, R.; Bodelle, B.; Smith, K.; et al. Comparison of machine learning algorithms to predict clinically significant prostate cancer of the peripheral zone with multiparametric MRI using clinical assessment categories and radiomic features. Eur. Radiol. 2020, 30, 6757–6769. [Google Scholar] [CrossRef]
- Castillo, T.J.M.; Arif, M.; Starmans, M.; Niessen, W.J.; Bangma, C.H.; Schoots, I.G.; Veenland, J.F. Classification of Clinically Significant Prostate Cancer on Multi-Parametric MRI, A Validation Study Comparing Deep Learning and Radiomics. Cancers 2021, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Ugga, L.; Romeo, V.; Radice, L.; Brunetti, A.; Imbriaco, M. Machine learning applications in prostate cancer magnetic resonance imaging. Eur. Radiol. Exp. 2019, 3, 35. [Google Scholar] [CrossRef]
- Purysko, A.S. Invited Commentary, Prostate Cancer Diagnosis-Challenges and Opportunities for Artificial Intelligence. Radiographics 2021, 41, E177–E178. [Google Scholar] [CrossRef]
- Sarma, K.V.; Harmon, S.; Sanford, T.; Roth, H.R.; Xu, Z.; Tetreault, J.; Xu, D.; Flores, M.G.; Raman, A.G.; Kulkarni, R.; et al. Federated learning improves site performance in multicenter deep learning without data sharing. J. Am. Med. Inform. Assoc. 2021, 28, 1259–1264. [Google Scholar] [CrossRef]
- Litjens, G.; Debats, O.; Barentsz, J.; Karssemeijer, N.; Huisman, H. ProstateX challenge data. Cancer Imaging Arch. 2017, 10, K9TCIA. [Google Scholar]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA), maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-scale machine learning on heterogeneous systems. 2015. Available online: tensorflow.org (accessed on 23 August 2021).
- Deng, J.; Dong, W.; Socher, R.; Li, L.; Li, K.; Li, F. ImageNet, A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Mongan, J.; Moy, L.; Kahn, C.E.J. Checklist for Artificial Intelligence in Medical Imaging (CLAIM), A Guide for Authors and Reviewers. Radiol Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef]
- Jarrel, C.Y.S.; Jennifer, S.N.T.; Kitchen, A. Detection of prostate cancer on multiparametric MRI. Proc. SPIE 2017, 10134, 585–588. [Google Scholar]
- Kitchen, A.; Seah, J. Support vector machines for prostate lesion classification. Proc. SPIE 2017, 10134, 577–580. [Google Scholar]
- Liu, S.; Zheng, H.; Feng, Y.; Li, W. Prostate cancer diagnosis using deep learning with 3D multiparametric MRI. Proc. SPIE 2017, 10134, 581–584. [Google Scholar]
- Mehrtash, A.; Sedghi, A.; Ghafoorian, M.; Taghipour, M.; Tempany, C.M.; Wells, W.M.; Kapur, T.; Mousavi, P.; Abolmaesumi, P.; Fedorov, A. Classification of Clinical Significance of MRI Prostate Findings Using 3D Convolutional Neural Networks. In Proceedings of the Medical Imaging 2017: Computer-Aided Diagnosis, Orlando, FL, USA, 3 March 2017. [Google Scholar]
- Park, S.H.; Han, K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology 2018, 286, 800–809. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L. The Potential of Radiomic-Based Phenotyping in Precision Medicine, A Review. JAMA Oncol. 2016, 2, 1636–1642. [Google Scholar] [CrossRef]
- Kohli, M.; Prevedello, L.M.; Filice, R.W.; Geis, J.R. Implementing Machine Learning in Radiology Practice and Research. AJR Am. J. Roentgenol. 2017, 208, 754–760. [Google Scholar] [CrossRef]
