Abstract
In low-dose computed tomography (LDCT), lung segmentation effectively improves the accuracy of lung cancer diagnosis. However, excessive noise is inevitable in LDCT, which can decrease lung segmentation accuracy. To address this problem, it is necessary to derive an optimized kernel size when using the median modified Wiener filter (MMWF) for noise reduction. Incorrect application of the kernel size can result in inadequate noise removal or blurring, degrading segmentation accuracy. Therefore, various kernel sizes of the MMWF were applied in this study, followed by region-growing-based segmentation and quantitative evaluation. In addition to evaluating the segmentation performance, we conducted a similarity assessment. Our results indicate that the greatest improvement in segmentation performance and similarity was at a kernel size 5 × 5. Compared with the noisy image, the accuracy, F1-score, intersection over union, root mean square error, and peak signal-to-noise ratio using the optimized MMWF were improved by factors of 1.38, 33.20, 64.86, 7.82, and 1.30 times, respectively. In conclusion, we have demonstrated that by applying the MMWF with an appropriate kernel size, the optimization of noise and blur reduction can enhance segmentation performance.
1. Introduction
According to the Global Cancer Report, lung cancer is responsible for approximately 1.8 million deaths annually, and the 5-year survival rate for patients with advanced lung cancer is only 15% [1]. Lung cancer survival rates increase dramatically with early detection; therefore, the ability to identify malignant lung nodules in their early stages is crucial [2]. Low-dose computed tomography (LDCT) can detect nodules as small as 1 cm, which are difficult to detect using simple chest radiography, and detect nodules in areas concealed by various organs, allowing for the early diagnosis of lung cancer. The national lung cancer screening trial (NLST) demonstrated that screening for lung cancer using LDCT reduced mortality by 20% [3]. In addition, LDCT has the advantage of reducing the exposure dose by approximately 90% compared with conventional CT examinations using a low dose; therefore, LDCT is widely used in lung examinations [4].
Lung segmentation using LDCT can effectively improve the accuracy of lung cancer diagnosis by accurately detecting lesions or nodules located on the interior aspects of the lung [5]. Furthermore, lung segmentation is essential for analyzing lung parenchymal density and quantitatively analyzing mechanisms related to the lung and diaphragm regions [6]. The region-growing-based segmentation algorithm is a commonly used approach for lung segmentation. It allows fast processing and exhibits excellent effectiveness in capturing subtle attenuation changes within tissues [7]. In addition, a region-growing-based segmentation algorithm can constrain a region by specifying an initial starting point. This capability allows for accurate region segmentation, even in areas without clear boundaries, such as the lungs [8].
However, in LDCT, the inevitable presence of excessive noise poses challenges to achieving accurate segmentation [9]. The presence of white Gaussian noise exhibits a biologically irrelevant segmentation area, leading to over-segmentation and the representation of rounded tissue surfaces, such as the lungs, in spiky form [10,11]. Additionally, as Gaussian noise increases, the variance between the pixel values of objects and noise increases, decreasing pixel-to-pixel similarity hindering proper region expansion when performing segmentation [12]. Moreover, when the section contains a large amount of noise with a negative value, no segmentation occurs.
Various noise-removal filters such as Gaussian, median, and Wiener filters have been developed to enhance the segmentation accuracy. However, Gaussian filters cannot effectively remove noise types other than high-frequency noise [13]. In addition, the median filter erodes the distortion of spot edges, whereas the Wiener filter can induce blurring and spikes in the spots. To address these limitations and complement their characteristics, a fusion of the two filters, called the median modified Wiener filter (MMWF), has been proposed [14].
The MMWF is an adaptive spatial filter, which relies on the kernel size as its sole parameter [15]. However, if the kernel size is too small, the noise may not be effectively removed, whereas a large kernel size can effectively remove noise but lead to pronounced blurring [16]. When excessive blurring occurs in an image, maintaining segmentation accuracy during the execution of region-growing-based segmentation poses a challenge. Setting the threshold values defensively is necessary to prevent over-segmentation but it can lead to a decrease in segmentation accuracy. Therefore, it is essential to derive an optimal kernel size that balances noise and blur for the effective application of the MMWF.
The current study’s goal was to apply an MMWF modeled by the kernel size to noisy lung CT images to determine the ideal MMWF kernel size for increasing the accuracy of lung segmentation. Subsequently, a comparative evaluation was conducted using quantitative evaluation factors.
2. Materials and Methods
2.1. The Acquisition of the Lung Computed Tomography Images
We obtained lung CT images with a tube voltage of 120 kV, tube current of 160 mA, and slice thickness of 2.5 mm using data from the NLST dataset. Subsequently, two images were selected before and after segmenting the slice image, and five slice images were summed and averaged to obtain a standard image. To simulate conditions similar to those of low-dose CT images, Gaussian noise with a mean of 0 and variance of 0.05 was added to the acquired standard image using MATLAB [17,18]. Figure 1 shows the standard images with and without the added Gaussian noise.
