A Multiscale Deep Encoder–Decoder with Phase Congruency Algorithm Based on Deep Learning for Improving Diagnostic Ultrasound Image Quality
Abstract
:1. Introduction
2. Materials and Methods
2.1. MSDEPC Model Based on Deep Learning for SISR in Ultrasound Imaging
2.2. Datasets
2.3. Quantitative Evaluations of Image Quality
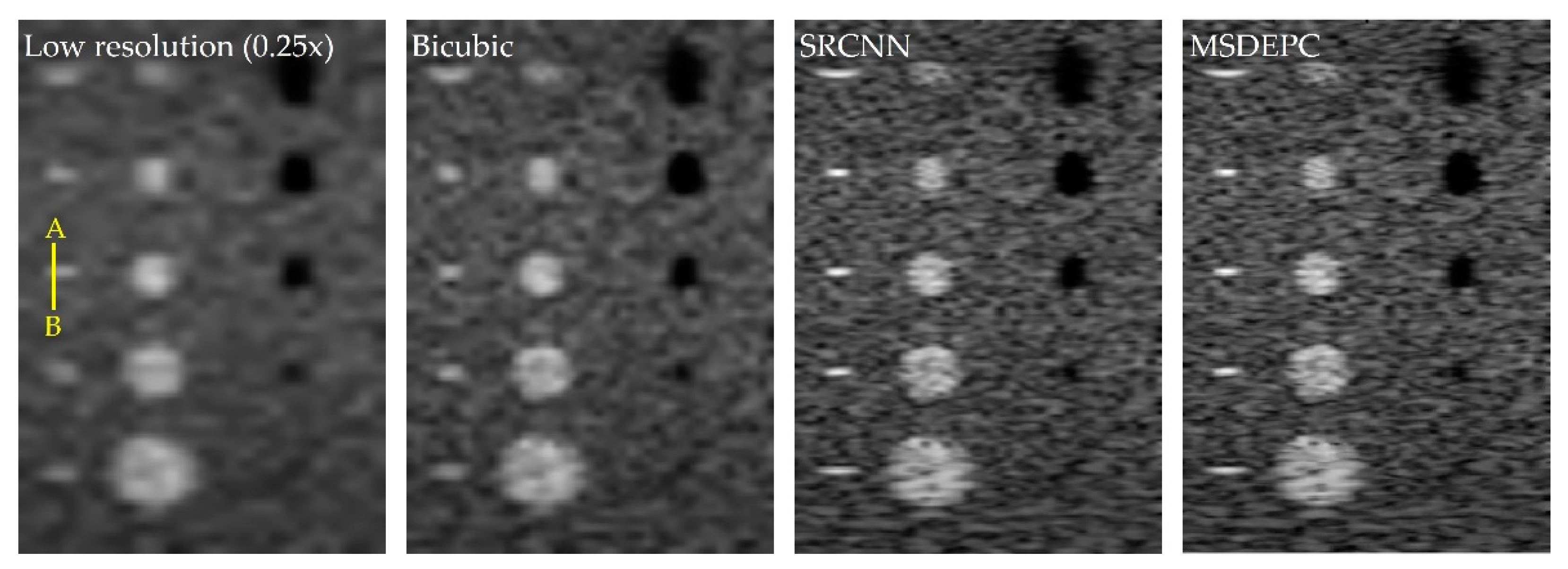
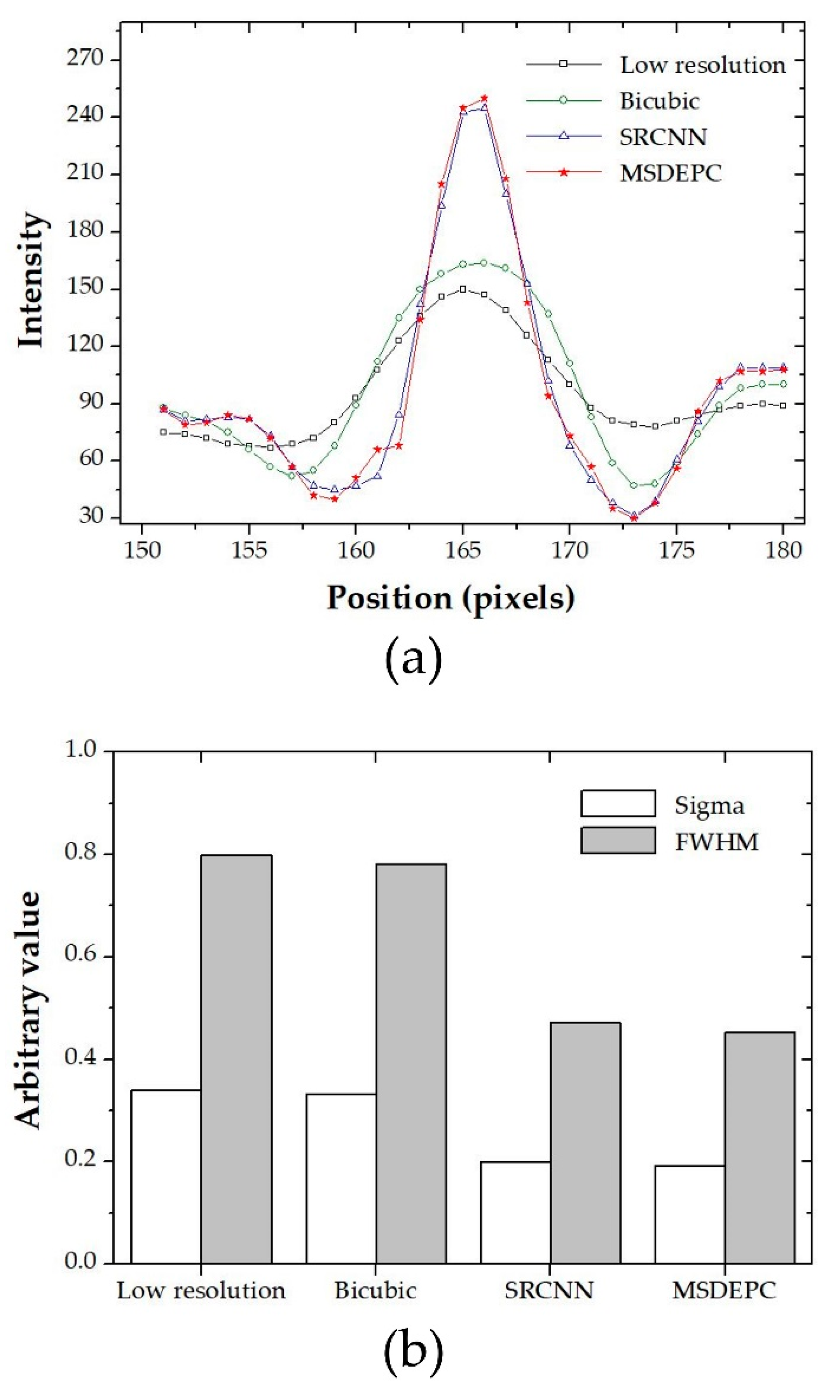
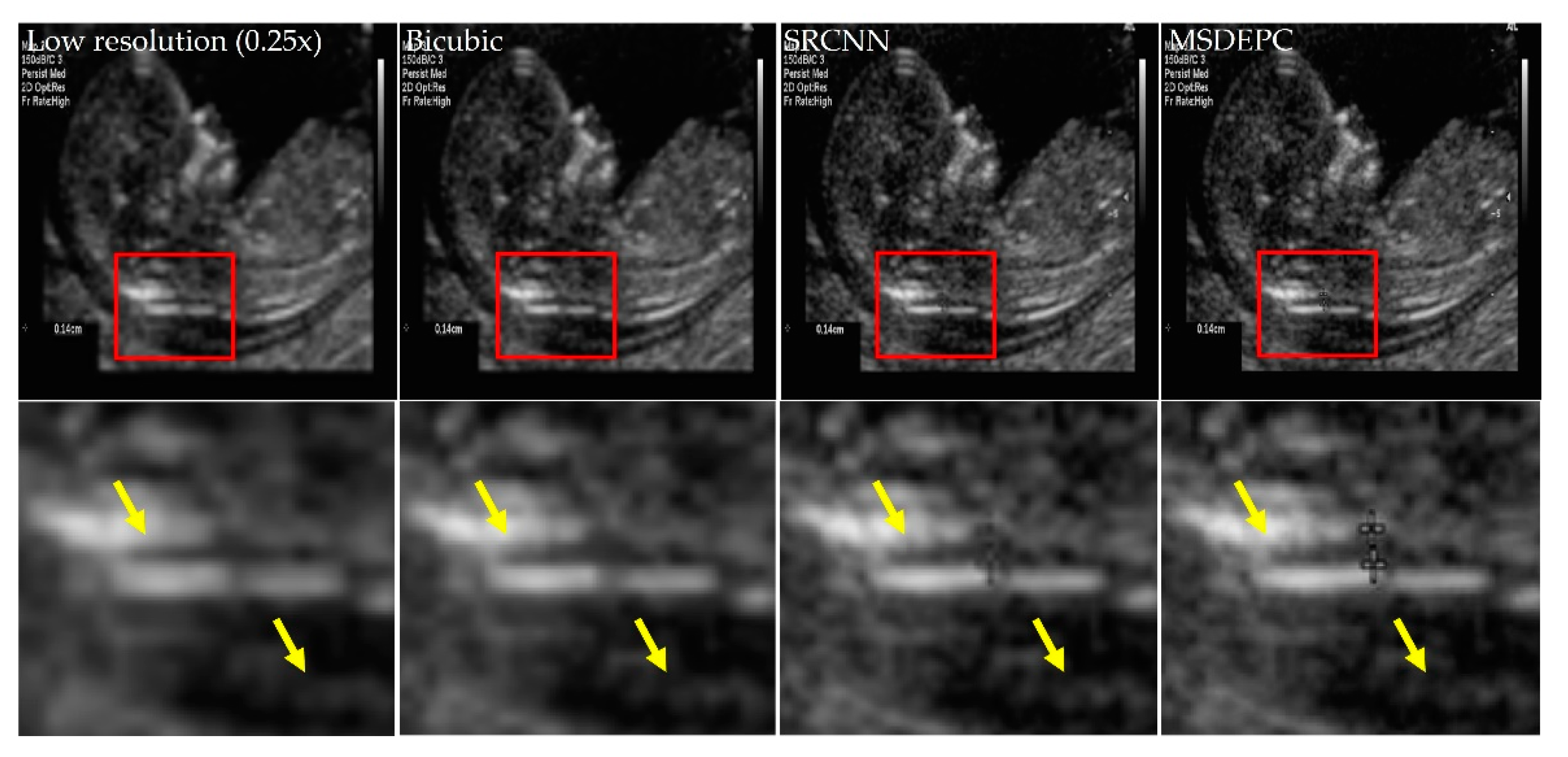
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carovac, A.; Smajlovic, F.; Junuzovic, D. Application of Ultrasound in Medicine. Acta Inform. Med. 2011, 19, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.J.; Lim, C.M.; Jeong, D.; Jeon, H.-R.; Cho, K.J.; Kim, S.Y. Efficacy of intraoperative wireless ultrasonography for uterine incision among patients with adherence findings in placenta previa. J. Obstet. Gynaecol. Res. 2020, 46, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.; Park, H.-C.; Lee, O.-J.; Yoon, C.; Choi, M.H.; Choi, C. Classification of Liver Fibrosis from Heterogeneous Ultrasound Image. IEEE Access 2023, 11, 9920–9930. [Google Scholar] [CrossRef]
- Kim, J.H.; Paik, N.-S.; Nam, S.Y.; Cho, Y.; Park, H.K. The Emerging Crisis of Stakeholders in Implant-based Augmentation Mammaplasty in Korea. J. Korean Med. Sci. 2020, 35, e103. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Ahn, Y.; Park, S.; Kong, H.-J.; Choi, J.Y.; Kim, K.; Nam, I.-C.; Lee, M.-C.; Masuoka, H.; et al. Investigation of optimal convolutional neural network conditions for thyroid ultrasound image analysis. Sci. Rep. 2023, 13, 1360. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.H.; Hwang, J.H.; Kim, J.H.; Park, S.; Lee, K.H.; Choi, S.T. Duplication of superficial femoral artery: Imaging findings and literature review. BMC Med. Imaging 2020, 20, 99. [Google Scholar] [CrossRef]
- Ng, A.; Swanevelder, J. Resolution in ultrasound imaging. Contin. Educ. Anaesth. Crit. Care Pain 2011, 11, 186–192. [Google Scholar] [CrossRef]