- Buch, K.; Kuno, H.; Qureshi, M.M.; Li, B.; Sakai, O. Quantitative variations in texture analysis features dependent on MRI scanning parameters, A phantom model. J. Appl. Clin. Med. Phys. 2018, 19, 253–264. [Google Scholar] [CrossRef]
- Nyul, L.G.; Udupa, J.K. On standardizing the MR image intensity scale. Magn. Reson. Med. 1999, 42, 1072–1081. [Google Scholar] [CrossRef]
- Isaksson, L.J.; Raimondi, S.; Botta, F.; Pepa, M.; Gugliandolo, S.G.; De Angelis, S.P.; Marvaso, G.; Petralia, G.; DE Cobelli, O.; Gandini, S.; et al. Effects of MRI image normalization techniques in prostate cancer radiomics. Phys. Medica 2020, 71, 7–13. [Google Scholar] [CrossRef]
- Sunoqrot, M.R.S.; Nketiah, G.A.; Selnaes, K.M.; Bathen, T.F.; Elschot, M. Automated reference tissue normalization of T2-weighted MR images of the prostate using object recognition. MAGMA 2021, 34, 309–321. [Google Scholar] [CrossRef]
- Castillo, T.J.M.; Arif, M.; Niessen, W.J.; Schoots, I.G.; Veenland, J.F. Automated Classification of Significant Prostate Cancer on MRI, A Systematic Review on the Performance of Machine Learning Applications. Cancers 2020, 12, 1606. [Google Scholar] [CrossRef]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Romeo, V.; Green, R.; Cantoni, V.; Ponsiglione, A.; Ugga, L.; Imbriaco, M. Machine learning for the identification of clinically significant prostate cancer on MRI, A meta-analysis. Eur. Radiol. 2020, 30, 6877–6887. [Google Scholar] [CrossRef]
- Mulherin, S.A.; Miller, W.C. Spectrum bias or spectrum effect? Subgroup variation in diagnostic test evaluation. Ann. Intern. Med. 2002, 137, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Leeflang, M.M.G.; Rutjes, A.W.S.; Reitsma, J.B.; Hooft, L.; Bossuyt, P.M.M. Variation of a test’s sensitivity and specificity with disease prevalence. CMAJ 2013, 185, E537–E544. [Google Scholar] [CrossRef]
- Koga, Y.; Song, H.; Chalmers, Z.R.; Newberg, J.; Kim, E.; Carrot-Zhang, J.; Piou, D.; Polak, P.; Abdulkadir, S.A.; Ziv, E.; et al. Genomic Profiling of Prostate Cancers from Men with African and European Ancestry. Clin. Cancer Res. 2020, 26, 4651–4660. [Google Scholar] [CrossRef] [PubMed]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Kostick-Quenet, K.M.; Cohen, I.G.; Gerke, S.; Lo, B.; Antaki, J.; Movahedi, F.; Njah, H.; Schoen, L.; Estep, J.E.; Blumenthal-Barby, J. Mitigating Racial Bias in Machine Learning. J. Law Med. Ethics 2022, 50, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine, facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef] [PubMed]
- NVIDIA Clara Imaging. Available online: https://developer.nvidia.com/clara-medical-imaging (accessed on 1 June 2022).
- Tensorflow. TensorFlow Federated, Machine Learning on Decentralized Data. Available online: https://www.tensorflow.org/federated (accessed on 1 June 2022).
- IBM. IBM Federated Learning. Available online: https://ibmfl.mybluemix.net/ (accessed on 1 June 2022).
- Meyer, A.; Chlebus, G.; Rak, M.; Schindele, D.; Schostak, M.; van Ginneken, B.; Schenk, A.; Meine, H.; Hahn, H.K.; Schreiber, A.; et al. Anisotropic 3D Multi-Stream CNN for Accurate Prostate Segmentation from Multi-Planar MRI. Comput. Methods Programs Biomed. 2021, 200, 105821. [Google Scholar] [CrossRef]
- Schimmöller, L.; Blondin, D.; Arsov, C.; Rabenalt, R.; Albers, P.; Antoch, G.; Quentin, M. MRI-Guided In-Bore Biopsy, Differences Between Prostate Cancer Detection and Localization in Primary and Secondary Biopsy Settings. AJR Am. J. Roentgenol. 2016, 206, 92–99. [Google Scholar] [CrossRef]