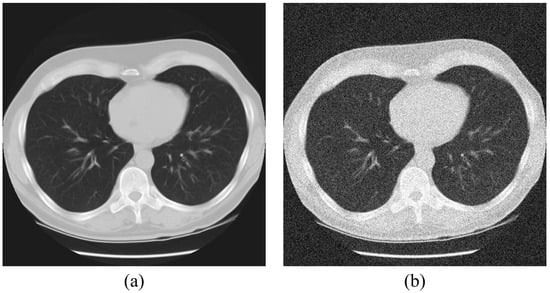
Figure 1.
Acquired images for (a) standard image (ground truth) and (b) noisy image.
2.2. Median Modified Wiener Filter (MMWF) Modeling
The MMWF is based on the Wiener filter. To minimize the average error by applying the median during Wiener filtering, it is modeled as follows [15]:
where, represents the coordinates of the median mask, is the median filter value of the local window, is the variance of the current slide window, and is the variance of the noise variance. Based on Equation (1), we modeled the kernel size of the MMWF as 3 × 3, 5 × 5, 7 × 7, 9 × 9, and 11 × 11 using MATLAB and applied them to the standard images with Gaussian noise.
2.3. Region-Growing-Based Segmentation Algorithm Modeling
We modeled a segmentation algorithm using a region-growing technique. The region-growing technique begins from a seed point and adds neighboring pixels individually, expanding the region by grouping pixels with similar characteristics into a single region. After selecting a seed point representing the organization to be segmented, the selected seed point was compared with all adjacent pixels with a threshold value. The pixels that exceeded the threshold value were grouped to form a single region. The region-growing process continued until every pixel value was grouped into a single region. The region-growing-based segmentation algorithm is modeled as follows:
where, denotes the segmented images, is the image before segmentation, is the average value of the segmented pixels, and is the set threshold. We fixed the threshold value to 160 to evaluate the segmentation performance under the same conditions. The threshold was set by trial-and-error based on the ground-truth image to a value just before over-segmentation occurs.
2.4. Quantitative Evaluation of the Acquired Image
2.4.1. Segmentation Performance Evaluation
We calculated the accuracy, F1-score, and intersection over union (IoU) values to quantitatively evaluate the performance of the region-growing-based segmentation algorithm. For all three factors, a value closer to one indicates a better segmentation performance. Accuracy is the most intuitive metric for evaluating segmentation performance, representing the degree to which it predicts true and false values. The formula for calculating accuracy is as follows [19]:
where, TP is a true-positive value that predicts a true value as true, and TN is a true- negative value that predicts a false value as false. FP is a false-positive value, predicting a false value as true, and FN is a false negative, predicting a true value as false.
F1-score is the harmonic average value of precision and recall. Precision is the ratio of the TP area to the sum of the TP and FP areas, and recall is the ratio of the TP area to the sum of the FN and TP areas. The formula for calculating the F1-score is shown in Equation (4) [20]:
IoU refers to the intersection of the original and comparison images divided by the union. The formula for calculating IoU is shown in Equation (5) [21].
2.4.2. Similarity Evaluation
The root mean square error (RMSE) and peak signal to noise ratio (PSNR) were measured to quantitatively evaluate the similarity between an image segmented from a standard image (referred to as the original image) and a filtered segmented image after adding noise (referred to as the comparison image). RMSEs measure the difference between the original and comparison image, and a smaller value indicates greater similarity between the two. RMSEs quantify the average difference between the pixel values of two images. PSNRs quantify how closely the signal of the comparison image approximates the signal distortion. A high PSNR indicates that the comparison image is closer to the original image in terms of signal fidelity. The formula for calculating RMSE and PSNR is as follows [22]:
3. Results
Gaussian noise was added to the standard lung CT image acquired using data from the NLST dataset, with MMWF kernel sizes of 3 × 3, 5 × 5, 7 × 7, 9 × 9, and 11 × 11. Figure 2 shows the MMWF applied to a lung CT image according to the kernel size, with Figure 2a as the ground-truth image and Figure 2b as the noisy image. The images from Figure 2c–g represent the application results of the MMWF with increasing kernel size.