- Kim, H.; Labropoulos, N. Image Optimization in Venous Ultrasound Examination. Ann. Phlebol. 2022, 20, 64–67. [Google Scholar] [CrossRef]
- Park, S.C.; Park, M.K.; Kang, M.G. Super-resolution image reconstruction: A technical overview. IEEE Signal Process. Mag. 2003, 20, 21–36. [Google Scholar] [CrossRef]
- Elad, M.; Feuer, A. Restoration of a single superresolution image from several blurred, noisy, and undersampled measured images. IEEE Trans. Image Process. 1997, 6, 1646–1658. [Google Scholar] [CrossRef]
- Kim, K.; Lee, Y. Improvement of signal and noise performance using single image super-resolution based on deep learning in single photon-emission computed tomography imaging system. Nucl. Eng. Technol. 2021, 53, 2341–2347. [Google Scholar] [CrossRef]
- Farsiu, S.; Robinson, M.D.; Elad, M.; Milanfar, P. Fast and robust multiframe super resolution. IEEE Trans. Image Process. 2004, 13, 1327–1344. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Shum, H.-Y. Fundamental limits of reconstruction-based super-resolution algorithms under local translation. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Freeman, W.T.; Jones, T.R.; Pasztor, E.C. Example-based super-resolution. IEEE Comput. Graph. Appl. 2002, 22, 56–65. [Google Scholar] [CrossRef]
- Geman, S.; Geman, D. Stochastic relaxation, gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Pattern Anal. Mach. Intell. 1984, PAMI-6, 721–741. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, Z.; Shum, H.-Y. Image super-resolution using gradient profile prior. In Proceedings of the 2008 IEEE Conference on Computer Vision and Pattern Recognition 2008, Anchorage, AK, USA, 24–26 June 2008. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Z.; Lin, Z.; Cohen, S.; Huang, T. Coupled dictionary training for image super-resolution. IEEE Trans. Image Process. 2012, 21, 3467–3478. [Google Scholar] [CrossRef]