- Allsbrook, W.C.; Mangold, K.; Johnson, M.H.; Lane, R.B.; Lane, C.G.; Amin, M.B.; Bostwick, D.G.; Humphrey, P.A.; Jones, E.C.; Reuter, V.E.; et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma, urologic pathologists. Hum. Pathol. 2001, 32, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Briganti, A.; Budäus, L.; Hellawell, G.; Hindley, R.G.; et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [CrossRef]
- Sonn, G.A.; Fan, R.E.; Ghanouni, P.; Wang, N.N.; Brooks, J.D.; Loening, A.M.; Daniel, B.L.; To’o, K.J.; Thong, A.E.; Leppert, J.L. Prostate Magnetic Resonance Imaging Interpretation Varies Substantially Across Radiologists. Eur. Urol. Focus 2019, 5, 592–599. [Google Scholar] [CrossRef]
- Westphalen, A.C.; McCulloch, C.E.; Anaokar, J.M.; Arora, S.; Barashi, N.S.; Barentsz, J.O.; Bathala, T.K.; Bittencourt, L.K.; Booker, M.T.; Braxton, V.G.; et al. Variability of the Positive Predictive Value of PI-RADS for Prostate MRI across 26 Centers, Experience of the Society of Abdominal Radiology Prostate Cancer Disease-focused Panel. Radiology 2020, 296, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Kulldorff, M.; Devesa, S.S.; Hayes, R.B.; Fraumeni, J.F.J. A geographic analysis of prostate cancer mortality in the United States, 1970–1989. Int. J. Cancer 2002, 101, 168–174. [Google Scholar] [CrossRef]
- Baade, P.D.; Youlden, D.R.; Krnjacki, L.J. International epidemiology of prostate cancer, geographical distribution and secular trends. Mol. Nutr. Food Res. 2009, 53, 171–184. [Google Scholar] [CrossRef]
- Ben Ammar, L.; Gasmi, K.; Ben Ltaifa, I. ViT-TB, Ensemble Learning Based ViT Model for Tuberculosis Recognition. Cybern. Systems 2022, 1–20. [Google Scholar] [CrossRef]
- Karim, G. Hybrid deep learning model for answering visual medical questions. J. Supercomput. 2022, 78, 15042–15059. [Google Scholar]

| ProstateX-2 | Local Institution | |||
|---|---|---|---|---|
| Patients | ||||
| Mean age | 66.0 | 63.6 | ||
| Range | 48–83 | 47–74 | ||
| Race | NA | 51% white, 44% AA, 5% Other | ||
| MRI exams | ||||
| Total | n = 112 | n = 63 | ||
| PIRADS 5 | NA | 25 | ||
| PIRADS 4 | NA | 14 | ||
| PIRADS 3 | NA | 2 | ||
| PIRADS 2 | 0 | 22 | ||
| Lesion pathology | ||||
| hrPCA | Total | n = 35 (31%) | n = 28 (44%) | |
| GG5 | 7 | 9 | ||
| GG4 | 8 | 4 | ||
| GG3 (4 + 3) | 20 | 15 | ||
| “Benign” | Total | n = 77 (69%) | n = 35 (56%) | |
| GG2 (3 + 4) | 41 | 11 | ||
| GG1 | 36 | 2 | ||
| <GG1 | - | 22 * | ||
| Lesion zonal distribution | ||||
| hrPCA | PZ | 15 | 22 | |
| TZ | 20 | 6 | ||
| “Benign” | PZ | 35 | 23 | |
| TZ | 42 | 12 |
| ResNet Algorithm | Training Image Source | Testing Image Source | Total Training Images | Total Testing Images |
|---|---|---|---|---|
| PX2 | 89 (80%) | 23 (20%) | ||
| ModelPX2 | PX2 | Local | 89 (80%) | 13 (20% of 63 initial) |
| PXL | 89 (80%) | 36 (20%) | ||
| Local | 50 (80%) | 13 (20%) | ||
| ModelLoc | Local | PX2 | 50 (80%) | 23 (20%) |
| PXL | 50 (80%) | 36 (20%) | ||
| PXL | 139 (80%) | 36 (20%) | ||
| ModelPXL | PXL | PX2 | 139 (80%) | 23 (20%) |
| Local | 139 (80%) | 13 (20%) |
| ResNet Model | Training Image Source | Testing Image Source | Training & Testing Image Sequence | |||||
|---|---|---|---|---|---|---|---|---|