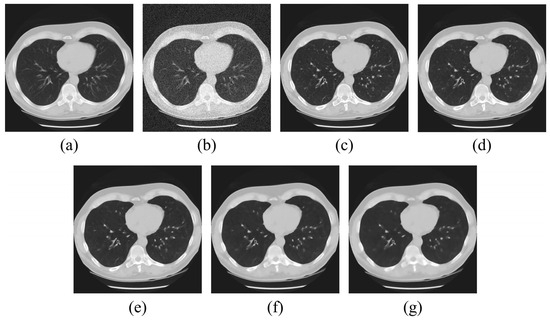
Figure 2.
Filtered lung CT images with (a) original, (b) noisy, (c–g) 3 × 3 MMWF, 5 × 5 MMWF, 7 × 7 MMWF, 9 × 9 MMWF, and 11 × 11 MMWF.
After applying the MMWF by kernel size, a region-growing-based segmentation algorithm was used to perform lung segmentation. The results are shown in Figure 3, with Figure 3a as the ground-truth of segmented image and Figure 3b showing that the accuracy of segmentation has significantly deteriorated due to the presence of noise. From Figure 3c–g, the images demonstrate the segmentation results as a function of increasing the MMWF kernel size.
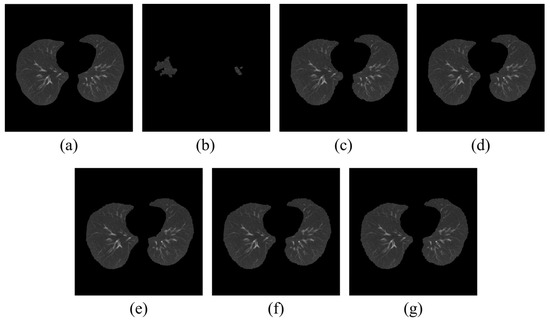
Figure 3.
Segmented lung image using region-growing-based segmentation algorithm with (a) original, (b) noisy, (c–g) 3 × 3 MMWF, 5 × 5 MMWF, 7 × 7 MMWF, 9 × 9 MMWF, and 11 × 11 MMWF.
Figure 4 shows the accuracy, F1-score, and IoU values obtained from the lung images with the MMWF with kernel sizes of 3 × 3, 5 × 5, 7 × 7, 9 × 9, and 11 × 11. The accuracy of the noisy image was 0.7210, whereas the accuracies for each kernel size were 0.9952, 0.9960, 0.9954, 0.9952, and 0.9949, respectively. When the kernel size was set to 5 × 5, the accuracy increased 1.38 times compared with that of the noisy image. The F1-score of the noisy image was 0.0299, while the F1-score for each kernel size was 0.9915, 0.9928, 0.9919, 0.9914, and 0.9909, respectively. Similar to the accuracy, the kernel size of 5 × 5 increased the F1-score 33.20-fold compared with that of the noisy image. The IoU value for the noisy image was 0.0152, whereas the values for each kernel size were 0.9831, 0.9858, 0.9839, 0.9829, and 0.9819, respectively. In line with previous evaluations, the kernel size of 5 × 5 increased the IoU value a 64.86 times in comparison with that of the noisy image. The accuracy, F1-score and IoU values demonstrated a decreasing trend as the kernel size increased beyond 5 × 5.
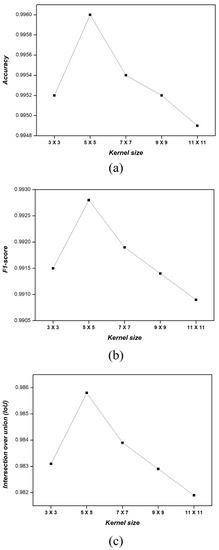
Figure 4.
(a) Accuracy, (b) F1-score, and (c) IoU results of the segmented image according to the kernel size of the applied MMWF.
Figure 5 shows the RMSEs and PSNRs obtained from the lung images when applying an MMWF with kernel sizes of 3 × 3, 5 × 5, 7 × 7, 9 × 9, and 11 × 11. The RMSE of the noisy image was 0.2229, whereas for each kernel size, it was 0.0292, 0.0271, 0.0285, 0.0294, and 0.0303, respectively. The kernel size of 5 × 5 yielded the lowest RMSE value, which was 7.82 times lower than that of the noisy image. The PSNR of the noisy image was 61.1689, whereas for each kernel size, it was 78.8171, 79.4585, 79.0361, 78.7585, and 78.5779, respectively. The kernel size of 5 × 5 generated the highest value, which was 1.30 times higher than that of the noisy image.
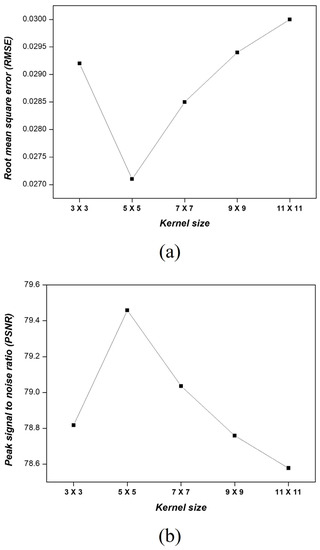
Figure 5.