- Yang, J.; Lin, Z.; Cohen, S. Fast image super-resolution based on in-place example regression. In Proceedings of the 2013 IEEE Conference on Computer Vision and Pattern Recognition 2013, Portland, OR, USA, 23–28 June 2013. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, X.; Tian, Y.; Wang, W.; Xue, J.-H.; Liao, Q. Deep learning for single image super-resolution: A brief review. IEEE Trans. Multimed. 2019, 21, 3106–3121. [Google Scholar] [CrossRef]
- Li, K.; Yang, S.; Dong, R.; Wang, X.; Huang, J. Survey of single image super-resolution reconstruction. IET Image Process. 2020, 14, 2273–2290. [Google Scholar] [CrossRef]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Image super-resolution using deep convolutional networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 38, 295–307. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.K.; Lee, K.M. Accurate image super-resolution using very deep convolutional networks. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef]
- Ledig, C.; Theis, L.; Huszár, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef]
- Anwar, S.; Khan, S.; Barnes, N. A deep journey into super-resolution: A survey. ACM Comput. Surv. 2020, 53, 1–34. [Google Scholar] [CrossRef]
- Liu, H.; Fu, Z.; Han, J.; Shao, L.; Hou, S.; Chu, Y. Single image super-resolution using multi-scale deep encoder-decoder with phase congruency edge map guidance. Inf. Sci. 2019, 473, 44–58. [Google Scholar] [CrossRef]
- Liu, H.; Liu, J.; Hou, S.; Tao, T.; Han, J. Perception consistency ultrasound image super-resolution via self-supervised CycleGAN. Neural. Comput. Appl. 2021, 35, 12331–12341. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, X.; Chu, Y. Super-resolution reconstruction algorithm for infrared image with double regular items based on sub-pixel convolution. Appl. Sci. 2020, 10, 1109. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Kesarwani, A.; Purohit, K.; Dalui, M.; Kisku, D.R. Measuring the degree of suitability of edge detection operators prior to an application. In Proceedings of the 2020 IEEE Applied Signal Processing Conference (ASPCON), Kolkata, India, 7–9 October 2020. [Google Scholar] [CrossRef]