| T2 | ADC | T2 & ADC | ||||||
| AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | |||
| PX2 * | 0.93 | 0.91 | 0.91 | 0.88 | 0.95 | 0.90 | ||
| ModelPX2 | PX2 (89) | Local † | 0.49 | 0.53 | 0.23 | 0.48 | 0.46 | 0.55 |
| PXL † | 0.87 | 0.79 | 0.80 | 0.78 | 0.78 | 0.80 | ||
| Local * | 0.96 | 0.89 | 0.82 | 0.82 | 0.98 | 0.92 | ||
| ModelLoc | Local | PX2 † | 0.50 | 0.54 | 0.49 | 0.49 | 0.41 | 0.51 |
| PXL † | 0.77 | 0.71 | 0.84 | 0.84 | 0.94 | 0.87 | ||
| PXL * | 0.83 | 0.89 | 0.98 | 0.92 | 0.96 | 0.93 | ||
| ModelPXL | PXL | PX2 † | 0.92 | 0.92 | 0.85 | 0.91 | 0.85 | 0.93 |
| Local † | 0.96 | 0.86 | 0.88 | 0.92 | 0.99 | 0.92 | ||
| ResNet Algorithm | Training Image Source | Testing Image Source | Training & Testing Image Sequence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2 | ADC | T2 & ADC | ||||||||||||
| AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | |||||||||
| PZ | TZ | PZ | TZ | PZ | TZ | PZ | TZ | PZ | TZ | PZ | TZ | |||
| PX2 * | 0.92 | 0.93 | 0.91 | 0.90 | 0.91 | 0.91 | 0.88 | 0.88 | 0.94 | 0.95 | 0.91 | 0.90 | ||
| ModelPX2 | PX2 | Local † | 0.44 | 0.61 | 0.53 | 0.55 | 0.23 | 0.25 | 0.48 | 0.48 | 0.45 | 0.31 | 0.44 | 0.44 |
| PXL † | 0.88 | 0.87 | 0.8 | 0.77 | 0.80 | 0.81 | 0.77 | 0.80 | 0.77 | 0.79 | 0.80 | 0.80 | ||
| Local * | 0.88 | 0.99 | 0.86 | 0.89 | 0.82 | 0.81 | 0.84 | 0.80 | 0.95 | 0.96 | 0.90 | 0.94 | ||
| ModelLoc | Local | PX2 † | 0.48 | 0.52 | 0.53 | 0.55 | 0.54 | 0.45 | 0.49 | 0.48 | 0.55 | 0.29 | 0.57 | 0.46 |
| PXL † | 0.79 | 0.69 | 0.79 | 0.70 | 0.94 | 0.72 | 0.84 | 0.80 | 0.89 | 0.94 | 0.83 | 0.88 | ||
| PXL * | 0.83 | 0.92 | 0.89 | 0.90 | 0.98 | 0.99 | 0.92 | 0.93 | 0.96 | 0.90 | 0.91 | 0.98 | ||
| ModelPXL | PXL | PX2 † | 0.93 | 0.92 | 0.92 | 0.92 | 0.88 | 0.85 | 0.92 | 0.90 | 0.85 | 0.86 | 0.93 | 0.92 |
| Local † | 0.96 | 0.95 | 0.94 | 0.78 | 0.90 | 0.88 | 0.92 | 0.91 | 0.99 | 0.97 | 0.92 | 0.92 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Provenzano, D.; Melnyk, O.; Imtiaz, D.; McSweeney, B.; Nemirovsky, D.; Wynne, M.; Whalen, M.; Rao, Y.J.; Loew, M.; Haji-Momenian, S. Machine Learning Algorithm Accuracy Using Single- versus Multi-Institutional Image Data in the Classification of Prostate MRI Lesions. Appl. Sci. 2023, 13, 1088. https://doi.org/10.3390/app13021088
Provenzano D, Melnyk O, Imtiaz D, McSweeney B, Nemirovsky D, Wynne M, Whalen M, Rao YJ, Loew M, Haji-Momenian S. Machine Learning Algorithm Accuracy Using Single- versus Multi-Institutional Image Data in the Classification of Prostate MRI Lesions. Applied Sciences. 2023; 13(2):1088. https://doi.org/10.3390/app13021088
Chicago/Turabian StyleProvenzano, Destie, Oleksiy Melnyk, Danish Imtiaz, Benjamin McSweeney, Daniel Nemirovsky, Michael Wynne, Michael Whalen, Yuan James Rao, Murray Loew, and Shawn Haji-Momenian. 2023. "Machine Learning Algorithm Accuracy Using Single- versus Multi-Institutional Image Data in the Classification of Prostate MRI Lesions" Applied Sciences 13, no. 2: 1088. https://doi.org/10.3390/app13021088
APA StyleProvenzano, D., Melnyk, O., Imtiaz, D., McSweeney, B., Nemirovsky, D., Wynne, M., Whalen, M., Rao, Y. J., Loew, M., & Haji-Momenian, S. (2023). Machine Learning Algorithm Accuracy Using Single- versus Multi-Institutional Image Data in the Classification of Prostate MRI Lesions. Applied Sciences, 13(2), 1088. https://doi.org/10.3390/app13021088