(a) RMSE and (b) PSNR results of the segmented image according to the kernel size of the MMWF.
4. Discussion
LDCT is widely used for the examination of diseases such as lung cancer, owing to its low radiation dose. However, the limited number of photons used in LDCT results in increased noise, which degrades image quality. The presence of noise in LDCT leads to a deterioration in the performance of lung segmentation algorithms used for detecting and extracting lung tissue, ultimately affecting diagnostic accuracy [9]. To address this issue, various filtering techniques such as Gaussian, median, and Wiener filters have been proposed. However, conventional filtering techniques have drawbacks such as erosion and distortion of the edges of structures or blurring. To overcome these limitations, a fusion algorithm called an MMWF has been proposed [14].
In the MMWF, the kernel size is a key parameter that determines the trade-off between noise removal and preserving signal details, including boundary edges [23]. However, if the kernel size is too small, the noise removal capability is reduced. In contrast, if the kernel size is excessively large, the image may become excessively blurred. When the image is blurred, the threshold must be set defensively to prevent over-segmentation, reducing segmentation accuracy. Therefore, deriving an optimal kernel size that can optimize noise and blur reduction is crucial for the effective application of the MMWF.
Hence, the current study aimed to determine the optimal kernel size for the MMWF to improve the performance of lung segmentation. The MMWF was modeled and applied with kernel sizes ranging from 3 × 3 to 11 × 11 at 2 × 2 intervals. Figure 4 shows that a kernel size of 5 × 5 yielded the highest accuracy, F1-score, and IoU values. RMSE and PSNR were measured to analyze the relationship between the image restoration rate and segmentation performance due to the MMWF. Figure 5 indicates that a kernel size of 5 × 5 had the highest PSNR and lowest RMSE values. Overall, a kernel size of 5 × 5 resulted in the best performance for all evaluation metrics. We found that the metrics of segmentation performance and similarity evaluation tends to be similar, indicating that optimizing the MMWF to remove only noise with minimal distortion is important to improve segmentation performance. Therefore, the optimal kernel size for the MMWF to achieve accurate segmentation was determined to be 5 × 5.
We observed that when the kernel size was too small, the noise removal was inadequate, leading to uneven lumen signals and a degradation in segmentation performance (Figure 6). Setting a high threshold value to overcome this problem can result in excessive tissue segmentation. Conversely, when the kernel size is too large, edge information is lost, making it difficult to distinguish the Hounsfield unit differences between tissues. Lowering the threshold value to address this problem resulted in improper tissue segmentation.
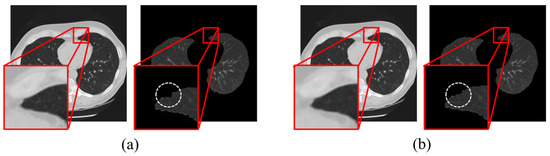
Figure 6.
Illustration of region-growing performance for lung segmentation using MMWF-based denoising with (a) 3 × 3 and (b) optimized kernel size 5 × 5.
Lung segmentation is a crucial initial step in image-based approaches for the early detection, diagnosis, and treatment of various respiratory diseases such as lung cancer and chronic bronchitis [24]. Numerous software tools are currently being developed for the effective detection and identification of pulmonary nodules in lung CT images [25]. Specifically, computer-aided detection (CAD) systems have significantly affected lung cancer diagnosis by improving nodule detection rates and distinguishing between benign and malignant nodules [26]. Lung segmentation serves as preprocessing step in CAD systems and influences their accuracy and precision [27]. Therefore, accurate lung segmentation is essential to improve the overall performance of CAD systems. Enhancing the accuracy of lung segmentation through denoising is vital for improving the effectiveness of CAD systems.
Various non-local algorithms, such as the fast non-local means (FNLM) algorithm are currently being developed for noise removal. These non-local algorithms utilize the intensity and relative distances of neighboring pixels to process images and achieve superior noise reduction compared with the MMWF [28]. We investigated whether the FNLM algorithm exhibited superior performance in segmentation tasks compared with the MMWF. Additional experiments were conducted by applying the median filter and the FNLM algorithm. In line with the evaluation methods employed in our study, we assessed the segmentation performance using the median filter and the FNLM algorithm. The results are presented in Figure 7. When using the median filter, the accuracy, F1-score, and IoU values were 0.9840, 0.9706, and 0.9430, respectively. In the case of the FNLM algorithm, these values were 0.9878, 0.9774, and 0.9559. When employing an MMWF, the corresponding values were 0.9960, 0.9928, and 0.9858. Accuracy, F1-score, and IoU were significantly improved when the MMWF was applied. To evaluate similarity while using the median filter and the FNLM algorithm, we measured RMSE and PSNR, as shown in Figure 8. For the median filter, the RMSE and PSNR values were 0.1264 and 66.0961, respectively. When applying the FNLM algorithm, these values were 0.0366 and 76.8610. When the MMWF was applied, the values were 0.0271 and 79.4585. The highest similarity was observed when the MMWF was used. Evidently, improvement in segmentation performance does not necessarily correlate with noise removal performance.