- Kovesi, P. Image features from phase congruency. Videre J. Comput. Vis. Res. 1999, 1, 1–26. [Google Scholar]
- Forero, M.G.; Jacanamejoy, C.A. Unified mathematical formulation of monogenic phase congruency. Mathematics 2021, 9, 3080. [Google Scholar] [CrossRef]
- Morrone, M.C.; Owens, R.A. Feature detection from local energy. Pattern Recognit. Lett. 1987, 6, 303–313. [Google Scholar] [CrossRef]
- Bounneche, M.D.; Boubchir, L.; Bouridane, A.; Nekhoul, B.; Ali-Chérif, A. Multi-spectral palmprint recognition based on oriented multiscale log-gabor filters. Neurocomputing 2016, 205, 274–286. [Google Scholar] [CrossRef]
- Xie, S.; Tu, Z. Holistically-nested edge detection. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015. [Google Scholar] [CrossRef]
- Shen, W.; Zhao, K.; Jiang, Y.; Wang, Y.; Zhang, Z.; Bai, X. Object skeleton extraction in natural images by fusing scale-associated deep side outputs. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Las Vegas, NV, USA, 7–13 December 2015. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on International Conference on Machine Learning (ICML’15), Lille, France, 6–11 July 2015; Volume 37, pp. 448–456. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2015, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Tupholme, G.E. Generation of acoustic pulses by baffled plane pistons. Mathematika 1969, 16, 209–224. [Google Scholar] [CrossRef]
- Jensen, J.A.; Munk, P. Computer Phantoms for Simulating Ultrasound B-Mode and CFM Images. Acoust. Imaging 1997, 23, 75–80. [Google Scholar] [CrossRef]
- Keys, R. Cubic convolution interpolation for digital image processing. IEEE Trans. Acoust. Speech Signal Process. 1981, 29, 1153–1160. [Google Scholar] [CrossRef]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted Boltzmann machines. In Proceedings of the 27th International Conference on International Conference on Machine Learning (ICML’10), Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Mastafiz, R.; Rahman, M.M.; Islam, A.K.M.K.; Belkasim, S. Focal Liver Lesion Detection in Ultrasound Image Using Deep Feature Fusions and Super Resolution. Mach. Learn. Knowl. Extr. 2020, 2, 172–191. [Google Scholar] [CrossRef]