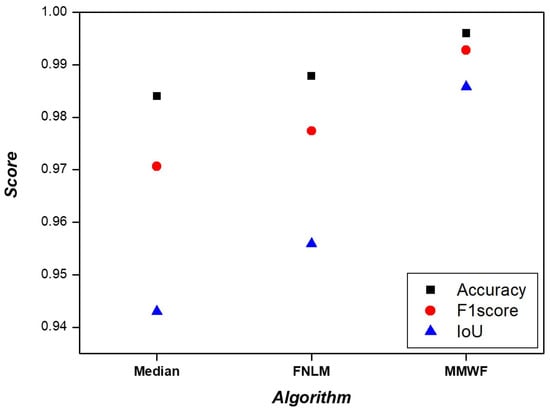
Figure 7.
Accuracy, F1-score, and IoU results of the segmented image according to median filter, FNLM algorithm, and MMWF.
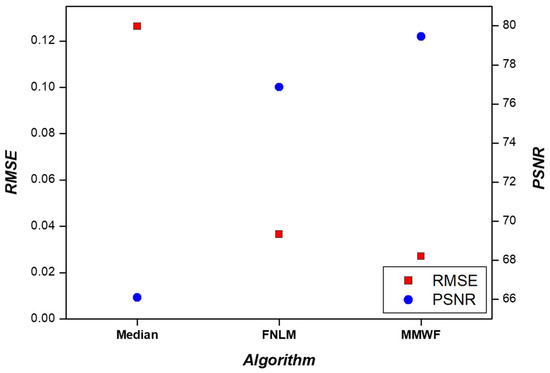
Figure 8.
Results of RMSE and PSNR with median filter, FNLM algorithm, and MMWF.
Furthermore, when calculating the time resolution, the median filter exhibited a value of 1.73 s, the MMWF showed 2.57 s, and the FNLM algorithm showed 28.92 s (Figure 9). MMWF demonstrated a time resolution improvement of 11.30 times over the FNLM algorithm. While the median filter provided the best time resolution, there was not a significant difference when compared to using the MMWF. Given that the MMWF outperforms significantly in terms of accuracy, F1-score, IoU, RMSE, and PSNR values, it is deemed most suitable for segmentation tasks.
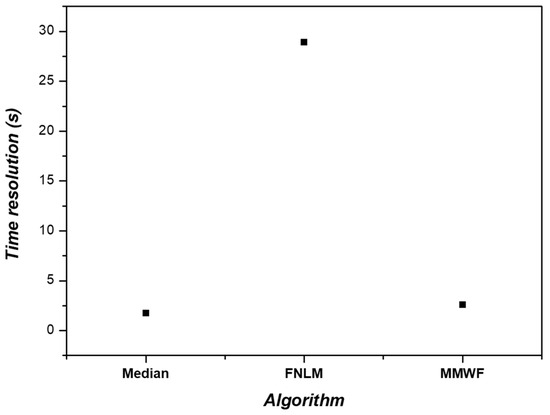
Figure 9.
Results of time resolution with median filter, FNLM algorithm, and MMWF.
In this study, our performance evaluation of segmentation employed metrics such as accuracy, F1-score, and IoU, with the assessment of similarity using RMSE and PSNR. However, evaluation metrics like asymmetric surface distance (ASSD), and relative volume difference (RVD) are also being employed for the evaluation of segmentation. The ASSD computes the difference in distance between the predicted segmentation image and the actual segmentation image using Euclidean distance. RVD represents the difference in volume between the predicted segmentation and the actual segmentation, primarily applied when predicting tumor sizes [29]. Thus, a variety of evaluation metrics exist, and it is suggested that future research should explore optimization strategies utilizing a broader range of evaluation factors. Furthermore, while this study focused on two-dimensional segmentation on individual slices, future evaluation on volume data should include three-dimensional segmentation across multiple slices.
The segmentation performance of the method used in this study greatly depends on the location and selection method of the seed point initially set [30]. If the seed point is set incorrectly, the segmentation accuracy is drastically reduced. In addition, segmentation will not be performed properly if the boundaries of the area are ambiguous. The region-growing-based method is inaccurate, especially in area boundaries. Therefore, further research is needed on segmentation methods that can compensate for these shortcomings. Furthermore, the region-growing-based segmentation method employed in this study exhibits limitations in cases of high anatomical variability within the organ or where diseases exist [31]. Particularly, it faces challenges when segmenting organs with ambiguous boundaries with surrounding structures. In scenarios with unclear organ boundaries, it becomes more sensitive to noise and blurring, making threshold setting difficult. Thus, additional optimization research is necessary to address these issues. There is a need to explore segmentation approaches other than region growing, such as model- and shape-based methods. Additionally, these image processing methods have been optimized for a single or limited number of images, which may limit their performance when handling large and variable volumes of images in clinical settings [23]. In addition, manual segmentation methods are time-consuming and rely on human interpretations and visual observation [29]. Hence, there is a potential risk of misdiagnosis in certain segmented regions as disease or tumors. Therefore, future research should focus on enhancing performance by establishing extensive CT image databases in the field of medical image analysis through the application of the MMWF approach. Utilizing deep learning techniques like convolutional neural networks and U-Net for automated segmentation methods rather than manual approaches becomes essential. The MMWF is expected to be used as a pre-processing algorithm for image augmentation based on fast computation times in deep learning. Furthermore, such network construction can be applied not only to lung segmentation, but also to various other areas’ segmentation, such as cortical bone, which is difficult to segment due to the intricate distinction between marrow and bone in the periosteal boundary and connective tissue and bone in the endosteal boundary [32].
Various preprocessing techniques are available to enhance segmentation performance. In addition to noise removal, techniques such as window adjustment, resolution enhancement, data standardization, and histogram equalization are employed [29]. Windowing is utilized to emphasize regions of interest in an image and remove unnecessary information, and is often applied to visualize specific areas more effectively by configuring appropriate window width and level settings. Resolution enhancement aims to sharpen indistinct boundaries and enable detailed observation, thus improving the spatial or temporal resolution of an image. Typically, resolution enhancement is achieved through techniques such as super-resolution, interpolation, and deep learning. Data standardization involves transforming pixel values in an image to a standard scale, enhancing the stability of segmentation algorithms. By adjusting the mean and standard deviation of the data, all images can be represented within a consistent range of pixel intensities. Histogram equalization adjusts the pixel intensity distribution of an image, enhancing the contrast between dark and bright regions. This process can improve boundary detection during segmentation. Consequently, in addition to noise removal, various preprocessing steps are performed, and optimization studies for each preprocessing step are deemed necessary.
Recently, no-reference-based parameters are being used as a method to evaluate the degree of improvement in medical image quality. Natural image quality evaluator (NIQE) and blind image spatial quality evaluator (BRISQUE) evaluation methods are used to measure the noise performance of medical images and have the advantage of quantitatively analyzing the overall image quality [33,34]. The factors that analyze the form of natural scene statistics in the acquired medical image are NIQE and BRISQUE evaluation parameters, and the principle of deriving these values is to analyze the degree of distortion of the shape of the Gaussian curve. In particular, NIQE and BRISQUE can be used as a method to comprehensively analyze the decrease in the spatial resolution or sharpness of medical images that inevitably occurs depending on the denoising method [35]. Accordingly, we expect that the above no-reference-based evaluation methods can be used as indicators to confirm the performance of segmentation using the MMWF technology.
5. Conclusions
Herein, we have aimed to determine the optimal kernel size for the MMWF to improve the accuracy of lung segmentation in LDCT images. By modeling and applying the MMWF with different kernel sizes, we have found that the kernel size of 5 × 5 improved the outcomes the most. Therefore, a kernel size of 5 × 5 would be appropriate when using the MMWF for pre-processing before performing segmentation. In conclusion, applying the MMWF with an appropriate kernel size enhances the segmentation performance by optimizing the noise and blur. The derived kernel size 5 × 5 is considered applicable not only to lung segmentation but also to CT images for the segmentation of other organs. The current study provides evidence that the MMWF, when applied with an appropriate kernel size, improves lung segmentation performance.