- Kim, K.W.; Kim, M.J.; Lee, S.S.; Kim, H.J.; Shin, Y.M.; Kim, P.-N.; Lee, M.-G. Sparing of Fatty Infiltration Around Focal Hepatic Lesions in Patients with Hepatic Steatosis: Sonographic Appearance with CT and MRI Correlation. Am. J. Roentgenol. 2008, 190, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Sandhu, P.; Arora, S.; Bedi, G. Role of high resolution ultrasound complementary to digital mammography. Ann. Afr. Med. 2018, 17, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Lei, Y.-M.; Li, N.; Yu, J.; Jiang, X.-Y.; Yu, M.-H.; Hu, H.-M.; Zeng, S.-E.; Cui, X.-W.; Ye, H.-R. Ultrasound super-resolution imaging for differential diagnosis of breast masses. Front. Oncol. 2022, 12, 1049991. [Google Scholar] [CrossRef]
- Acharya, U.R.; Faust, O.; Sree, S.V.; Molinari, F.; Suri, J.S. ThyroScreen system: High resolution ultrasound thyroid image characterization into benign and malignant classes using novel combination of texture and discrete wavelet transform. Comput. Methods Programs Biomed. 2012, 107, 233–241. [Google Scholar] [CrossRef]
- Guraya, S.S. The Associations of Nuchal Translucency and Fetal Abnormalities; Significance and Implications. J. Clin. Diagn. Res. 2013, 7, 936–941. [Google Scholar] [CrossRef]
- Bulat, A.B.; Tzimiropoulos, G. To learn image super-resolution, use a GAN to learn how to do image degradation first. In Proceedings of the ECCV 2018: Computer Vision–ECCV, Munich, Germany, 8–14 September 2018; pp. 187–202. [Google Scholar] [CrossRef]
- Liu, J.; Tang, J.; Wu, G. AdaDM: Enabling normalization for image super-resolution. arXiv 2021, arXiv:2111.13905. [Google Scholar] [CrossRef]
- Lepcha, D.C.; Goyal, B.; Dogra, A.; Goyal, V. Image super-resolution: A comprehensive review, recent trends, challenges and applications. Inf. Fusion 2023, 91, 230–260. [Google Scholar] [CrossRef]