Author Contributions
Conceptualization, S.L. and Y.L.; methodology, S.-H.K. and K.K.; software, S.L.; validation, Y.L. and K.K.; formal analysis, S.L.; funding acquisition, S.L. and K.K.; investigation, S.L., H.K. and M.P.; data curation, S.L. and M.P.; writing—original draft preparation, S.L., H.K., M.P. and S.-H.K.; writing—review and editing, K.K. and Y.L.; project administration, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2023-00239193) and by the Women Graduate Student Engineering Research Team Program through Women in Science, Engineering and Technology (WISET) funded by the Ministry of Science and ICT (2023-02920001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This study was conducted using the National Lung Screening Trial (NLST) (Data set), which is an open data source provided by the National Lung Screening Trial Research Team. Additional information can be found at https://doi.org/10.7937/TCIA.HMQ8-J677 (accessed on 23 May 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schillebeeckx, E.; Lamote, K. Lung cancer screening by volume computed tomography: Thriving to high performance. Breathe 2021, 17, 210063. [Google Scholar] [CrossRef] [PubMed]
- Bjerager, M.; Palshof, T.; Dahl, R.; Vedsted, P.; Olesen, F. Delay in diagnosis of lung cancer in general practice. Br. J. Gen. Pract. 2006, 56, 863–868. [Google Scholar] [PubMed]
- Vansteenkiste, J.; Dooms, C.; Mascaux, C.; Nackaerts, K. Screening and early detection of lung cancer. Ann. Oncol. 2012, 23, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Malkawi, I.M.; Han, E.; Atalla, C.S.; Santucci, R.A.; O’Neil, B.; Wynberg, J.B. Low-Dose (10%) Computed Tomography May Be Inferior to Standard-Dose CT in the Evaluation of Acute Renal Colic in the Emergency Room Setting. J. Endourol. 2016, 30, 493–496. [Google Scholar] [CrossRef]
- Li, Z.; Yang, L.; Shu, L.; Yu, Z.; Huang, J.; Li, J.; Chen, L.; Hu, S.; Shu, T.; Yu, G. Research on CT Lung Segmentation Method Preschool Children based on Traditional Image Processing and ResUnet. Comput. Math. Methods Med. 2022, 2022, 7321330. [Google Scholar] [CrossRef]
- Hu, S.; Hoffman, E.A.; Reinhardt, J.M. Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans. Med. Imaging 2001, 20, 490–498. [Google Scholar] [CrossRef]
- Qian, Y.; Guirong, W. Lung Nodule Segmentation Using EM Algorithm. Sixth Int. Conf. Intell. Hum. -Mach. Syst. Cybern. 2014, 1, 20–23. [Google Scholar] [CrossRef]
- Yang, X.; Xu, G.; Zhou, T. An effective approach for CT lung segmentation using region growing. J. Phys. 2021, 2082, 012001. [Google Scholar] [CrossRef]
- Yoo, S.J.; Yoon, S.H.; Lee, J.H.; Kim, K.H.; Choi, H.I.; Park, S.J.; Goo, J.M. Automated Lung Segmentation on Chest Computed Tomography Images with Extensive Lung Parenchymal Abnormalities Using a Deep Neural Network. Korean J. Radiol. 2021, 22, 476–488. [Google Scholar] [CrossRef]
- Diwakar, J.; Kumar, M. A review on CT image noise and its denoising. Biomed. Signal Process. Control. 2018, 42, 73–88. [Google Scholar] [CrossRef]
- Chalfoun, J.; Majurski, M.; Dima, A.; Stuelten, C.; Peskin, A.; Brady, M. FogBank: A single cell segmentation across multiple cell lines and image modalities. BMC Bioinform. 2014, 15, 431. [Google Scholar] [CrossRef]
- Selamin, A.M.A.; Fadhil, A.F. A study of the effects of Gaussian noise on image features. Kirkuk Univ. J.-Sci. Stud. 2016, 11, 152–169. [Google Scholar] [CrossRef]
- Jia, Y.; Rong, C.; Wu, C.; Yang, Y. Research on the decomposition and fusion method for the infrared and visible images based on the guided image filtering and Gaussian filter. In Proceedings of the 2017 3rd IEEE International Conference on Computer and Communications (ICCC), Chengdu, China, 13–16 December 2017; pp. 1797–1802. [Google Scholar]
- Cannistraci, C.V.; Montevecchi, F.M.; Alessio, M. Median-modified Wiener filter provides efficient denoising, preserving spot edge and morphology in 2-DE image processing. Proteomics 2009, 9, 4908–4919. [Google Scholar] [CrossRef] [PubMed]
- Cannistraci, C.V.; Abbas, A.; Gao, X. Median Modified Wiener Filter for nonlinear adaptive spatial denoising of protein NMR multidimensional spectra. Sci. Rep. 2015, 5, srep08017. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.; Kang, S.; Lee, Y. Optimization of mask size for median-modified Wiener filter according to matrix size of computed tomography images. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2021, 1010, 165508. [Google Scholar] [CrossRef]