- Ma, W.; Wu, Y.; Liu, S.; Su, Q.; Zhong, Y. Remote sensing image registration based on phase congruency feature detection and spatial constraint matching. IEEE Access 2018, 6, 77554–77567. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, W.; Qin, J.; Wong, K.-H.; Choi, K.-S.; Heng, P.-A. Fast feature-preserving speckle reduction for ultrasound images via phase congruency. Signal Process. 2017, 134, 275–284. [Google Scholar] [CrossRef]
- Habibi, M.; Foroughi, S.; Karamzadeh, V.; Packirisamy, M. Direct sound printing. Nat. Commun. 2022, 13, 1800. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Han, D.-K.; Lee, Y. Near-field clutter artifact reduction algorithm based on wavelet thresholding method in echocardiography using 3D printed cardiac phantom. J. Korean Phys. Soc. 2022, 81, 441–449. [Google Scholar] [CrossRef]







| Computation Time (s) | Bicubic | SRCNN | MSDEPC |
|---|---|---|---|
| GPU (Titan Xp) | <0.01 | 0.21 | 0.29 |
| CPU (Intel Xeon) | <0.01 | 12.42 | 15.49 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, R.; Kim, K.; Lee, Y. A Multiscale Deep Encoder–Decoder with Phase Congruency Algorithm Based on Deep Learning for Improving Diagnostic Ultrasound Image Quality. Appl. Sci. 2023, 13, 12928. https://doi.org/10.3390/app132312928
Kim R, Kim K, Lee Y. A Multiscale Deep Encoder–Decoder with Phase Congruency Algorithm Based on Deep Learning for Improving Diagnostic Ultrasound Image Quality. Applied Sciences. 2023; 13(23):12928. https://doi.org/10.3390/app132312928
Chicago/Turabian StyleKim, Ryeonhui, Kyuseok Kim, and Youngjin Lee. 2023. "A Multiscale Deep Encoder–Decoder with Phase Congruency Algorithm Based on Deep Learning for Improving Diagnostic Ultrasound Image Quality" Applied Sciences 13, no. 23: 12928. https://doi.org/10.3390/app132312928
APA StyleKim, R., Kim, K., & Lee, Y. (2023). A Multiscale Deep Encoder–Decoder with Phase Congruency Algorithm Based on Deep Learning for Improving Diagnostic Ultrasound Image Quality. Applied Sciences, 13(23), 12928. https://doi.org/10.3390/app132312928