- Talebi, M.; Abbasi-Rad, S.; Malekzadeh, M.; Shahgholi, M.; Ardakani, A.A.; Foudeh, K.; Rad, H.S. Cortical Bone Mechanical Assessment via Free Water Relaxometry at 3 T. J. Magn. Reson. Imaging 2021, 54, 1744–1751. [Google Scholar] [CrossRef]
- Jain, A.; Bhateja, V. A versatile denoising method for images contaminated with Gaussian noise. In Proceedings of the CUBE International Information Technology Conference, Pune, India, 3–5 September 2012. [Google Scholar] [CrossRef]
- Dumakude, A.; Ezugwu, A.E. Automated COVID-19 detection with convolutional neural networks. Sci. Rep. 2023, 13, 10607. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Zhou, Z.; Wu, X.; Li, Y.; Wang, S.; Liao, W.; Ying, S.; Zhao, Z. Artificial intelligence for caries and periapical periodontitis detection. J. Densitry 2022, 122, 104107. [Google Scholar] [CrossRef]
- Yu, Y.; Jiang, H.; Zhang, X.; Chen, Y. Identifying Irregular Potatoes Using Hausdorff Distance and Intersection over Union. Sensors 2022, 22, 5740. [Google Scholar] [CrossRef]
- Kang, S.H.; Kim, M.K.; Lee, Y.J. The study on reuction for near field clutter (NFC) artifact based on wavelet thresholding method in ultrasound image using Field Ⅱ program. Optik 2018, 162, 220–227. [Google Scholar] [CrossRef]
- Dehmeshki, J.; Amin, H.; Valdovieso, M.; Ye, X. Segmentation of pulmonary nodules in thoracic CT scans: A region growing approach. IEEE Trans. Med. Imaging 2008, 27, 467–480. [Google Scholar] [CrossRef]
- Osadebey, M.; Andersen, H.K.; Waaler, D.; Fossaa, K.; Martinsen, A.C.T.; Pedersen, M. Three-stage segmentation of lung region from CT images using deep neural networks. BMC Med. Imaging 2021, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Nesbitt, J.C.; Putnam, J.B.; Walsh, G.L.; Roth, J.A.; Mountrain, C.F. Survival in early-stage non-small cell lung cancer. Ann. Thorac. Surg. 1995, 60, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, L.L.; Teutsch, S.; Johnson, M. Lung Cancer Screening with Sputum Cytologic Examination, Chest Radiography, and Computed Tomography: An Update for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2004, 140, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Shariaty, F.; Hosseinlou, S.; Rud, V.Y. Automatic lung segmentation method in computed tomography scans. J. Phys. Conf. Ser. 2019, 1236, 012028. [Google Scholar] [CrossRef]
- Buades, A.; Coll, B.; Morel, J.M. A non-local algorithm for image denoising. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–25 June 2005; Volume 2, pp. 60–65. [Google Scholar] [CrossRef]
- Gul, S.; Khan, M.S.; Bibi, A.; Khandakar, A.; Ayari, M.A.; Chowdhury, M.E.H. Deep learning techniques for liver and liver tumor segmentation: A review. Comput. Biol. Med. 2022, 147, 105620. [Google Scholar] [CrossRef]
- Malek, A.; Rahman, W.E.Z.W.A.; Yasiran, S.S.; Jumaat, A.K.; Jalil, U.M.A. Seed point selection for seed-based region growing in segmenting microcalcifications. In Proceedings of the 2012 International Conference on Statistics in Science, Business and Engineering (ICSSBE), Langkawi, Malaysia, 10–12 September 2012. [Google Scholar] [CrossRef]
- Li, J.; Lin, X.; Che, H.; Li, H.; Qian, X. Pancreas segmentation with probabilistic map guided bi-directional recurrent UNet. Phys. Med. Biol. 2021, 66, 115010. [Google Scholar] [CrossRef]
- Abbasi-Rad, S.; Akbari, A.; Malekzadeh, M.; Shahgholi, M.; Arabalibeik, H.; Rad, H.S. Quantifying cortical bone free water using short echo time (STE-MRI) at 1.5 T. Magn. Reson. Imaging 2020, 71, 17–24. [Google Scholar] [CrossRef]
- Mittal, A.; Soundararajan, R.; Bovik, A.C. Making a “Completely Blind” Image Quality Analyzer. IEEE Signal Process. Lett. 2013, 20, 209–212. [Google Scholar] [CrossRef]
- Mittal, A.; Moorthy, A.K.; Bovik, A.C. No-Reference Image Quality Assessment in the Spatial Domain. IEEE Trans. Image Process. 2012, 21, 4695–4708. [Google Scholar] [CrossRef]
- Lee, M.-H.; Yun, C.-S.; Kim, K.; Lee, Y. Image restoration algorithm incorporating methods to remove noise and blurring from positron emission tomography imaging for Alzheimer’s disease diagnosis. Phys. Medica 2022, 103, 181–189. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).